Abstract
Intrinsically disordered proteins are abundant in the eukaryotic proteome, and they are implicated in a range of different diseases. However, there is a paucity of experimental data on molecular details of the coupled binding and folding of such proteins. Two interacting and relatively well studied disordered protein domains are the activation domain from the p160 transcriptional co-activator ACTR and the nuclear co-activator binding domain (NCBD) of CREB binding protein. We have analyzed the transition state for their coupled binding and folding by protein engineering and kinetic experiments (Φ-value analysis) and found that it involves weak native interactions between the N-terminal helices of ACTR and NCBD, but is otherwise "disordered-like". Most native hydrophobic interactions in the interface between the two domains form later, after the rate-limiting barrier for association. Linear free energy relationships suggest a cooperative formation of native interactions, reminiscent of the nucleation-condensation mechanism in protein folding.
Intrinsically disordered proteins (IDPs) and disordered regions of proteins are crucial for central cellular processes such as receptor signalling, cell-cycle control, and transcription1,2,3,4,5. It is believed that the flexibility of IDPs gives a functional advantage, for example, it enables binding of the polypeptide in an extended conformation with a larger interface area per residue than globular proteins6 and many specific contacts. The flexibility is also likely the reason that IDPs often can evolve to interact with multiple partners like, for example, p537 and NCBD8,9,10,11 (see below). However, experimental data on details of the binding reaction mechanisms of IDPs are scarce and many of the hypotheses as to why disorder is beneficial remain untested by experiment.
Two disordered domains that participate in the formation of a multicomponent protein assembly that is involved in the activation and regulation of gene expression12,13 are the activation domain from the p160 transcriptional co-activator for thyroid hormone and retinoid receptors (ACTR) and the nuclear co-activator binding domain (NCBD) of CREB binding protein (CBP). These protein domains and their interaction have been characterized in detail by NMR8,14,15, SAXS14,16 and stopped-flow spectroscopy17. ACTR is highly disordered with some degree of secondary structure in the free state15,16, while NCBD has molten globule characteristics with a small hydrophobic core and low stability14,15,18,19. The bimolecular complex of the two domains displays a well-defined tertiary structure8 (Fig. 1) and is a classical example of coupled binding and folding of IDPs.
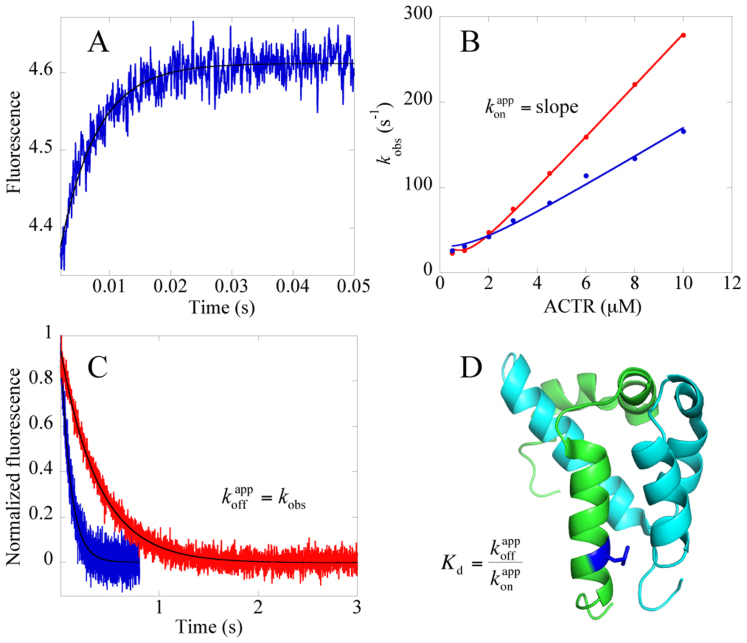
Figure 1. The experimental system.
(A) Example of an experimental binding trace for NCBDY2108W and an ACTR mutant, L1048A. Shown here is the main (fast) phase and protein concentrations are 1 μM NCBDY2108W and 10 μM L1048A-ACTR. (B) kobs values for the main phase obtained from binding traces (such as in panel A) plotted versus [ACTR] to obtain association rate constants, konapp by fitting the data to the general equation for the reversible association of two molecules under second order conditions46 (Eq. 4). At high concentrations of ACTR the equation approaches a linear function with a slope equal to kon. Red, NCBDY2108W and ACTRWT; blue, NCBDY2108W and ACTRL1048A. (C) Dissociation rate constants, koffapp, were determined in displacement experiments, in which NCBDWT was used to compete out NCBDY2108W from the complex. The solid line is a fit to a single exponential equation. At high concentration of NCBDWT the dissociation of NCBDY2108W/ACTR is virtually irreversible and kobs from the curve fitting is equal to koffapp. Red, NCBDY2108W/ACTRWT; blue, NCBDY2108W/ACTRL1048A. (D) Structure of the complex between ACTR (green) and NCBD (cyan), with L1048 highlighted in dark blue. The figure was generated using PyMol (The PyMol Molecular Graphics System, Version 1.3 Schrödinger, LLC).
To shed light on the molecular details of the coupled binding and folding reaction of ACTR and NCBD we have here used protein engineering in conjunction with kinetic experiments (Φ value analysis20,21) to determine the structure of the transition state in terms of formation of native hydrophobic tertiary contacts in the binding interface. Our study, which is the first one reported on a system, in which one of the components is completely disordered (ACTR) and the other one has molten globule characteristics (NCBD), demonstrates that very few native interactions are in the process of forming in the transition state for binding. However, a nucleus with partial formation of native hydrophobic interactions was found to be present between the N-terminal helices of both IDP domains.
Results
Design of site-directed mutants
In a previous study we characterized the kinetic binding reaction of ACTR and NCBD using engineered Trp residues as fluorescent probes17. In the current study we made ten deletion mutations (purportedly non-disruptive side chain truncations) at hydrophobic residues in ACTRWT and an additional ten in a pseudo wild type of NCBD (NCBDY2108W). The mutations were made in the binding interface between the two domains based on a published NMR structure of the complex8. ACTRWT is highly disordered in solution8,15 and mutations are not expected to disrupt any hydrophobic core. NCBD, on the other hand, displays significant residual structure as shown by NMR and circular dichroism (CD)11,14,15,19. Far-UV CD was used to assess the effect of the mutations on the secondary structure of NCBD (Supplementary Fig. S1). Six of the mutants displayed a CD spectrum identical to that of NCBDY2108W, whereas four appeared to have lost some α-helical structure. Trimethylamine N-oxide (TMAO) is known to shift the equilibrium towards the folded state22 and 0.7 M partially or fully restored the CD spectra for these four mutants (Supplementary Fig. S1).
Binding kinetics of mutants of ACTR and NCBD
The binding kinetics of the ten ACTR mutants was measured with NCBDY2108W and the kinetics of the ten NCBDY2108W mutants measured with ACTRWT, using the stopped-flow technique (Fig. 1 and Supplementary Table S1, Fig. S2 and Fig. S3). Two of the mutations (L1064A and L1071A in ACTR) were highly destabilizing for the bimolecular complex and their kinetics were measured in the presence of 0.7 M TMAO to reduce the observed rate constants (kobs) to a magnitude accessible by the stopped-flow instrument. Likewise, the four mutants with altered CD spectra were measured in the presence of 0.7 M TMAO to avoid ground state effects on the observed kinetics. Two more mutants, L2090A NCBD and L1052A ACTR, were also purified but did not yield reliable kinetics data, due to elevated kobs values, even in the presence of TMAO.
The binding kinetics of almost all mutants was biphasic, with a fast phase, which was linear with increasing concentration of ACTR and a slow phase, which appeared rather constant throughout the measured concentration interval. Similar kinetics was observed previously for NCBDY2108W and ACTRWT, and a detailed analysis has been published17 (see also text in Supplementary Information for further discussion on the slow phase). From the fast phase we extracted the apparent association rate constant (konapp) as the slope of the observed rate constant kobs versus ACTR concentration (Fig. 1, Supplementary Table S1, Fig. S2). The apparent dissociation rate constant (koffapp) for mutant ACTR/NCBD complexes was determined in separate displacement experiments (Fig. 1, Supplementary Table S1) as detailed in the Materials section.
In the experiments with TMAO, an additional phase of intermediate magnitude was clearly visible for one mutant, L2067A (kobs~19 s−1). Interestingly, a similar phase was previously detected for a double mutant involving a buried salt bridge, but also for the wild type at high ionic strength17. Careful analyses of binding traces for NCBDY2108W and wild-type ACTR (i.e., our wild type pair) in 0.7 M TMAO revealed that a similar intermediate phase could be fitted to the data. This result may be explained by an induced-fit scenario, in which the population of an intermediate is promoted by TMAO, high salt, and certain mutations. Thus, TMAO and salt may be used to tune the shape of the energy landscape for coupled binding and folding for ACTR/NCBD. However, while the presence of TMAO may shift the distribution of populations of free ACTR and NCBD, and the height of energetic barriers, it is unlikely to affect the transition state structure and overall mechanism as shown by the linear free energy diagrams (Brønsted plot), in which data collected in presence and absence of TMAO fall on the same line (Fig. 2 and Supplementary Fig. S4B).
Figure 2. Linear free energy relationships of rate and equilibrium binding constants upon mutation.
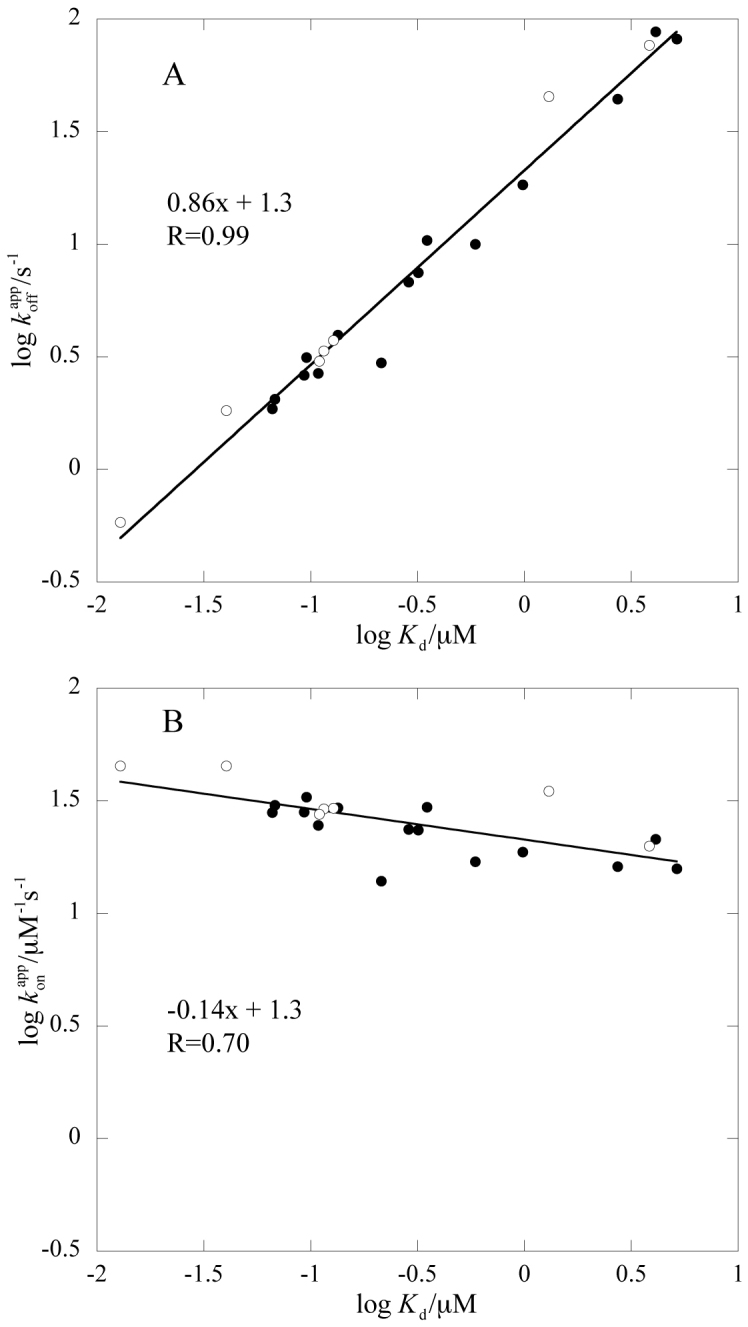
(A) log koffapp versus log Kd and (B) log konapp versus log Kd. Solid circles represent data that were obtained in 20 mM phosphate (pH = 7.4), 150 mM NaCl, whereas constants that were obtained in the same buffer but supplemented with 0.7 M TMAO are shown as open circles.
For three out of the four mutants, TMAO did not restore the magnitude of the CD signal completely, but they are close, having 90%, 88%, and 81% of the wild-type CD signal at 222 nm. However, as judged from Fig. 2, these small differences are not enough to result in a significant deviation from the overall linear trend in the Brønsted plot. Thus, for these mutants as well as the other ones measured in the presence of TMAO, the data report on the same transition state.
Structural details of the interface between ACTR and NCBD
The 3D structure of the complex between ACTR and NCBD8 shows that the leucine rich binding interface of NCBD/ACTR is well packed and contains specific hydrophobic interactions. Accordingly, several of the conservative deletion mutations resulted in significantly reduced binding affinities (Table 1 and Supplementary Table S1), in particular those from Leu to Ala in LXXLL/LLXXL motifs. These results agree well with previous mutational studies of the interaction between NCBD and ACTR19 or TIF2, an ACTR homolog23, as well as other NCBD binding proteins24,25. On the other end, the mutations V2109A NCBD and V1077A ACTR were not destabilizing but displayed even lower koffapp and Kd values than the wild type (Supplementary Table S1). Both of these mutated residues are positioned at the C-terminal helices of the respective domain, and as shown in the NCBD/ACTR structure8 they also interact with each other. While the thermodynamic origin of these changes is not clear, it has previously been shown that the C-terminal part of helix three in unbound NCBD displays significant fast backbone dynamics15 compared to the rest of NCBD, and the region in bound ACTR that forms the C-terminal helix, has very little helical content in unbound ACTR15,16. One may speculate that the disorder that is present in the respective helix is modulated by mutation, for example through stabilization of the helix, such that a slightly higher affinity is obtained, compared to the wild type.
Table 1. Φbinding values for the interaction between wild type and mutants of ACTRWT and NCBDY2108W, respectively, measured in 20 mM phosphate (pH = 7.4), 150 mM NaCl, and 277 K.
| ACTRWT | NCBDY2108W | ||||
|---|---|---|---|---|---|
| NCBDY2108W mutant | ΔΔGEq kcal/mol | Φbinding | ACTR mutant | ΔΔGEq kcal/mol | Φbinding |
| I2062V α1 | 0.20 ± 0.01 | −0.12 ± 0.07 | L1048A α1 | 1.02 ± 0.02 | 0.27 ± 0.02 |
| V2086A α2 | 0.73 ± 0.02 | −0.04 ± 0.02 | L1049A α1 | 1.30 ± 0.02 | 0.17 ± 0.02 |
| L2087A α2 | 2.2 ± 0.1 | 0.14 ± 0.03 | L1055A α1 | 0.46 ± 0.05 | 0.85 ± 0.10 |
| L2096A α3 | 1.9 ± 0.1 | 0.16 ± 0.03 | L1056A α1 | 2.09 ± 0.1 | 0.07 ± 0.02 |
| I2101Va α3 | 0.01 ± 0.02 | – | A1061Ga loop | 0.08 ± 0.06 | – |
| V2109A α3 | −0.17 ± 0.02 | 0.22 ± 0.09 | I1067V α2 | 0.62 ± 0.02 | 0.16 ± 0.02 |
| I1073V α3 | 0.68 ± 0.03 | 0.15 ± 0.05 | |||
| V1077A α3 | −0.19 ± 0.02 | −0.01 ± 0.13 | |||
aToo low ΔΔGEq to calculate a reliable Φbinding value.
Calculation of Φbinding values
Linear free energy relationships clearly demonstrate that the effect of the mutations is largely in the dissociation rate constant (Fig. 2A). However, a few mutations affected the association rate constant (Supplementary Fig. S2), suggesting that some side-chains have begun to form native-like interactions in the rate limiting transition state for binding. In order to quantify the degree of native contact formation and thus get a picture of the structure of the transition state for the coupled binding and folding of ACTR/NCBD, we calculated Φ values for binding, Φbinding21,26 (Table 1, see Supplementary Information for a detailed discussion of the Φbinding values in relation to konapp, koffapp and the slow phase for each plausible reaction scheme) and mapped the values onto the structure of the complex between the two proteins (Fig. 3). Φbinding values were calculated by relating the change in free energy for the rate limiting barrier for binding (ΔΔGTS, calculated from kon values) with the total change in free energy for the binding reaction at equilibrium (ΔΔGEq, calculated from Kd values, which in turn were calculated from koffapp divided by konapp) (Equations 1–3, Supplementary Information).
The wild type in the calculations refers to the interaction between NCBDY2108W and ACTRWT. A Φbinding value of zero would imply that the mutated residue is not making any native interactions in the transition state of the binding reaction. A Φbinding value of 1, on the other hand, implies that the residue makes a full native interaction in the transition state. Intermediate values are subject to a number of caveats20, but are usually interpreted as partial formation of native interactions in the transition state. We calculated a Φbinding value if the absolute value of ΔΔGEq > 0.16 kcal/mol. This may appear as a very low cut-off value, but is based on the very accurate determinations of konapp and koffapp. For most mutants, the largest source of error is the concentration determination of ACTR variants, which directly affects konapp and which we estimate to about 5%. The Φbinding values in Table 1 report on the transition state that is rate limiting for binding in 20 mM sodium phosphate (pH = 7.4), 150 mM NaCl. In the presence of TMAO, it is likely that the same transition state has been stabilized resulting in a higher konapp value. An alternative explanation is that an earlier transition state becomes (partially) rate limiting. konapp for NCBDY2108W and wild-type ACTR in presence of 0.7 M TMAO, was determined to be around 45 μM−1s−1, which was used to calculate Φbinding values in Table 2.
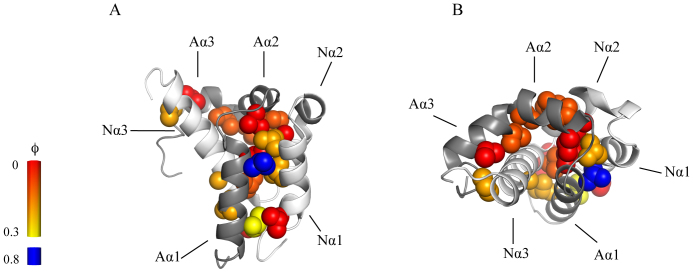
Figure 3. A structural model of the transition state for coupled binding and folding.
Φbinding values from Table 1 and 2 mapped onto the structure of the complex between ACTR and NCBD. ACTR is shown in dark grey and NCBD in light grey. Residues are colour coded by the magnitude of their Φbinding values, with a gradient ranging from 0 (red) to 0.3 (yellow). ACTRL1055A, which has the highest Φbinding value, is blue colored. The negative Φbinding values were close to zero and are colored red. Panel A and B show the same structure from two different angles. Images were generated using PyMol (The PyMol Molecular Graphics System, Version 1.3 Schrödinger, LLC).
Table 2. Φbinding values for the interaction between wild type and mutants of ACTRWT and NCBDY2108W, respectively, measured in 20 mM phosphate (pH = 7.4), 150 mM NaCl, 0.7 M TMAO, and 277 K.
| ACTRWT | NCBDY2108W | ||||
|---|---|---|---|---|---|
| NCBDY2108W mutant | ΔΔGEq kcal/mol | Φbinding | ACTR mutant | ΔΔGEq kcal/mol | Φbinding |
| L2067A α1 | 0.63 ± 0.08 | 0.0 ± 0.1 | L1064A α2 | 2.5 ± 0.1 | 0.06 ± 0.02 |
| L2070A α1 | 1.2 ± 0.1 | 0.20 ± 0.04 | L1071A loop/α2 | 3.1 ± 0.2 | 0.14 ± 0.03 |
| L2074A α1 | 1.3 ± 0.1 | 0.19 ± 0.04 | |||
| A2099G α3 | 1.2 ± 0.1 | 0.23 ± 0.04 | |||
The structure of the transition state for coupled binding and folding
Following the guidelines of Fersht20, it is useful to categorize Φ values as low, intermediate and high. The low Φbinding values (<0.2) at almost all positions show that most of the native hydrophobic interactions in the interface between ACTR and NCBD have not formed in the transition state for the binding reaction, suggesting that most of the initial encounter complex does not contain native-like hydrophobic interactions. However, a few ACTR mutations situated in helix 1 make direct interactions with NCBD and displayed intermediate or even high Φbinding values. Although Φbinding values for the NCBD mutants were low, two mutations in helix 1 gave Φbinding values of 0.2, suggesting that the N-terminal helices of ACTR and NCBD form the initial native intermolecular contacts.
Discussion
Protein engineering together with detailed kinetic analyses allowed us to determine a structural model of the rate-limiting transition state in the coupled binding and folding involving the IDP, ACTR, and the molten globule, NCBD (Fig. 3). In terms of hydrophobic interactions in the interface between the two disordered domains, the transition state for the initial association is more disordered-like than native-like. Overall, native interactions in the binding interface are formed and consolidated subsequent to the rate-limiting barrier for association. This is in agreement with previous studies on the kinetic binding mechanism of IDPs and disordered regions using NMR27, MD28 or stopped-flow spectroscopy17,29,30,31,32, in which data also suggest that native interactions form late on the reaction pathway. It is usually very difficult to prove multistep binding reactions and there are a number of possible binding mechanisms for the association of ACTR and NCBD. Importantly, our Φbinding values report on the transition state for the productive binding pathway for either of the four mechanisms (Scheme 1–4 in Supplementary Fig. S5), which are overall consistent with the data set, as discussed in detail in Supplementary Information. Recent data on NCBD show that it exists in at least two different conformations in the free state33,34, which lends support to model 4 in Fig. S5. Thus, in their free states, there may be several different forms of ACTR and NCBD, with different degrees of structure and multiple pathways for binding, with external conditions determining the flux through each pathway35. However, the pathways converge after their initial association and ACTR and NCBD will experience a cooperative formation of native bonds as the barrier is crossed as shown by the linear free energy relationship (Fig. 2).
Looking at the transition state in more detail, a few native contacts have begun to form between the N-terminal helices of the respective protein in the transition state for binding. In fact, the linear free energy relationship (Fig. 2) is reminiscent of those obtained for proteins obeying the nucleation-condensation mechanism in protein folding36. Thus, following formation of a few native interactions during the crossing of the main rate-limiting barrier, the remaining native contacts form after the main barrier. Binding of IDPs often involves extended protein conformations with a binding surface containing hydrophilic as well as hydrophobic interactions. Interestingly, the region with highest Φbinding values contains the LXXLL/LLXXL recognition motifs, where X stands for any amino acid. These motifs, in which any bulky hydrophobic residue can replace Leu, are known to mediate protein-protein interactions in transcriptional regulation37.
Furthermore, NMR and CD studies15,16 have shown that the region that constitutes helix one in bound ACTR in fact displays transient helix formation in the free state. Previous studies38,39,40,41 have emphasized the functional importance of preformed structural elements and our observation that such elements form the initial native contacts during the binding between ACTR and NCBD lends support to this hypothesis.
In our initial study on ACTR/NCBD17 we characterized a buried salt bridge by mutagenesis (R2104L in NCBD and D1068L in ACTR). It is clear that the effect on the association rate constant was large (~20 fold) while the koffapp was similar to that of the wild type proteins. This would imply a Φbinding value of one, however, the R2104L mutation has clear effects on the ground state of NCBD19 and the two Leu residues that replaced the salt bridge will most probably contribute to binding by forming new hydrophobic interactions. Thus, while mutation of the salt bridge strongly affects konapp, suggesting that the salt bridge stabilizes the transition state for binding, we cannot estimate a Φbinding value for this interaction. Previous studies on interactions involving IDPs42 or folded proteins43 have demonstrated that mutation of charged residues may affect the association rate constant such that high Φbinding values are obtained. Thus, charged residues have a higher tendency than hydrophobic amino acids to form native-like interactions in the transition state for binding, due to long-range electrostatic effects21,44.
A recent molecular dynamics simulation study38 concluded that the end helices α1 and α3 in NCBD, which dominate the number of contacts it makes with ACTR in the complex, are most readily formed in the free state, having a conformation similar to the bound state. Here we show that α1 in NCBD forms weak native-like interactions with ACTR in the transition state, and two out of four mutations in α3 in NCBD has an intermediate Φ value (Table 1). It should be noted that these mutations involve hydrophobic substitutions, whereas α3 in NCBD also contains charged residues, which, as previously discussed, tend to adopt higher Φ values than hydrophobic positions21. Thus, it is possible that this region may make long-range native-like electrostatic interactions in the rate-limiting transition state.
In summary, we propose, based on available experimental data, the following scenario for the productive interaction between ACTR and NCBD. Helix one of the highly disordered ACTR forms transiently15,16, with folding and unfolding probably on the ns-μs timescale45. In a helix-like state it makes initial weak native-like interactions with helix one of NCBD that are rate limiting for the binding reaction. The initial association is also promoted by R2104 in NCBD and D1068 in ACTR17, which may steer the two protein domains into the correct orientation as the native salt bridge is formed. Formation of native hydrophobic contacts in the binding interface proceeds in a cooperative fashion following crossing of the rate-limiting transition state for association. It is likely that this mechanism is common for coupled binding and folding reactions of IDPs.
Methods
Protein expression and purification
Human NCBD and ACTR were expressed and purified as described previously17. Briefly, BL21(DE3) pLysS cells were used to express NCBD or ACTR in fusion with an N-terminal His-tagged lipoyl fusion protein with a thrombin site separating the lipoyl and NCBD or ACTR sequences. The fusion protein was first subjected to a Ni-sepharose fast flow (GE Healthcare) column purification step followed by thrombin cleavage, after which a second Ni-sepharose fast flow purification step was utilized to remove the lipoyl protein and other impurities. This was followed by reversed phase chromatography using a C-8 (Grace Davison Discovery) column. Mutations were generated by inverted PCR and mutants were expressed and purified as described for the wild-type proteins17. The identity of each mutant was verified with MALDI-TOF. The concentration of NCBD was determined by measuring the absorbance at 280 nm, whereas the concentration of ACTR, which lacks Tyr and Trp residues, was determined by measuring the absorbance at 205 nm and using an extinction coefficient of 31.9 mL mg−1 cm−1 obtained from amino acid analysis.
Binding kinetics
Binding kinetics of NCBD/ACTR was measured using an upgraded SX-17MV stopped-flow spectrometer (Applied Photophysics, Leatherhead, U.K.) and monitoring Trp fluorescence17. Excitation was at 280 nm and a 320 nm long pass filter was used to monitor the emission. Measurements were done at 277 K in either 20 mM sodium phosphate (pH = 7.4), 150 mM NaCl, or the same buffer supplemented with 0.7 M TMAO. In order to obtain association rate constants (konapp) for NCBDY2108W/ACTRmutant and NCBDY2108Wmutant/ACTRWT, the concentration of ACTR was varied while keeping the concentration of NCBD constant at 1 μM. Observed rate constants were plotted versus [ACTR] and the data were fitted to the general equation for the reversible association of two molecules46 to determine konapp (Fig. 1) (Eq. 4).
[NCBD]0 and [ACTR]0 are the total concentrations of the respective species. Note that the equation breaks down to a linear function under pseudo-first order conditions, that is when [ACTR]0 ≫ [NCBD]0.
Apparent dissociation rate constants were determined through displacement experiments, by mixing a pre-formed NCBDY2108W/ACTRmutant or NCBDY2108Wmutant/ACTRWT complex (1.1–2.5 μM NCBD mixed with 1–2 μM ACTR) with an excess of [NCBDWT], such that ACTR is trapped in a spectroscopically silent complex. Higher concentration of mutant NCBD/ACTR complex was used for mutants with low affinity, to improve the amplitude of the kinetic trace. The observed rate constant at high [NCBDWT] is equal to koffapp.
Circular dichroism spectroscopy
CD spectra were recorded using a JASCO-810 spectrometer with a peltier temperature control system. Spectra were recorded between 260 nm and 200 nm, at 298 K or 277 K, and 20 mM phosphate (pH = 7.4), 150 mM NaCl, either with or without 0.7 M TMAO.
Author Contributions
J.D. and P.J. planned and designed the experiments. J.D., X.M. and Å.E. performed the experiments. J.D. and P.J. analyzed data and wrote the paper.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by the Swedish Research Council (NT) and the Human Frontiers Young Investigator Science Program (to P. J.) and the Lars Hierta Foundation (to J. D.) and the Magnus Bergvall Foundation (to J. D.).
References
- Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M. & Obradovic Z. Intrinsic disorder and protein function. Biochemistry 41, 6573–82 (2002). [DOI] [PubMed] [Google Scholar]
- Wright P. E. & Dyson H. J. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 (2005). [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 27, 527–533 (2002). [DOI] [PubMed] [Google Scholar]
- Liu J. et al. Intrinsic disorder in transcription factors. Biochemistry 45, 6873–6888 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H. J. Expanding the proteome: disordered and alternatively folded proteins. Q. Rev. Biophys., 1–52 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros B., Tompa P., Simon I. & Dosztanyi Z. Molecular principles of the interactions of disordered proteins. J. Mol. Biol. 372, 549–561 (2007). [DOI] [PubMed] [Google Scholar]
- Vousden K. H. & Lane D. P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 8, 275–283 (2007). [DOI] [PubMed] [Google Scholar]
- Demarest S. J. et al. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415, 549–553 (2002). [DOI] [PubMed] [Google Scholar]
- Waters L. et al. Structural diversity in p160/CREB-binding protein coactivator complexes. J. Biol. Chem. 281, 14787–14795 (2006). [DOI] [PubMed] [Google Scholar]
- Qin B. Y. et al. Crystal structure of IRF-3 in complex with CBP. Structure 13, 1269–1277 (2005). [DOI] [PubMed] [Google Scholar]
- Lee C. W., Martinez-Yamout M. A., Dyson H. J. & Wright P. E. Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein. Biochemistry 49, 9964–9971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90, 569–580 (1997). [DOI] [PubMed] [Google Scholar]
- Goodman R. H. & Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14, 1553–1577 (2000). [PubMed] [Google Scholar]
- Kjaergaard M., Teilum K. & Poulsen F. M. Conformational selection in the molten globule state of the nuclear coactivator binding domain of CBP. Proc. Natl. Acad. Sci. USA 107, 12535–12540 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M. O., Bae S. H., Dyson H. J. & Wright P. E. NMR relaxation study of the complex formed between CBP and the activation domain of the nuclear hormone receptor coactivator ACTR. Biochemistry 47, 1299–1308 (2008). [DOI] [PubMed] [Google Scholar]
- Kjaergaard M. et al. Temperature-dependent structural changes in intrinsically disordered proteins: formation of alpha-helices or loss of polyproline II? Protein Sci. 19, 1555–1564 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan J., Schmidt T., Mu X., Engström Å. & Jemth P. Fast Association and Slow Transitions in the Interaction between Two Intrinsically Disordered Protein Domains. J. Biol. Chem. 287, 34316–34324 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaergaard M., Poulsen F. M. & Teilum K. Is a malleable protein necessarily highly dynamic? The hydrophobic core of the nuclear coactivator binding domain is well ordered. Biophys. J. 102, 1627–1635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarest S. J., Deechongkit S., Dyson H. J., Evans R. M. & Wright P. E. Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB-binding protein. Protein Sci. 13, 203–210 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R. & Sato S. Phi-value analysis and the nature of protein-folding transition states. Proc. Natl. Acad. Sci. USA 101, 7976–7981 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G., Haran G. & Zhou H. X. Fundamental aspects of protein-protein association kinetics. Chem. Rev. 109, 839–860 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskakov I. & Bolen D. W. Forcing thermodynamically unfolded proteins to fold. J. Biol. Chem. 273, 4831–4834 (1998). [DOI] [PubMed] [Google Scholar]
- Voegel J. J. et al. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17, 507–519 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S. et al. A Conserved alpha-helical motif mediates the binding of diverse nuclear proteins to the SRC1 interaction domain of CBP. J. Biol. Chem. 279, 14055–14064 (2004). [DOI] [PubMed] [Google Scholar]
- Ryan C. M., Harries J. C., Kindle K. B., Collins H. M. & Heery D. M. Functional interaction of CREB binding protein (CBP) with nuclear transport proteins and modulation by HDAC inhibitors. Cell cycle 5, 2146–2152 (2006). [DOI] [PubMed] [Google Scholar]
- Taylor M. G., Rajpal A. & Kirsch J. F. Kinetic epitope mapping of the chicken lysozyme. HyHEL-10 Fab complex: delineation of docking trajectories. Protein Sci. 7, 1857–1867 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugase K., Dyson H. J. & Wright P. E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1026 (2007). [DOI] [PubMed] [Google Scholar]
- Turjanski A. G., Gutkind J. S., Best R. B. & Hummer G. Binding-induced folding of a natively unstructured transcription factor. PLoS Comp. Biol. 4, e1000060 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq S. R. et al. Side-chain interactions form late and cooperatively in the binding reaction between disordered peptides and PDZ domains. J. Am. Chem. Soc. 134, 599–605 (2012). [DOI] [PubMed] [Google Scholar]
- Karlsson O. A., Chi C. N., Engström Å. & Jemth P. The transition state of coupled folding and binding for a flexible beta-finger. J. Mol. Biol. 417, 253–261 (2012). [DOI] [PubMed] [Google Scholar]
- Bachmann A., Wildemann D., Praetorius F., Fischer G. & Kiefhaber T. Mapping backbone and side-chain interactions in the transition state of a coupled protein folding and binding reaction. Proc. Natl. Acad. Sci. USA 108, 3952–3957 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni S., Morrone A., Giri R. & Brunori M. A folding-after-binding mechanism describes the recognition between the transactivation domain of c-Myb and the KIX domain of the CREB-binding protein. Biochem. Biophys. Res. Commun. 428, 205–209 (2012). [DOI] [PubMed] [Google Scholar]
- Kjaergaard M., Andersen L., Nielsen L. D. & Teilum K. A Folded Excited State of Ligand-Free Nuclear Coactivator Binding Domain (NCBD) Underlies Plasticity in Ligand Recognition. Biochemistry In press, doi:10.1021/bi4001062 (2013). [DOI] [PubMed] [Google Scholar]
- Japrung D. et al. Single-molecule studies of intrinsically disordered proteins using solid-state nanopores. Anal. Chem. 85, 2449–2456 (2013). [DOI] [PubMed] [Google Scholar]
- Hammes G. G., Chang Y. C. & Oas T. G. Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. U S A 106, 13737–13741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki L. S., Otzen D. E. & Fersht A. R. The structure of the transition state for folding of chymotrypsin inhibitor 2 analysed by protein engineering methods: evidence for a nucleation-condensation mechanism for protein folding. J. Mol. Biol. 254, 260–288 (1995). [DOI] [PubMed] [Google Scholar]
- Plevin M. J., Mills M. M. & Ikura M. The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 30, 66–69 (2005). [DOI] [PubMed] [Google Scholar]
- Knott M. & Best R. B. A preformed binding interface in the unbound ensemble of an intrinsically disordered protein: evidence from molecular simulations. PLoS Comp. Biol. 8, e1002605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M., Simon I., Friedrich P. & Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 338, 1015–1026 (2004). [DOI] [PubMed] [Google Scholar]
- Sivakolundu S. G., Bashford D. & Kriwacki R. W. Disordered p27Kip1 exhibits intrinsic structure resembling the Cdk2/cyclin A-bound conformation. J. Mol. Biol. 353, 1118–1128 (2005). [DOI] [PubMed] [Google Scholar]
- Cino E. A., Wong-ekkabut J., Karttunen M. & Choy W. Y. Microsecond molecular dynamics simulations of intrinsically disordered proteins involved in the oxidative stress response. PLOS ONE 6, e27371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsath L., Dvorsky R., Fiegen D., Carlier M. F. & Ahmadian M. R. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol. Cell 20, 313–324 (2005). [DOI] [PubMed] [Google Scholar]
- Selzer T., Albeck S. & Schreiber G. Rational design of faster associating and tighter binding protein complexes. Nat. Struct. Biol. 7, 537–541 (2000). [DOI] [PubMed] [Google Scholar]
- Kiel C., Selzer T., Shaul Y., Schreiber G. & Herrmann C. Electrostatically optimized Ras-binding Ral guanine dissociation stimulator mutants increase the rate of association by stabilizing the encounter complex. Proc. Natl. Acad. Sci. USA 101, 9223–9228 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuweiler H., Johnson C. M. & Fersht A. R. Direct observation of ultrafast folding and denatured state dynamics in single protein molecules. Proc. Natl. Acad. Sci. USA 106, 18569–18574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta F. The study of bimolecular reactions under non-pseudo-first order conditions. Biophys. Chem. 116, 251–256 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information