Abstract
We present a method that uses fluorescent cellular barcodes to increase the number of unique samples that can be analyzed simultaneously by microengraving—a nanowell array-based technique for quantifying the secretory responses of thousands of single cells in parallel. By using n different fluorescent dyes to generate 2n unique cellular barcodes, we achieved a 2n-fold reduction in the number of arrays and quantity of reagents required per sample. The utility of this approach was demonstrated in three applications of interest in clinical and experimental immunology. Using barcoded human peripheral blood mononuclear cells and T cells, we constructed dose-response curves, profiled the secretory behavior of cells treated with mechanistically distinct stimuli, and tracked the secretory behaviors of different lineages of CD4+ T helper cells. In addition to increasing the number of samples analyzed by generating secretory profiles of single cells from multiple populations in a time- and reagent-efficient manner, we expect that cellular barcoding in combination with microengraving will facilitate unique experimental opportunities for quantitatively analyzing interactions among heterogeneous cells isolated in small groups (~2–5 cells).
INTRODUCTION
Immune cells secrete cytokines to coordinate intercellular communication within the immune network.1 There is great interest in profiling the secretory activity of immune cells because the cytokines they secrete play a central role in the maintenance of immune homeostasis, the elimination of infectious pathogens, and the induction of allergic and autoimmune responses.2,3 The considerable cell-to-cell variability present within populations of immune cells underscores the importance of analytical techniques that enable high-throughput, single-cell secretory measurements.
We previously developed a technique called microengraving that uses dense arrays of subnanoliter wells (nanowells) to quantify the secretion of multiple cytokines from thousands of individual cells in parallel.4–8 Cells are isolated in an array of nanowells, and a glass slide bearing cytokine-specific antibodies is compressed on the array to capture the cytokines secreted by the cells in each well. Single-cell secretory profiles are created by registering the spatial address of each spot on the resulting microarray of secreted proteins back to the corresponding nanowell, and hence the cell(s), that produced the cytokines. Microengraving can be repeatedly performed on the same cells in a non-destructive manner, enabling analytical processes that are not feasible using destructive or end-point single-cell measurements of cytokine production (e.g., intracellular cytokine staining or ELISPOT). These processes include the retrieval of viable cytokine-secreting cells5,6 and longitudinal tracking of single-cell secretory profiles.4
To date, microengraving has been performed with a throughput of one sample of cells per process. In many cases, however, the analysis of multiple samples in parallel would increase experimental efficiency. One strategy for multiplexing is to use unique combinations of fluorescent dyes to identify distinct groups of cells. This strategy, known as fluorescent cellular barcoding, has been used to increase the throughput of flow cytometry9,10 and cell-based assays for drug screening,11 as well as to track the behavior of specific cells within complex populations.12–14 In this Technical Note, we describe the development and validation of three sets of cellular barcodes that are compatible with microengraving. Application of these cellular barcodes enables the simultaneous analysis of multiple samples of cells by microengraving on a single array of nanowells, and thus increases sample throughput, minimizes sample-to-sample technical variability, and reduces both the number of arrays and the quantity of reagents used. Moreover, cellular barcoding opens the door to new applications of microengraving, such as the quantitative analysis of secretory networks governing cell-cell interactions in multi-celled wells.
EXPERIMENTAL SECTION
Fabrication of Arrays of Nanowells
Poly(dimethylsiloxane) (PDMS) (Sylgard 184 Silicone Elastomer Kit; Dow Corning, Midland, MI) arrays of nanowells comprising 50 µm cubic wells (84,672 wells/array) were prepared on 75 × 25 mm2 glass slides (Corning, Lowell, MA) following previously reported protocols15 with minor adaptations. Details can be found in the Supporting Information (SI).
Cells
Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation using Ficoll-Paque PLUS (GE Healthcare, Piscataway, NJ) from the whole blood of healthy donors (Research Blood Components, Boston, MA) and either used fresh or cryopreserved. Before use, cryopreserved PBMCs were thawed, washed with complete media (RPMI-1640 (Mediatech, Manassas, VA) supplemented with 10% heat inactivated fetal bovine serum (FBS; PAA Laboratories, New Bedford, MA), 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, and 10 mM HEPES (all from Mediatech)), and then rested overnight (37°C, 5% CO2) in complete media. T cells were isolated from PBMCs by negative selection (EasySep Human T Cell Enrichment Kit; STEMCELL Technologies, Vancouver, BC, Canada) and incubated in complete media until use. Details on T helper (Th) cell polarization can be found in the SI.
Stimulations
Prior to performing the dye swap experiment, T cells (106 cells/mL) were stimulated in a conical tube for 3 h with 25 ng/mL phorbol 12-myristate 13-acetate (PMA) and 1 µM ionomycin (both from Sigma-Aldrich, St. Louis, MO). To evaluate the effect of dose on the functional responses to PMA/ionomycin, T cells (75,000 cells/well) were stimulated in a 96-well flat-bottom plate for 5 h with 10 ng/mL PMA and 0, 0.25, 0.5, or 1 µg/mL ionomycin. To compare diverse stimulation conditions, PBMCs (106 cells/well) were incubated in a 96-well U-bottom plate in media only or stimulated for 4 h with PMA/ionomycin (10 ng/mL PMA, 1 µM ionomycin), the Toll-like receptor 4 (TLR4) agonist lipopolysaccharide (LPS-EK, 1 µg/mL; InvivoGen, San Diego, CA), or the TLR7/8 agonist R848 (1 µg/mL; InvivoGen). To test CD4+ Th cell lineages, Th0-, Th1-, or Th2-polarized cells (105 cells/well) were incubated in a 96-well U-bottom plate in media only or stimulated for 4 h with PMA/ionomycin (10 ng/mL PMA, 1 µg/mL ionomycin) as indicated.
Cellular Barcoding
Three sets of cellular barcodes were applied as described below. Cells were then washed twice with media and either loaded into a 96-well plate to determine classification accuracy or mixed and loaded onto an array of nanowells for imaging and microengraving.
Antibody-based barcoding
Groups of cells were barcoded by the combinatorial application of anti-CD45-quantum dot (QD) 705 (20 nM; Invitrogen) and anti-CD45-QD800 (20 nM; Invitrogen) in media for 15 min at room temperature. Four (22) unique barcodes were defined based on QD705 and QD800 staining.
Cytosolic barcoding
Groups of cells were barcoded by the combinatorial application of the membrane-permeable dyes carboxyfluorescein diacetate succinimidyl ester (CFSE, 1 µM; Invitrogen, Grand Island, NY) and CellTracker Red (CTR, 2.5 µM; Invitrogen). Four (22) unique barcodes were defined based on CFSE and CTR staining. Cells were labeled by directly adding the appropriate combination of dyes to cell suspensions during the last 30 min of stimulation.
Streptavidin-based barcoding
Cells were suspended in Hank’s Balanced Salt Solution (HBSS) at 106 cells/mL in 15 mL conical tubes that were previously blocked with 0.5% polyvinyl alcohol to prevent the adhesion of cells. The cells were labeled with sulfo-NHS-LC-biotin (0.1 mg/mL; Thermo Scientific, Waltham, MA) for 30 min at 4°C. After one wash, groups of cells were barcoded by the combinatorial application of 40 nM of streptavidin-phycoerythrin (PE)-Cy7 (BioLegend), streptavidin-ITK-QD705 (Invitrogen), and streptavidin-ITK-QD800 (Invitrogen) in HBSS for 15 min at room temperature in a 96-well plate that was pre-blocked with 10% FBS. Eight (23) unique barcodes were defined based on PECy7, QD705, and QD800 staining.
Loading of Cells onto Arrays of Nanowells
Cells originating from different groups were combined into a single suspension (5 × 105 cells/mL) after each group had been uniquely barcoded. Then, 300 µL of cell suspension was deposited onto the array. To minimize the chance of cell-cell interactions occurring in the mixed cell suspension, cells were deposited on the array within minutes of being combined together. Cells were allowed to settle by gravity for 5 min before the array was washed gently with media.
Detection of Secreted Proteins by Microengraving
Immediately after labeling and loading the cells, microengraving was performed for 1 h to detect interferon-γ (IFN-γ), macrophage inflammatory protein-1β (MIP-1β), interleukin-2 (IL-2), IL-4, IL-6, and tumor necrosis factor-α (TNF) using previously reported protocols5,15 with minor adaptations. The specific cytokines analyzed in each experiment are indicated in the text. Additional details can be found in the SI.
Staining for Viability and Surface Marker Expression
Where indicated, cells were stained on the arrays with anti-CD8-AlexaFluor647 (2 µg/mL; BioLegend) or anti-CD3-PerCP-eFluor710 (2 test volumes; eBioscience, San Diego, CA). Staining solutions were applied to the arrays for 30 min at room temperature or 4°C and then washed with media. Shortly before imaging, each array was covered with 200 µL of the viability dye calcein violet (2 µM; Invitrogen). Lifter slips (Electron Microscopy Sciences, Hatfield, PA) were placed on top of the arrays to prevent drying during imaging.
Imaging Cytometry
The arrays were imaged using an automated, inverted epifluorescence microscope (Axio Observer, 10×/0.3 objective; Carl Zeiss, Jena, Germany; or, Eclipse Ti, 10×/0.45 objective; Nikon Instruments, Tokyo, Japan) with an EM-CCD camera (ImagEM; Hamamatsu Photonics, Hamamatsu, Japan; or, iXon3; Andor Technology, South Windsor, CT). A custom-written MATLAB script (Enumerator) was used to analyze the images. This script returned each cell’s nanowell “address” and its intensity in each fluorescent channel (Figure S-1).
Data Analysis
Custom-written scripts (MATLAB R2010b; MathWorks, Natick, MA) were used to assign well occupancies and correlate imaging cytometry data with secreted protein data for each well. Only viable cells (calcein violet+) were included in the analysis. Statistical tests were performed in Prism 5 (GraphPad Software, La Jolla, CA). Additional details can be found in the SI.
RESULTS AND DISCUSSION
Overview of Cellular Barcoding Applied to Multiplex Single-Cell Secretory Measurements
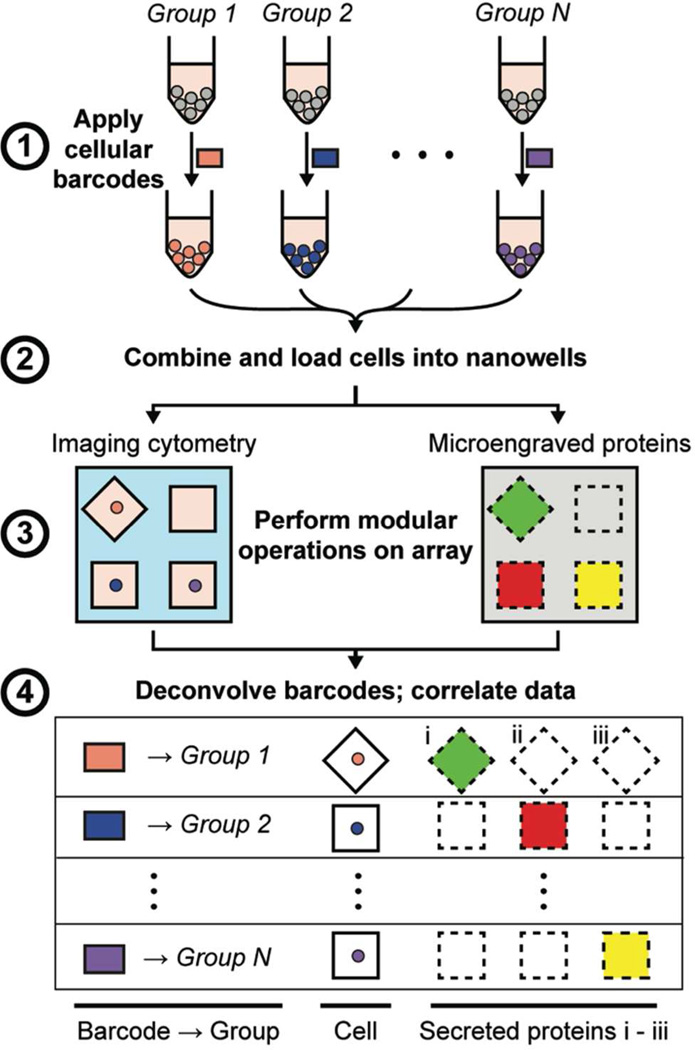
To implement cellular barcoding in conjunction with microengraving, live cells originating from different populations were labeled with unique combinations of fluorescent dyes, combined, and then loaded onto a single array of nanowells (Figure 1). Imaging cytometry was used to determine each cell’s barcode (corresponding to the cell’s population of origin), viability, and expression of surface markers, while microengraving was used to measure the proteins secreted by the cells in each nanowell (Figure S-2).
Figure 1.
Schematic for using cellular barcodes to increase the throughput of secretory measurements from single cells. (1) Distinct groups of cells (e.g., from different treatment conditions) are labeled with unique combinations of fluorescent dyes (barcodes). (2) The cells are combined and loaded onto the array of nanowells. (3) Viability and surface marker expression (labeled on-chip), as well as the barcodes of each cell, are determined by imaging cytometry. Microengraving is performed to measure the factors secreted by the cells in each well. (4) Barcodes are deconvolved during image analysis to identify each cell’s group of origin. Data from imaging cytometry and microengraving are matched on a per-well basis.
Development and Validation of Cellular Barcodes
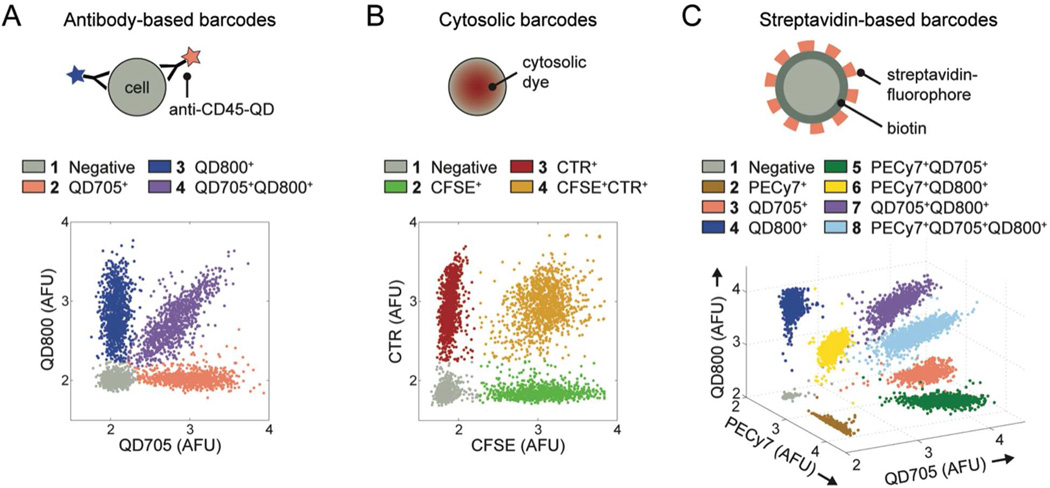
We developed and validated three sets of cellular barcodes for use in nanowell-based assays (Figure 2). The first set targeted hematopoietic cells (such as immune cells) expressing CD45 on their surface. Four antibody-based barcodes were created by labeling CD45+ cells with combinations of two different fluorophore-conjugated antibodies against CD45 (Figure 2A). This strategy of barcoding is extendable to other surface-expressed markers common to all populations of cells of interest, but cannot be applied if cells express surface proteins heterogeneously or if suitable antibodies are unavailable. We therefore established a second set of cellular barcodes to label cells with four combinations of two fluorescent, cytosolic dyes (CFSE and CTR) (Figure 2B). Unlike antibody-based barcodes, cytosolic barcodes are suitable for general use with all cell types and are durably retained in cells for hours to days. These features make cytosolic barcodes useful for experiments that involve mixed cell types or that require deconvolution of populations after prolonged culture. Finally, to extend the depth of barcoding, we created streptavidin-based barcodes by biotinylating cells and then labeling them with eight combinations of three different fluorophore-conjugated streptavidins (streptavidin-PE-Cy7, streptavidin-QD705, and streptavidin-QD800) (Figure 2C). The high efficiency of cellular labeling with biotin-streptavidin and the large spectral selection of commercially available streptavidin-fluorophore conjugates make this approach useful when a high depth of barcoding is needed (e.g., parallel analysis of disaggregated tissue biopsies).
Figure 2.
Barcoded T cells. Cells were labeled with (A) antibody-based barcodes, (B) cytosolic barcodes, or (C) streptavidin-based barcodes. The “Negative” population denotes cells that were not labeled. AFU, arbitrary fluorescence units (logicle transformation).
All three sets of barcodes produced uniform, unambiguous cellular staining (Figure 2) and a reproducibly high accuracy of objective classification (Figure S-3 and Table S-1). We note that factors such as the resolution of the microscope, size of the cells, and density of the cells in the nanowells can influence the accuracy of classification; thus, it is important to measure the accuracy for each different experimental system in which barcodes are used.
As with any perturbation, the application of barcodes could potentially affect the biology of the cell. For example, functionalization of the cell surface for labeling might affect the cell’s ability to respond to autocrine cues. It was therefore necessary to assess whether barcoding perturbed the cellular functions measured over the timescale of interest here (hours). Accordingly, we performed a dye-swap experiment to validate that barcoding did not affect the short-term secretory responses of the cells. Primary human T cells were stimulated with PMA and ionomycin, divided into aliquots, barcoded, and loaded together onto an array of nanowells. Microengraving was then performed to quantify the secretion of IFN-γ, MIP-1β, and IL-2 from the cells in each barcoded group. The percentage of single cells that secreted each cytokine was uniform across all barcoded groups within a given set of barcodes (Figure S-4). The inter-barcode coefficients of variation (CVs) were 3–14% (IFN-γ), 3–14% (MIP-1β), and 2–12% (IL-2). These CVs are comparable to the inter-assay variability of other single-cell assays measuring the production of cytokines, including intracellular cytokine staining16,17 and ELISPOT.18 Furthermore, the median fluorescence intensities (MFIs) for positive secretion events were similar across barcoded groups (Figure S-5). In each set of barcodes, one group of cells was not labeled until after microengraving, and thus served as an internal, unperturbed control. The secretory response of this group was not significantly different from the responses of the other barcoded groups, further indicating that the application of the barcodes did not affect the short-term secretory biology of the cells. We therefore proceeded to use cellular barcoding in three applications relevant to experimental and clinical immunology.
Application 1: Efficient Construction of Dose-Response Curves
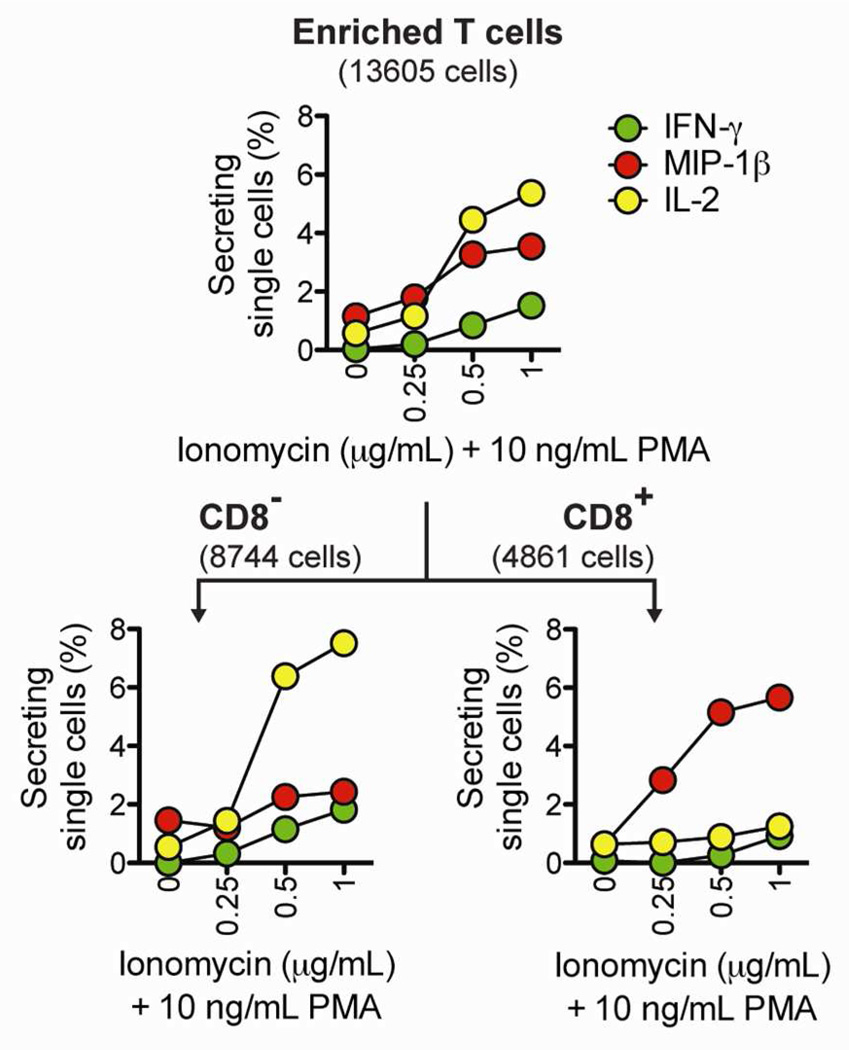
Many immunological assays rely on stimulating cells with a chemical stimulant or an antigen, and then measuring the resulting secretion of cytokines. In these assays, it is often beneficial to test a range of doses to determine the optimal concentration of the stimulating reagent. We used cytosolic barcodes to efficiently measure, on a single array of nanowells, the secretory response of human T cells activated with 10 ng/mL PMA in combination with four doses of ionomycin (0, 0.25, 0.5, and 1 µg/mL). After microengraving and deconvolution of barcoded single cells, we observed a dose-dependent increase in secretion (Figure 3, top panel). By staining for CD8 (a surface marker that distinguishes subsets of T cells), we identified subset-specific differences in secretion (Figure 3, bottom panel). In response to increasing doses of ionomycin, the percentage of CD8− T cells secreting IL-2 increased sharply, but the percentage of CD8+ T cells secreting IL-2 remained at basal levels. In contrast, the percentage of CD8+ T cells secreting MIP-1β increased strongly in response to increasing doses of ionomycin, but the percentage of CD8− T cells secreting MIP-1β remained close to basal levels. These observations are consistent with previous studies.19,20 Together, these results demonstrate that cellular barcoding allows dose-response curves to be efficiently constructed from a single microengraving process.
Figure 3.
Application of cellular barcodes to construct dose-response curves. Barcoded T cells were stimulated with 0, 0.25, 0.5, or 1 µg/mL ionomycin and 10 ng/mL PMA. Microengraving was used to measure single-cell secretory responses, and surface phenotype was distinguished by on-chip labeling with anti-CD8. The numbers of single cells analyzed are indicated in parentheses.
Application 2: Profiles of Secretory Responses Induced by Diverse Stimuli
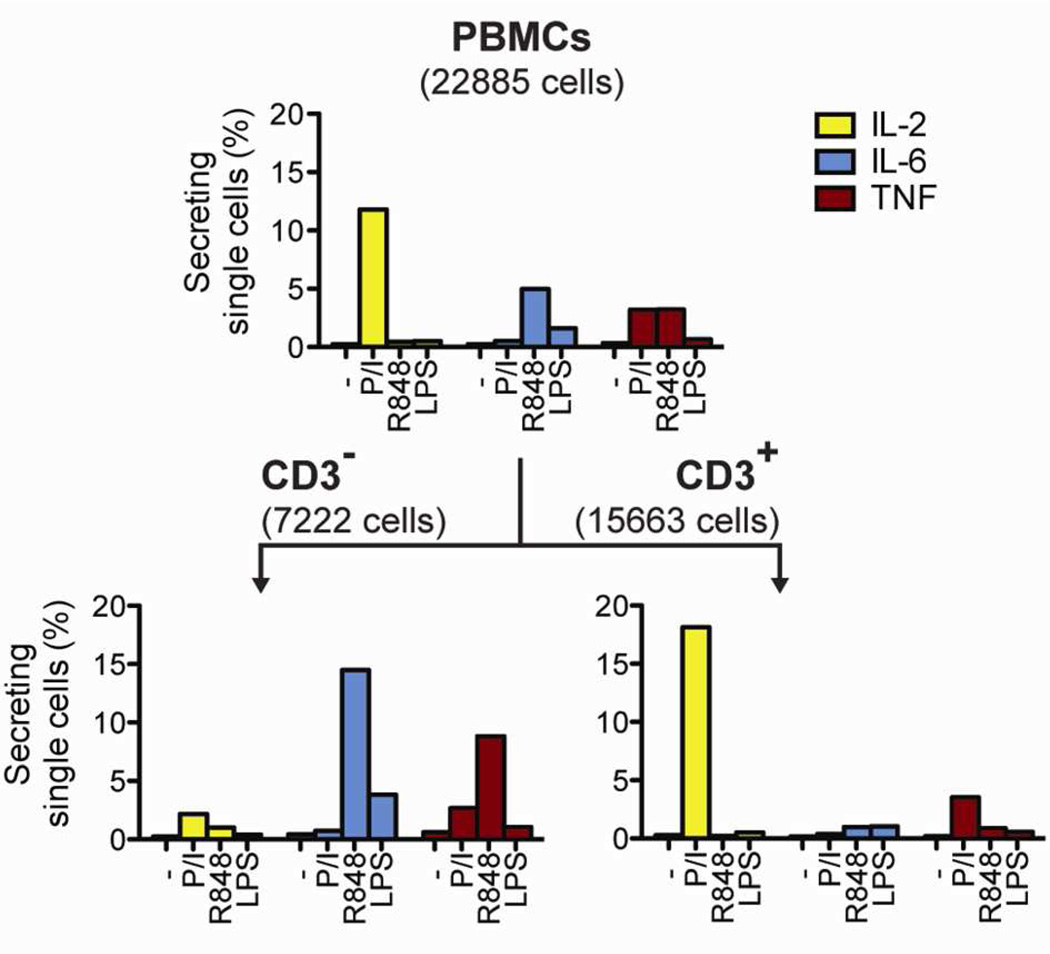
Clinical studies are currently trying to establish robust methods to monitor human immune responses.21,22 One approach is to test how immune cells respond to diverse stimuli that trigger distinct signaling pathways.21 To test this scenario, we applied cytosolic barcodes to PBMCs treated with four different stimulation conditions and measured their secretory responses by microengraving. We observed secretion profiles consistent with the class of stimuli applied (Figure 4, top panel). Unstimulated cells had low numbers of secreting single cells for all cytokines. Treatment with PMA and ionomycin stimulated the secretion of IL-2 and TNF, whereas treatment with TLR agonists LPS or R848 induced the secretion of IL-6 and TNF. By staining for the T cell-specific surface marker CD3, we identified CD3+ T cells as the dominant population responding to stimulation with PMA and ionomycin (Figure 4, bottom panel). In contrast, the majority of secretory responses to both TLR stimuli came from CD3− cells. These findings are consistent with existing knowledge on the secretory responses induced by each of these stimuli23,24 and show that cellular barcoding can be used to multiplex the analysis of diverse stimulatory conditions for applications of microengraving in immune monitoring, such as the rapid evaluation of immunological responses to disease states or vaccination.
Figure 4.
Application of cellular barcodes to measure the percentage of single PBMCs secreting cytokines after treatment with mechanistically distinct stimuli. Barcoded PBMCs were stimulated with PMA/ionomycin (P/I), R848, LPS, or left unstimulated (−). Microengraving was used to measure single-cell secretory responses, and surface phenotype was distinguished by onchip labeling with anti-CD3. The numbers of single cells analyzed are indicated in parentheses.
Application 3: Profiles of Lineage-Dependent Secretory Responses
Different lineages of CD4+ T helper (Th) cells have distinct transcription factors and secretory profiles,25 but cannot be distinguished by their surface markers. To measure the secretory profiles of multiple lineages on a single array of nanowells, we used cytosolic barcodes to label PMA/ionomycin-stimulated Th cells that had been cultured under Th0-, Th1-, or Th2-polarizing conditions, which each promote distinct secretory patterns. Th1 cells can secrete IFN-γ and IL-2, Th2 cells can secrete IL-4 and IL-2, and Th0 cells can secrete both Th1 and Th2 cytokines,25–28 although in vitro polarization does not produce 100% conversion.29 After microengraving and deconvolution of barcoded single cells, we found that the percentages of single cells from each lineage that secreted IL-2, IFN-γ, or IL-4 (Figure S-6A), as well as the rates at which secretion-positive cells from each lineage secreted each cytokine (Figure S-6B), were both consistent with the expected lineage-specific secretory patterns. Together, these results show that cellular barcoding in combination with microengraving enables efficient tracking of the identities of T cells that have distinct secretory profiles but indistinguishable sets of surface-expressed markers.
CONCLUSIONS
Here we have demonstrated three sets of cellular barcodes that can be used to increase the throughput of single-cell secretory measurements by 2n-fold, where n is the number of dyes used to generate the barcodes. Although we focused on single cells, the platform is also well suited for measuring the secretory profiles from small groups of cells (~1–5 cells/nanowell) with precisely defined demographics. We anticipate that the combination of cellular barcoding and microengraving will enable the quantitative analysis of cell-cell interactions with a resolution that has not been possible using traditional experimental systems and will yield novel insights into the mechanisms governing the behavior of complex cellular systems.
Supplementary Material
ACKNOWLEDGMENT
This work was funded in part by grants from the Ragon Institute of MGH, MIT, and Harvard, the National Institute of Allergy and Infectious Diseases (Grants 1R56AI104274, 1U19AI089992 and 5U01AI068618), and the W.M. Keck Foundation. Y.J.Y. was funded in part by a fellowship from the National Science Foundation and the Collamore-Rogers Fellowship. J.C.L is a Latham Family Career Development Professor and Camille Dreyfus Teacher-Scholar. D.J.I. is an investigator of the Howard Hughes Medical Institute.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
J.C.L. is a founder, shareholder, and consultant for Enumeral Biomedical.
REFERENCES
- 1.Frankenstein Z, Alon U, Cohen IR. Biol Direct. 2006;1:32. doi: 10.1186/1745-6150-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Shea JJ, Ma A, Lipsky P. Nat Rev Immunol. 2002;2:37–45. doi: 10.1038/nri702. [DOI] [PubMed] [Google Scholar]
- 3.Seder RA, Darrah PA, Roederer M. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 4.Han Q, Bagheri N, Bradshaw EM, Hafler DA, Lauffenburger DA, Love JC. Proc Natl Acad Sci U S A. 2012;109:1607–1612. doi: 10.1073/pnas.1117194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Lab Chip. 2010;10:1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varadarajan N, Kwon DS, Law KM, Ogunniyi AO, Anahtar MN, Richter JM, Walker BD, Love JC. Proc Natl Acad Sci U S A. 2012;109:3885–3890. doi: 10.1073/pnas.1111205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi J, Love KR, Gong Y, Gierahn TM, Love JC. Anal Chem. 2011;83:6890–6895. doi: 10.1021/ac2013916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Q, Han Q, Bradshaw EM, Kent SC, Raddassi K, Nilsson B, Nepom GT, Hafler DA, Love JC. Anal Chem. 2010;82:473–477. doi: 10.1021/ac9024363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krutzik PO, Clutter MR, Trejo A, Nolan GP. Chapter 6. Curr Protoc Cytom. 2011:31. doi: 10.1002/0471142956.cy0631s55. Unit 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krutzik PO, Nolan GP. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 11.Mattheakis LC, Dias JM, Choi YJ, Gong J, Bruchez MP, Liu J, Wang E. Anal Biochem. 2004;327:200–208. doi: 10.1016/j.ab.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 12.Kulesa PM, Teddy JM, Smith M, Alexander R, Cooper CH, Lansford R, McLennan R. BMC Dev Biol. 2010;10:101. doi: 10.1186/1471-213X-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gan WB, Grutzendler J, Wong WT, Wong RO, Lichtman JW. Neuron. 2000;27:219–225. doi: 10.1016/s0896-6273(00)00031-3. [DOI] [PubMed] [Google Scholar]
- 14.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 15.Ogunniyi AO, Story CM, Papa E, Guillen E, Love JC. Nat Protoc. 2009;4:767–782. doi: 10.1038/nprot.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nomura LE, Walker JM, Maecker HT. Cytometry. 2000;40:60–68. doi: 10.1002/(sici)1097-0320(20000501)40:1<60::aid-cyto8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 17.Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. J Immunol Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulet S, Ndongala ML, Peretz Y, Boisvert MP, Boulassel MR, Tremblay C, Routy JP, Sekaly RP, Bernard NF. J Immunol Methods. 2007;320:18–29. doi: 10.1016/j.jim.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podtschaske M, Benary U, Zwinger S, Hofer T, Radbruch A, Baumgrass R. PLoS One. 2007;2:e935. doi: 10.1371/journal.pone.0000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grob M, Schmid-Grendelmeier P, Joller-Jemelka HI, Ludwig E, Dubs RW, Grob PJ, Wuthrich B, Bisset LR. Allergy. 2003;58:239–245. doi: 10.1034/j.1398-9995.2003.00035.x. [DOI] [PubMed] [Google Scholar]
- 21.Longo DM, Louie B, Putta S, Evensen E, Ptacek J, Cordeiro J, Wang E, Pos Z, Hawtin RE, Marincola FM, Cesano A. J Immunol. 2012;188:1717–1725. doi: 10.4049/jimmunol.1102514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, Kennedy K, Wu H, Bennouna S, Oluoch H, Miller J, Vencio RZ, Mulligan M, Aderem A, Ahmed R, Pulendran B. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner TL, Ahonen CL, Couture AM, Gibson SJ, Miller RL, Smith RM, Reiter MJ, Vasilakos JP, Tomai MA. Cell Immunol. 1999;191:10–19. doi: 10.1006/cimm.1998.1406. [DOI] [PubMed] [Google Scholar]
- 24.Muzio M, Bosisio D, Polentarutti N, D'Amico G, Stoppacciaro A, Mancinelli R, van't Veer C, Penton-Rol G, Ruco LP, Allavena P, Mantovani A. J Immunol. 2000;164:5998–6004. doi: 10.4049/jimmunol.164.11.5998. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J, Yamane H, Paul WE. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gajewski TF, Joyce J, Fitch FW. J Immunol. 1989;143:15–22. [PubMed] [Google Scholar]
- 27.Gajewski TF, Lancki DW, Stack R, Fitch FW. J Exp Med. 1994;179:481–491. doi: 10.1084/jem.179.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luchansky MS, Bailey RC. J Am Chem Soc. 2011;133:20500–20506. doi: 10.1021/ja2087618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy E, Shibuya K, Hosken N, Openshaw P, Maino V, Davis K, Murphy K, O'Garra A. J Exp Med. 1996;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.