Abstract
EPA Method 1600 and Enterolert are used interchangeably to measure Enterococcus for fecal contamination of public beaches, but the methods occasionally produce different results. Here we assess whether these differences are attributable to the selectivity for certain species within the Enterococcus group. Both methods were used to obtain 1279 isolates from 17 environmental samples, including influent and effluent of four wastewater treatment plants, ambient marine water from seven different beaches, and freshwater urban runoff from two stream systems. The isolates were identified to species level. Detection of non-Enterococcus species was slightly higher using Enterolert (8.4%) than for EPA Method 1600 (5.1%). E. faecalis and E. faecium, commonly associated with human fecal waste, were predominant in wastewater; however, Enterolert had greater selectivity for E. faecalis, which was also shown using a laboratory-created sample. The same species selectivity was not observed for most beach water and urban runoff samples. These samples had relatively higher proportions of plant associated species, E. casseliflavus (18.5%) and E. mundtii (5.7%), compared to wastewater, suggesting environmental inputs to beaches and runoff. The potential for species selectivity among water testing methods should be considered when assessing the sanitary quality of beaches so that public health warnings are based on indicators representative of fecal sources.
1. Introduction
EPA Method 1600 and Enterolert (IDEXX, Westbrook, ME, USA) are two EPA-approved methods that are often used to measure Enterococcus for recreational bathing water quality assessments [1]. The methods have been used interchangeably for (1) regulatory monitoring to detect possible fecal contamination of water; (2) epidemiology studies to correlate swimmer's illness rates with densities of Enterococci in water, and (3) microbial source tracking studies to reduce fecal inputs to protect public health. A number of studies have found that these two methods generally produce comparable results [2–5]. However, several authors have found that the results may be markedly different [6–9].
There are several reasons, other than sample variability, that may explain the inconsistency between methods. EPA Method 1600 is a membrane filtration approach, where water is passed through a membrane that is subsequently placed atop Enterococcus Indoxyl-β-D-glucoside (mEI) agar and, following incubation, examined for colonies with blue halos. Enterolert is a defined substrate methodology that measures a fluorescent endpoint based on enterococci metabolizing 4-methylumbelliferone-β-D-glucoside in liquid media.
Differences in the combinations of growth-controlling substrates in these media could lead to selectivity of enterococcal species and detection of nonenterococcal bacteria, including Streptococcus spp., Aerococcus spp., and Lactococcus spp. [10]. False positive rates have been found to vary between 4% and 26% for Enterolert [11] and between 11% and 26% for mEI agar [10].
Another possible reason for the occasional differences between methods would be differential selectivity for species within the Enterococcus group, particularly since Enterolert uses a liquid broth culture media and EPA Method 1600 uses a solid agar that will affect the growth kinetics of cells. In liquid media, faster growing bacteria can outgrow their slower counterparts, even with diluted samples [12]. In contrast, bacterial cells growing on a membrane placed atop agar media are spatially separated, providing less opportunity for competition.
Knowledge of Entercoccus species distribution in urban runoff and wastewater may be useful for assessing recreational waters deemed unsafe for swimming based on enterococci water quality standards. E. faecalis and E. faecium are the two most prevalent species in human feces [12]; E. casseliflavus and E. mundtii are associated with plants and soil [13]; these species are not considered typical members of the human intestinal microflora [14]. Thus, characterizing the distribution of enterococcal species representative of fecal contamination should be conducted using methods that are not subject to species selectivity.
Here, we test the hypothesis that EPA Method 1600 and Enterolert differ in species selectivity by examining the species composition of their isolates when both methods were used to process a common set of samples.
2. Methods
Differences in species selectivity were determined using 18 environmental samples and a laboratory-created sample with known species composition. The environmental samples were collected from seven marine beach sites, two freshwater urban runoff sites, influent from four wastewater treatment plants (WWTPs), and secondary effluent from four WWTPs (Table 1). The beach water was collected using 100 mL plastic bottles at ankle depth upon an incoming wave. Urban runoff was collected as 1 L samples just below the water surface. Wastewater was collected as 1 L samples from influent or effluent pipes.
Table 1.
Sources of samples.
| Beach water | |
| Imperial Beach, San Diego | |
| San Mateo Beach, San Clemente | |
| Doheny State Beach, Dana Point | |
| Cabrillo Beach, Los Angeles | |
| Surfrider Beach, Malibu | |
| Paradise Cove, Malibu | |
| Big Sycamore, Malibu | |
| Urban runoff | |
| Dominguez Channel, Los Angeles | |
| Tijuana River, San Diego | |
| Wastewater treatment plant (WWTP) | |
| Joint Water Pollution Control Plant of the Los Angeles County Sanitation District |
|
| Orange County Sanitation District, Huntington Beach | |
| South Orange County Wastewater Authority, Dana Point | |
| Encina Wastewater Authority, Carlsbad |
The laboratory-created sample consisted of clean seawater inoculated with approximately 1,000 colony forming units per 100 mL each of E. faecium and E. faecalis. The seawater was collected 18 kilometers offshore, at 10-meter depth with no measurable enterococci present. The enterococci cultures were prepared using strains from environmental samples that were enumerated using EPA Method 1600 and Enterolert and identified to species using the Vitek microbial identification system (bioMérieux, St. Louis, MO, USA).
All samples were analyzed within six hours of collection following EPA standards [15] and the Enterolert manufacturer's instructions. For EPA Method 1600, 10–50 mL volumes of sample were filtered onto mEI, and presumptive enterococci isolates were obtained by selecting up to five colonies (per sample) with blue halos from mEI agar (Northeast Laboratory, Waterville, ME, USA) and subculturing them onto tryptic soy agar (TSA) with 5% sheep blood (Northeast Laboratory, Waterville, ME, USA). After 24 h incubation at 35°C, the blood agar plates (BAPs) were examined to ensure they were pure cultures. Isolates from BAPs were subcultured onto TSA slants (Northeast Laboratory, Waterville, ME, USA) and incubated as before. The TSA slants were stored at 4°C until speciation was performed.
Ten mL of sample were used for the Enterolert Quanti-Tray method and enterococci isolates following the method of Kinzelman et al. [6]. The back of the Quanti-Tray was disinfected with 70% alcohol, and media from up to five fluorescing (positive) wells was withdrawn using sterile syringes. The media was then inoculated into brain heart infusion (BHI) broth (BD, Franklin Lakes, New Jersey, USA) containing 6.5% NaCl at a 1 : 20 dilution. Inoculated broth was then incubated at 41°C for 48 h. Cultures that had growth were subcultured onto BAPs, and then colonies from the BAPs were subcultured onto TSA slants, incubated, and stored at 4°C.
2.1. Isolate Identification
Approximately 80 isolates each from 17 environmental samples that were obtained using EPA Method 1600 and Enterolert were identified to species using the Vitek microbial identification system (bioMérieux, St. Louis, MO, USA) (Table 2). Isolates identified as Enterococcus species with discrimination of <80% confidence were categorized as “indeterminant”. Isolates identified as species other than Enterococcus were categorized as “non-Enterococcus”. E. casseliflavus/E. gallinarum isolates that could not be discriminated using Vitek were tested for motility and pigment production following Ferguson et al. [10] to differentiate these species. Since E. mundtii is not identified by Vitek [16], a total of 107 isolates from beach water, wastewater and freshwater were screened for this species using published biochemical tests, including motility; pigment production; and fermentation of arabinose, sucrose, and mannitol [17].
Table 2.
Sources of samples and numbers of isolates analyzed for speciation using EPA Method 1600 and Enterolert.
| Source (no. of samples) | EPA Method 1600 | Enterolert | No. of isolates |
|---|---|---|---|
| Beaches (7) | 275 | 303 | 578 |
| Urban runoff (2) | 91 | 99 | 290 |
| Wastewater influent (4) | 126 | 130 | 256 |
| Wastewater effluent (4) | 129 | 126 | 255 |
| Culture (1) | 20 | 46 | 66 |
|
| |||
| Total | 621 | 658 | 1279 |
3. Results and Discussion
3.1. Enterococcus Species Assemblages
1279 presumptive enterococci isolates from environmental samples (beach water, urban runoff, and wastewater treatment plant influent and effluent) (Table 2) using EPA Method 1600 and Enterolert were examined and found to include nine species of Enterococcus. E. faecium and E. faecalis were the most frequent species identified among isolates obtained using Enterolert, with each species comprising about one third of isolates from all samples (Figure 1). These were also the most frequent species obtained using EPA Method 1600, though E. faecium was more common (44%) than E. faecalis (13%). E. gallinarum and E. casseliflavus were the next most frequently identified species by both methods. Non-Enterococcus species comprised 8% of the isolates derived from Enterolert and 5% from EPA Method 1600. The most common nonenterococcal bacteria isolated were Proteus mirabilis from Enterolert wells and Aerococcus viridans from EPA Method 1600. Other species identified by both methods included Streptococcus bovis, S. uberis, S. mutans, and S. pneumonia. Five percent of the isolates were identified with a low level of certainty and classified as “indeterminant.”
Figure 1.
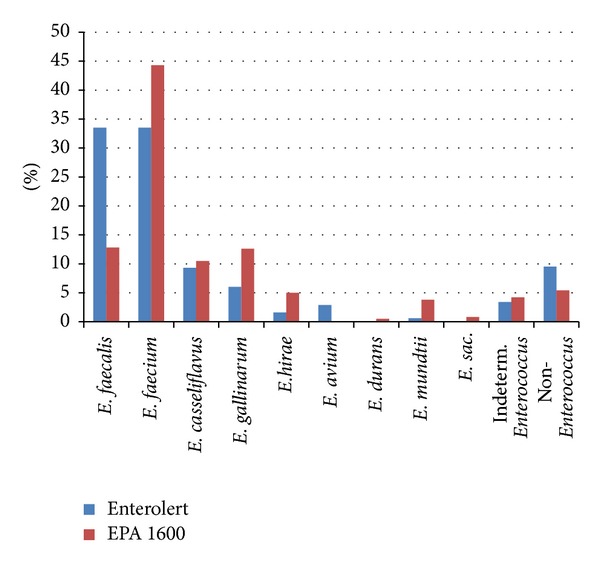
Enterococcus species found overall using Enterolert versus EPA Method 1600.
Overall relative abundance of species among all the environmental samples was not significantly different between methods (χ 2, (1, N = 1104) = 47.4, P < 0.001). However, the relative proportions of E. faecium and E. faecalis were significantly different between methods for the wastewater samples, with E. faecalis the most frequently observed species for Enterolert and E. faecium dominant for EPA Method 1600 (Figure 2). The proportions of E. faecalis and E. faecium were generally similar among two urban runoff samples and eight beach water samples (Fisher's exact test, P = 0.31).
Figure 2.
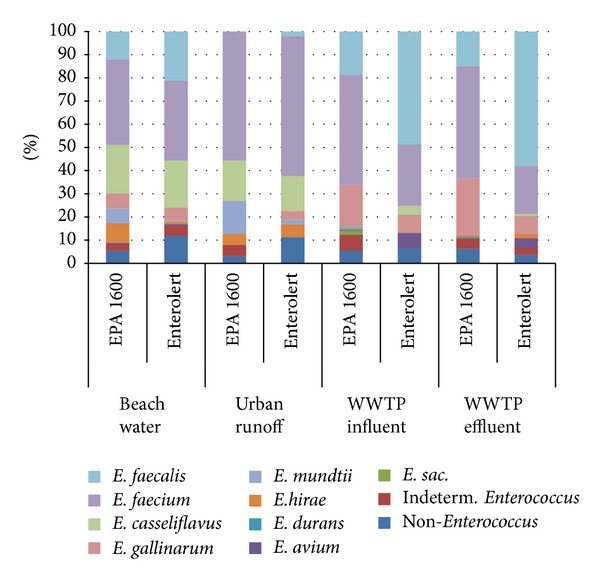
Distribution of predominant Enterococcus species found among beach water, urban runoff, and wastewater treatment plant (WWTP) influent (untreated) and effluent (treated) samples.
The same selectivity observed in the wastewater samples using Enterolert was also observed in the laboratory sample spiked with similar concentrations of E. faecalis and E. faecium (Figure 3). EPA Method 1600 identified 65% of the isolates as E. faecalis, 30% as E. faecium, and 5% as “indeterminant.” In contrast, 98% of the Enterolert isolates were identified as E. faecalis.
Figure 3.
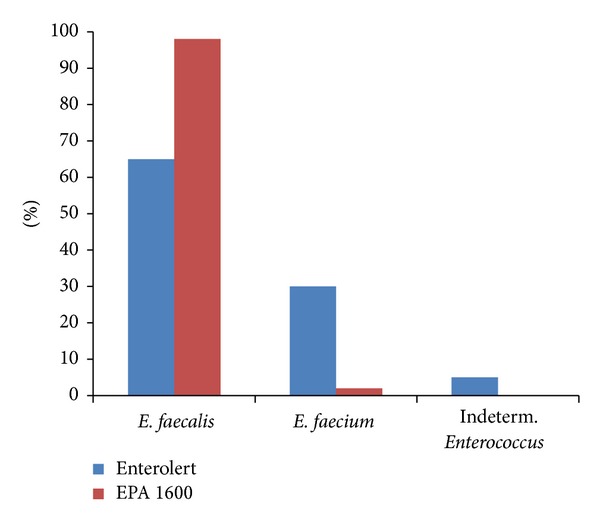
Percent E. faecalis and E. faecium isolates from culture sample containing 1 : 1 ratio of both species obtained using Enterolert and EPA Method 1600.
Enterococcal species identification using Vitek was supplemented with pigment production and motility testing to identify E. mundtii and E. casseliflavus that may be misidentified as E. gallinarum by Vitek alone [16]. Of the 107 isolates screened for E. mundtii, twelve isolates were misidentified as E. gallinarum by Vitek; six of these were confirmed as E. mundtii, four as non-Enterococcus species, and two as E. casseliflavus based on additional biochemical tests, pigmentation, and motility. Vitek identifications of E. faecalis and E. faecium were much more accurate; all but one isolate each of E. faecalis and E. faecium were identified similarly by Vitek and conventional biochemical tests.
3.2. Enterococcus Species Selectivity
While the two methods generally yielded the same species assemblage, Enterolert identified a higher proportion of E. faecalis than EPA Method 1600 did across all sample types. There are a number of reasons why this might occur, one of which is the difference between liquid and solid growth media discussed earlier. Another difference is media formulation. The mEI media used in EPA Method 1600 contain additives, such as triphenyltetrazolium chloride to differentiate Enterococcus from other Gram-positive cocci, sodium azide and nalidixic acid to inhibit the growth of Gram-negative bacteria, and cycloheximide to suppress the growth of fungi; Enterolert media are proprietary, and it is unknown whether similar additives to increase specificity are present. The media also differ in the reporter molecules they use to detect enterococci. While mEI media rely on the ability of Enterococcus to metabolize indoxyl-β-D-glucoside to produce a blue compound, Enterolert media rely on 4-methylumbelliferone-β-D-glucoside to produce a fluorescent metabolite.
Another potential mechanism for the observed differences is oxygen availability. Enterolert media are heat-sealed within a Quanti-Tray, creating a more anaerobic incubation condition compared to mEI agar in a petri dish. Enterococci are facultative anaerobes [17], but it is unknown whether growth rates vary among species and strains under differing oxygen levels. Differences in growth rates among Enterococcus species could also account for differing results between methods. Delayed growth rates and positive fluorescence reactions (after 24 hours) of certain enterococcal strains in Quanti-Trays raised concerns that Enterolert may underestimate enterococci densities [9, 18]. Species selection bias could have been introduced during the BHI broth (with 6.5% NaCl) subculture step that we used to facilitate isolation of enterococci from Enterolert. This enrichment step was used to reduce non-Enterococcus species in samples and also because previous attempts to subculture directly from Enterolert onto nonselective media resulted in nondetection of enterococci. To assess potential selection bias, we compared the growth rates of E. faecalis (ATCC 29212), E. faecium (ATCC 35667), and E. casseliflavus (ATCC 700527) in BHI broth with 6.5% NaCl versus Enterolert media, determining the doubling time of each species using optical density (600 nanometers; UV160U Spectrophotometer, Shimadzu Scientific Instruments, Columbia, MD, USA) and plate counts on mEI. E. faecium grew faster than E. faecalis in Enterolert and in BHI broth with 6.5% NaCl. However, Enterolert was not selective for E. faecium, and E. faecium was not predominant in the actual samples suggesting that other factors likely accounted for the Enterolert selectivity for E. faecalis observed in the wastewater samples. Enterolert also showed the same selectivity for E. faecalis among isolates subcultured directly from a laboratory-created sample without using BHI step.
3.3. Speciation Method Limitations
We used the Vitek system, which is routinely used by environmental laboratories, to verify presumptive enterococci on mEI, for most of our species identification (APHA 2000). One limitation of Vitek is that the database does not include E. mundtii, which is a species that is present in environmental waters [10]. In a previous study, Moore et al. [16] found that 14% of the isolates speciated by Vitek without supplementary testing were misidentified. In our study, 17% of the isolates identified by Vitek had discrepant identifications compared to conventional biochemical testing. The majority of these were isolates that had been misidentified by Vitek as E. gallinarum that were later identified as E. mundtii, E. casseliflavus, or non-Enterococcus based on supplementary testing including pigment, motility, and additional biochemical tests. Thus, the overall percentages of E. gallinarum identified in this study using Vitek alone may be somewhat inflated.
Molecular methods that could have been used to identify Enterococcus isolates include 16S rRNA gene sequencing [18] and genus and species specific multiplex-PCR [19]. These methods are rapid, cost-effective and allow high throughput. In some cases, conventional biochemical testing may be useful when used in conjunction with molecular methods, such as 16S rRNA sequencing [20, 21]. For example, pigment production and motility can be used to discriminate E. casseliflavus and E. gallinarum with identical 16S rRNA gene sequences [16]. Since E. casseliflavus is associated with plants, discriminating these two species could be important for assessing natural sources. Multiplex-PCR is another molecular-based speciation method that was shown as having 90% concordance with Vitek and conventional biochemical methods for identifying Enterococcal species from environmental and fecal isolates [19]. However, the sensitivity of multiplex PCR may be a concern; Layton et al. [22] used a modified version of Jackson's multiplex PCR method and identified eight Enterococcus species among isolates from animal fecal samples but found that samples with less than 30 colony forming units (CFU) required a culture enrichment step; following enrichment, ~70% of potential enterococcal species were detected using multiplex PCR.
3.4. Enterococcus Species Distribution in Wastewater, Beach Water, and Urban Runoff
While there were some differences in species selectivity between Enterolert and EPA Method 1600, these differences were much smaller than the species composition differences that both methods found among the different sample types. For instance, both methods found much higher percentages of E. casseliflavus in beach samples (~20%) than in wastewater samples (<4%). Both methods also found similar enterococcal species distribution among sewage influent from the four wastewater treatment plants, despite differences in size, treatment processes, and the nature of service areas. The dominance of E. faecium and E. faecalis in the wastewater systems is consistent with previous studies that have examined species distributions of Enterococcus in wastewater [23–25]. Higher percentages of E. faecalis and E. faecium in wastewater streams are also consistent with clinical studies that have established that these two species comprise a significant fraction of the enterococci found in human and animal feces [14, 22, 26]. Similarly, the prevalence of E. casseliflavus in beaches and runoff samples with minimal human fecal sources and its lower occurrence in wastewater samples is also consistent with clinical human fecal samples [27, 28] and its known association with natural sources, such as plants [29, 30].
4. Conclusions
EPA Method 1600 and Enterolert detected similar proportions of Enterococcus species in marine and spiked samples; however, Enterolert was more selective for E. faecalis in wastewater samples. Also, Enterolert yielded higher percentages of non-Enterococcus organisms in beach water and runoff samples, which could account for occasional differences in water quality assessments using both methods. Further insights on the diversity of Enterococcus species in environmental waters may help to improve studies on health risk assessments. For species identification of Enterococcus using culture methods, EPA Method 1600 is recommended to obtain a more accurate characterization.
Conflict of Interests
The authors wish to confirm that there is no known conflict of interests or financial support associated with this work that could have influenced the study findings.
Acknowledgments
The authors thank Matthew Hamilton, Melissa Madison, Linda Tran, and Andriana Gallegos-Amparo, as well as the Los Angeles County Sanitation District, Orange County Sanitation District, South Orange County Wastewater Authority, Encina Wastewater Authority, and Orange County Public Health Laboratory, for their assistance in collecting and processing the samples.
References
- 1.Weisberg SB, Noble RT, Griffith JF. Microbial indicators of marine recreational water quality. In: Hurst CJ, Crawford RL, Garland JL, Lipson DA, Miles AL, Stetzenbach LD, editors. Manual of Environmental Microbiology. 3rd edition. Washington, DC, USA: ASM Press; 2007. pp. 280–289. [Google Scholar]
- 2.Budnick GE, Howard RT, Mayo DR. Evaluation of enterolert for enumeration of enterococci in recreational waters. Applied and Environmental Microbiology. 1996;62(10):3881–3884. doi: 10.1128/aem.62.10.3881-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckner KF. Comparison of membrane filtration and multiple-tube fermentation by the Colilert and Enterolert methods for detection of waterborne coliform bacteria, Escherichia coli, and enterococci used in drinking and bathing water quality monitoring in southern Sweden. Applied and Environmental Microbiology. 1998;64(8):3079–3083. doi: 10.1128/aem.64.8.3079-3083.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noble RT, Leecaster MK, McGee CD, Weisberg SB, Ritter K. Comparison of bacterial indicator analysis methods in stormwater-affected coastal waters. Water Research. 2004;38(5):1183–1188. doi: 10.1016/j.watres.2003.11.038. [DOI] [PubMed] [Google Scholar]
- 5.Griffith JF, Aumand LA, Lee IM, et al. Comparison and verification of bacterial water quality indicator measurement methods using ambient coastal water samples. Environmental Monitoring and Assessment. 2006;116(1–3):335–344. doi: 10.1007/s10661-006-7571-z. [DOI] [PubMed] [Google Scholar]
- 6.Kinzelman J, Ng C, Jackson E, Gradus S, Bagley R. Enterococci as indicators of Lake Michigan recreational water quality: comparison of two methodologies and their impacts on public health regulatory events. Applied and Environmental Microbiology. 2003;69(1):92–96. doi: 10.1128/AEM.69.1.92-96.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maheux AF, Picard FJ, Boissinot M, et al. Analytical limits of three β-glucosidase-based commercial culture methods used in environmental microbiology, to detect enterococci. Water Science and Technology. 2009;60(4):943–954. doi: 10.2166/wst.2009.428. [DOI] [PubMed] [Google Scholar]
- 8.Shibata T, Solo-Gabriele HM, Sinigalliano CD, et al. Evaluation of conventional and alternative monitoring methods for a recreational marine beach with nonpoint source of fecal contamination. Environmental Science and Technology. 2010;44(21):8175–8181. doi: 10.1021/es100884w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valente MS, Pedro P, Alonso MC, Borrego JJ, Dionísio L. Are the defined substrate-based methods adequate to determine the microbiological quality of natural recreational waters? Journal of Water and Health. 2010;8(1):11–19. doi: 10.2166/wh.2009.220. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson DM, Moore DF, Getrich MA, Zhowandai MH. Enumeration and speciation of enterococci found in marine and intertidal sediments and coastal water in southern California. Journal of Applied Microbiology. 2005;99(3):598–608. doi: 10.1111/j.1365-2672.2005.02660.x. [DOI] [PubMed] [Google Scholar]
- 11.Sercu B, van de Werfhorst LC, Murray JLS, Holden PA. Cultivation-independent analysis of bacteria in IDEXX quanti-tray/2000 fecal indicator assays. Applied and Environmental Microbiology. 2011;77(2):627–633. doi: 10.1128/AEM.01113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aarestrup FM, Agerso Y, Gerner-Smidt P, Madsen M, Jensen LB. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagnostic Microbiology and Infectious Disease. 2000;37(2):127–137. doi: 10.1016/s0732-8893(00)00130-9. [DOI] [PubMed] [Google Scholar]
- 13.Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. Enterococci in the environment. Microbiology and Molecular Biology Reviews. 2012;76(4):685–706. doi: 10.1128/MMBR.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tannock GW, Cook G. Enterococci as members of the intestinal microflora of humans. In: Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB, editors. The Enterococci: Pathogenesis, Molecular Biology and Antibiotic Resistance. Washington, DC, USA: ASM Press; 2002. pp. 104–105. [Google Scholar]
- 15.USEPA. EPA. 821-r-02-022. Washington, DC, USA: United States Environmental Protection Agency; 2002. Enterococci in water by membrane filtration using membrane-Enterococcus indoxyl-β-D-glucoside Agar (mEI) [Google Scholar]
- 16.Moore DF, Zhowandai MH, Ferguson DM, McGee C, Mott JB, Stewart JC. Comparison of 16S rRNA sequencing with conventional and commercial phenotypic techniques for identification of enterococci from the marine environment. Journal of Applied Microbiology. 2006;100(6):1272–1281. doi: 10.1111/j.1365-2672.2006.02879.x. [DOI] [PubMed] [Google Scholar]
- 17.Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. Journal of Clinical Microbiology. 1989;27(4):731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harwood VJ, Delahoya NC, Ulrich RM, et al. Molecular confirmation of Enterococcus faecalis and E. faecium from clinical, faecal and environmental sources. Letters in Applied Microbiology. 2004;38(6):476–482. doi: 10.1111/j.1472-765X.2004.01518.x. [DOI] [PubMed] [Google Scholar]
- 19.Jackson CR, Fedorka-Cray PJ, Barrett JB. Use of a genus- and species-specific multiplex PCR for identification of enterococci. Journal of Clinical Microbiology. 2004;42(8):3558–3565. doi: 10.1128/JCM.42.8.3558-3565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angeletti S, Lorino G, Gherardi G, Battistoni F, de Cesaris M, Dicuonzo G. Routine molecular identification of enterococci by gene-specific PCR and 16s ribosomal DNA sequencing. Journal of Clinical Microbiology. 2001;39(2):794–797. doi: 10.1128/JCM.39.2.794-797.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domig KJ, Mayer HK, Kneifel W. Methods used for the isolation, enumeration, characterisation and identification of Enterococcus spp. International Journal of Food Microbiology. 2003;88(2-3):165–188. doi: 10.1016/s0168-1605(03)00178-8. [DOI] [PubMed] [Google Scholar]
- 22.Layton BA, Walters SP, Lam LH, Boehm AB. Enterococcus species distribution among human and animal hosts using multiplex PCR. Journal of Applied Microbiology. 2010;109(2):539–547. doi: 10.1111/j.1365-2672.2010.04675.x. [DOI] [PubMed] [Google Scholar]
- 23.Sinton LW, Donnison AM. Characterisation of faecal streptococci from some New Zealand effluents and receiving waters. New Zealand Journal of Marine and Freshwater Research. 1994;28(2):145–158. [Google Scholar]
- 24.Blanch AR, Caplin JL, Iversen A, et al. Comparison of enterococcal populations related to urban and hospital wastewater in various climatic and geographic European regions. Journal of Applied Microbiology. 2003;94(6):994–1002. doi: 10.1046/j.1365-2672.2003.01919.x. [DOI] [PubMed] [Google Scholar]
- 25.Moore DF, Guzman JA, McGee C. Species distribution and antimicrobial resistance of enterococci isolated from surface and ocean water. Journal of Applied Microbiology. 2008;105(4):1017–1025. doi: 10.1111/j.1365-2672.2008.03828.x. [DOI] [PubMed] [Google Scholar]
- 26.Murray BE. The life and times of the Enterococcus . Clinical Microbiology Reviews. 1990;3(1):46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruoff KL, De La Maza L, Murtagh MJ, Spargo JD, Ferraro MJ. Species identities of enterococci isolated from clinical specimens. Journal of Clinical Microbiology. 1990;28(3):435–437. doi: 10.1128/jcm.28.3.435-437.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern CS, Carvalho MDGS, Teixeira LM. Characterization of enterococci isolated from human and nonhuman sources in Brazil. Diagnostic Microbiology and Infectious Disease. 1994;20(2):61–67. doi: 10.1016/0732-8893(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 29.Ott E-M, Müller T, Müller M, et al. Population dynamics and antagonistic potential of enterococci colonizing the phyllosphere of grasses. Journal of Applied Microbiology. 2001;91(1):54–66. doi: 10.1046/j.1365-2672.2001.01334.x. [DOI] [PubMed] [Google Scholar]
- 30.Aarestrupp FM, Butaye P, Wolfgang W. Nonhuman reservoirs of enterococci. In: Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB, editors. The Enterococci: Pathogenesis, Molecular Biology and Antibiotic Resistance. Washington, DC, USA: ASM Press; 2002. pp. 104–105. [Google Scholar]


