Abstract
The most common translocation in childhood T-cell acute lymphoblastic leukemia (T-ALL) involves the LMO2 locus, resulting in ectopic expression of the LMO2 gene in human thymocytes. The LMO2 gene was also activated in patients with X-linked Severe Combined Immune Deficiency treated with gene therapy because of retroviral insertion in the LMO2 locus. The LMO2 insertions predisposed these children to T-ALL, yet how LMO2 contributes to T cell transformation remains unclear. The LIM (Lin 11, Isl-1, Mec-3) domain containing LMO2 protein regulates erythropoiesis as part of a large transcriptional complex consisting of LMO2, TAL1, E47, GATA1 and LDB1 that recognizes bipartite E-box-GATA1 sites on target genes. Similarly, a TAL1/E47/LMO2/LDB1 complex is observed in human T-ALL and Tal1 and Lmo2 expression in mice results in disease acceleration. To address the mechanism(s) of Tal1/Lmo2 synergy in leukemia, we generated Lmo2 transgenic mice and mated them with mice that express wild-type Tal1 or a DNA-binding mutant of TAL1. Tal1/Lmo2 and MutTAL1/Lmo2 bitransgenic mice exhibit perturbations in thymocyte development due to reduced E47/HEB transcriptional activity and develop leukemia with identical kinetics. These data demonstrate that the DNA-binding activity of Tal1 is not required to cooperate with Lmo2 to cause leukemia in mice and suggest that Lmo2 may cooperate with Tal1 to interfere with E47/HEB function(s).
Keywords: TAL1, Lmo2, E47, HEB, T cell acute lymphoblastic leukemia (T-ALL)
Introduction
LMO2 was first implicated in leukemogenesis in 1992 when found associated with a t(11;14)(p13;q11) and t(7;11)(q34;p13) chromosomal translocations (Boehm et al., 1990; Garcia et al., 1991). The LMO1 and LMO2 proteins are found expressed in human T-cell acute lymphoblastic leukemia (T-ALL) and LMO proteins are co-expressed in approximately 80% of TAL1 + human T-ALL patients (Rabbitts, 1994; Wadman et al., 1994; Ferrando et al., 2002). Interest in LMO2-mediated leukemogenesis was reignited in 2002, when children in two X-linked Severe Combined Immune Deficiency gene therapy trials developed T-ALL because of retroviral insertion in the LMO2 locus (Hacein-Bey-Abina et al., 2003a, b). Two leukemic patients exhibited evidence of TAL1 activation and three patients developed NOTCH1 mutations (Hacein-Bey-Abina et al., 2008; Howe et al., 2008). Studies in mouse T-ALL models revealed that Tal1 and Lmo proteins cooperate to cause leukemia in mice (Wadman et al., 1994; Larson et al., 1996; Aplan et al., 1997), but precisely how LMO proteins contribute to leukemogenesis remains unclear.
LMO2 is part of the ‘LIM only’ family of proteins that are thought to serve as bridging factors in transcriptional complexes (Wadman et al., 1997). LIM-domains are cysteine-rich zinc-binding domains that are structurally similar to DNA-binding GATA finger domains; however, currently there is no evidence to suggest that LMO proteins bind DNA. LMOs are thought to mediate protein:protein interactions through their LIM domains and evidence suggests that LMOs act as transcriptional co-regulators. There are four human LMO proteins (LMO1-4) and all four LMO proteins have been associated with oncogenesis (Fisch et al., 1992; Visvader et al., 2001; Aoyama et al., 2005).
LMO2 is a nuclear protein that in erythroid cells forms a multi-protein complex, which includes TAL1, E47, GATA-1 and LDB1 (Valge-Archer et al., 1994). These proteins form a functional transcriptional complex and recognize a bipartite DNA sequence consisting of an E-box and a GATA site separated by one helix turn (Wadman et al., 1997). The Tal1/E47/Lmo2/Ldb1/ GATA-1 complex regulates the expression of genes important in erythroid or megakaryocytic differentiation, including p4.2, glycophorin A, c-kit, p21CIP and the transcription factors Gfi-1b and eklf (Krosl et al., 1998; Lecuyer et al., 2002; Xu et al., 2003; Lahlil et al., 2004; Goardon et al., 2006). Lmo2 is thought to bridge the TAL1/E47 heterodimer to GATA-1, and by mechanisms not fully understood, enhance transcription (Xu et al., 2003). Based on their roles in regulating gene expression during hematopoiesis, Tal1 and Lmo2 have been thought to contribute to leukemia by transactivating the expression of genes such as retinaldehyde dehydrogenase, the transmembrane protein TALLA1, the receptor tyrosine phosphatase IA2, cyclin D2, heat shock cognate 73 and a bub-like gene (Ono et al., 1997; Davenport et al., 2000).
An alternative model posits that in leukemic cells the Tal1/Lmo2 complex interferes with the transcriptional activity of E47/HEB, by sequestering E47 and HEB proteins and/or by recruiting co-repressors to E-box regulated loci (Herblot et al., 2000; O’Neil et al., 2004). The E47/HEB complex directs the expression of several genes essential for proper lymphoid development including the preTα chain of the pre-T cell receptor (pre-TCR), the Rag1 and Rag2 recombinases, CD3, CD4, CD5 and the T cell receptor α/β genes (Greenbaum and Zhuang, 2002). A HEB or E47 deficiency disrupts thymocyte development and E47-deficient mice are predisposed to the development of T cell leukemia (Bain et al., 1997; Yan et al., 1997). Our published work provides genetic evidence to support the inhibition model of TAL1-mediated leukemogenesis. We demonstrate that a DNA-binding mutant form of TAL1 (R188G;R189G) (referred to here as mutTAL1) is able to induce T-ALL-like disease in mice and T cell leukemogenesis is accelerated when Tal1 is expressed on an E2A or HEB heterozygous background (O’Neil et al., 2001, 2004). Moreover, the functions of Tal1 in early hematopoietic development also do not appear to require the DNA-binding properties of Tal1 (Schlaeger et al., 2004). More recently, the ETO-2 repressor has been identified as a novel component of the TAL1/LMO2 complex in erythroid cells (Goardon et al., 2006). Thus, Tal1/E47 or HEB/Lmo2 complexes may also actively repress the expression of E-box containing genes during normal erythroid differentiation and in leukemia.
To determine the molecular basis of the observed Tal1/ Lmo2 synergy and to distinguish between the transcriptional activation and inhibition models, we asked whether the DNA-binding properties of Tal1 were required to collaborate with Lmo2 to cause T cell leukemia in mice. To test this model, we generated bitransgenic mice co-expressing Lmo2 with wild-type Tal1 or with a DNA-binding TAL1 mutant. We observed disease acceleration in both the Tal1/Lmo2 and mutTAL1/Lmo2 transgenic mouse lines, indicating that the DNA-binding properties of TAL1 are not required to cooperate with Lmo2 to cause disease in mice. Moreover, thymocyte development was inhibited in pre-leukemic Tal1/Lmo2 and mutTAL1/Lmo2 mice because of reduced expression of E47/HEB-regulated genes important for thymocyte differentiation. These data indicate that Lmo2 may accelerate Tal1-mediated leukemogenesis by enhancing the sequestration and/or inhibition of the E47/HEB heterodimer.
Results and discussion
Generation of transgenic mice expressing a DNA-binding mutant of TAL1 and Lmo2
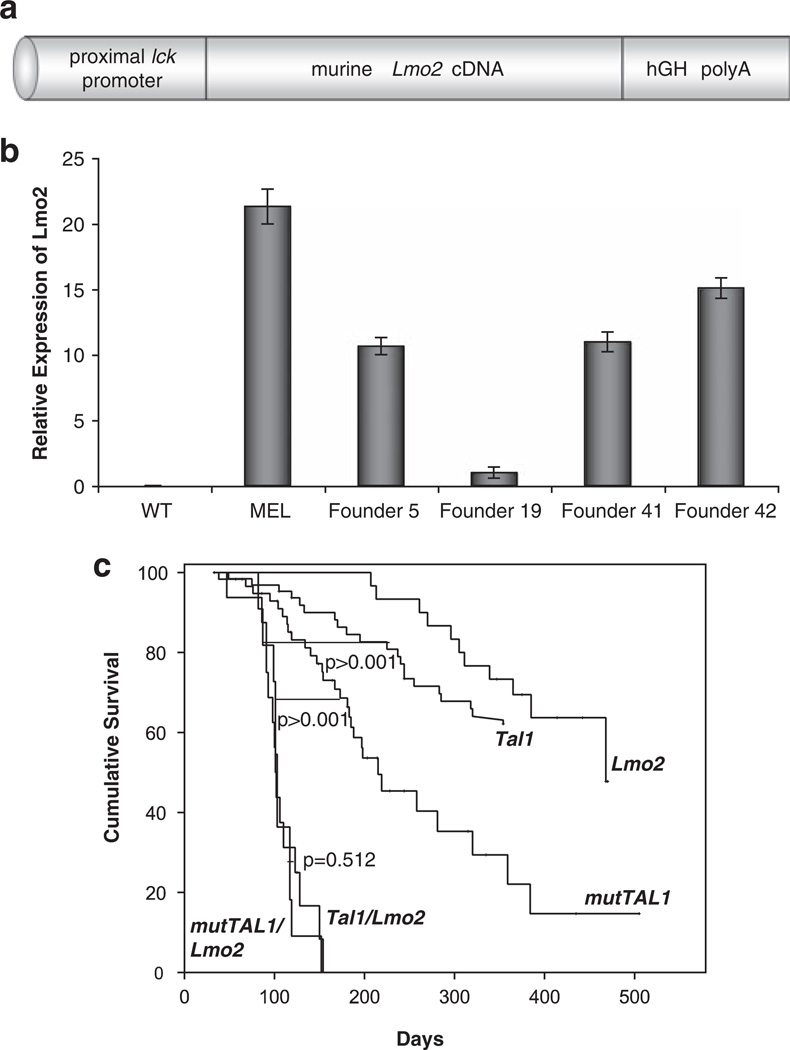
To determine how Lmo2 contributes to TAL1-mediated leukemogenesis and to understand the basis for the TAL1 and LMO oncogene cooperativity, we asked whether the DNA-binding properties of TAL1 were required to cooperate with Lmo2 to cause disease in mice. To express both oncogenes in developing mouse thymocytes, we generated transgenic mouse lines where the mouse Lmo2 complementary DNA was expressed under the control of the proximal lck promoter (Figure 1a). The 3′ untranslated regions of this construct contain introns, exons and the poly A addition site of the human growth hormone gene (Abraham et al., 1991). The prox lck Lmo2 construct was microinjected into the pronuclei of fertilized FVB/N oocytes. A total of 16 founder mice were identified initially by Southern blot analysis and two founder lines (Fo5 and 19) with variable expression levels were expanded for further study (Figure 1b).
Figure 1.
The DNA-binding properties of TAL1 are not required to cooperate with Lmo2 to cause leukemia in mice. (a) Schematic representation of the prox lck/Lmo2 transgene. The murine Lmo2 cDNA is expressed under control of the proximal lck promoter and human growth hormone (hGH) splice and poly(A)+ addition sequences. (b) Expression of the Lmo2 transgene. RNA was isolated from a murine erythroleukemic cell line (MEL) and from thymocytes isolated from wild-type (wt) or Lmo2 founder mice designated F5, F19, F41 and F42. Lmo2 mRNA levels were determine using quantitative PCR with β-actin serving as an internal control. (c) Survival curves for the Tal1, mutTAL1, Lmo2, Tal1/Lmo2 and mutTAL1/Lmo2 mice. Mice were monitored daily for evidence of disease, upon which the mice were sacrificed and a post-mortem examination performed. The cohort of Tal1/Lmo2 mice consisted of 15 animals and the cohort of mutTAL1/Lmo2 mice consisted of 11 animals. The Lck-Lmo2 only cohort consisted of 26 mice. Tal1 and mutTAL1 survival curves have been published previously (Kelliher et al., 1996; O’Neil et al., 2001).
The Lmo2 transgenic mouse lines were mated with transgenic mice expressing wild-type Tal1 or a DNA-binding mutant of TAL1. The DNA-binding mutant mice express a form of TAL1 where the contact residues of the basic DNA-binding domain, arginine 188 and 189 have been mutated to glycines (designated R188G;R189G) (Hsu et al., 1994). This mutant form of TAL1 heterodimerizes with E47 or HEB proteins but fails to stably bind E-box binding sequences in vitro and in vivo (Hsu et al., 1994; O’Neil et al., 2001). Like Tal1 transgenic mice, expression of a TAL1 DNA-binding mutant perturbs thymocyte development and approximately half of the mice develop a T-ALL-like disease (O’Neil et al., 2001).
To determine whether the DNA-binding activities of TAL1 were required to cooperate with Lmo2 to cause disease in mice, we generated a cohort of 15 Tal1/Lmo2 and 11 mutTAL1/Lmo2 bitransgenic mice and monitored the animals for development of disease. The Lck-Lmo2 transgenic lines developed disease following a long latency and cooperated with Tal1 and the DNA-binding mutant to accelerate leukemogenesis in mice (P<0.0001) (Figure 1c). Surprisingly, Tal1/Lmo2 and mutTAL1/Lmo2 transgenic mouse lines developed disease with nearly identical kinetics (Figure 1c), in spite of the fact that the DNA-binding mutant TAL1 transgenic mice develop disease more rapidly than wild-type Tal1 transgenic mice (O’Neil et al., 2001). Approximately 30% of Tal1 transgenic mice develop leukemia after a long latency, whereas mutTAL1 mice develop disease with a median survival of 215 days (Kelliher et al., 1996; O’Neil et al., 2001). In contrast, when Lmo2 is co-expressed with Tal1 or mutTAL1, 100% of the mice develop T-ALL-like disease with a median survival of approximately 100 days. No significant difference was observed when Tal1/Lmo2 and mutTAL1/Lmo2 survival curves were compared (P<0.581). Histopathologic examination of tumors isolated from the bitransgenic mice revealed a range of tumor immunophenotypes (Table 1), with tumors predominantly containing DN and DP or CD8SP cells observed in both Tal1/Lmo2 and mutTAL1/Lmo2 mice (Table 1). This study reveals that Tal1/Lmo2 synergy does not require the DNA-binding properties of TAL1, suggesting that Lmo2 does not contribute to leukemia by enhancing or altering the transcriptional activity of the Tal1/E47 or HEB heterodimer.
Table 1.
Immunophenotype of Tal1/Lmo2 and mutTAL1/Lmo2 tumors
| Animal number | Genotype | CD3 | CD25 | DN | CD4 | CD8 | DP | Phenotype | Survival (days) |
|---|---|---|---|---|---|---|---|---|---|
| 5624 | Tal1/Lmo2 | 31.6 | 55.7 | 27.9 | 0.1 | 71.2 | 0.8 | DN, CD8-SP | 102 |
| 5629 | Tal1/Lmo2 | 71.3 | 0.4 | 30.9 | 0.2 | 68.1 | 0.7 | DN, CD8lo | 91 |
| 5632 | Tal1/Lmo2 | 94.2 | 7.7 | 1.7 | 0.2 | 82.6 | 15.5 | CD8-SP | 91 |
| 5666 | Tal1/Lmo2 | 95.1 | 95.3 | 3.3 | 2.0 | 2.1 | 92.6 | DP | 98 |
| 5354 | Tal1/Lmo2 | 29.3 | 62.9 | 30.1 | 0.6 | 4.3 | 65 | DN, DP | 47 |
| 5338 | Tal1/Lmo2 | 69.5 | 70.6 | 25.8 | 0.1 | 42.8 | 31.1 | DN, CD8-SP, DP | 86 |
| 5351 | Tal1/Lmo2 | 23.7 | 97.9 | 0.3 | 0.1 | 0.5 | 99.1 | DP | 106 |
| 5313 | Tal1/Lmo2 | 96.2 | 3.3 | 0.8 | 93.4 | 1.1 | 4.7 | CD4-SP | 154 |
| 5353 | Tal1/Lmo2 | 27.5 | 4.6 | 9.7 | 80.1 | 0.4 | 9.8 | CD4-SP | 123 |
| 6540 | mutTal1/Lmo2 | 52.7 | 13.8 | 53.7 | 8.2 | 10.9 | 27.1 | DN, DP-lo | 99 |
| 5984 | mutTal1/Lmo2 | 50.0 | 95.5 | 51.0 | 1.0 | 39.5 | 8.5 | DN, CD8-SP | 101 |
Tumors were stained with antibodies to CD3, CD4, CD8 and CD25 and analyzed by flow cytometry.
The percentage of stained cells is indicated.
Thymocyte developmental arrest in Tal1/Lmo2 and mutTAL1/Lmo2 mice
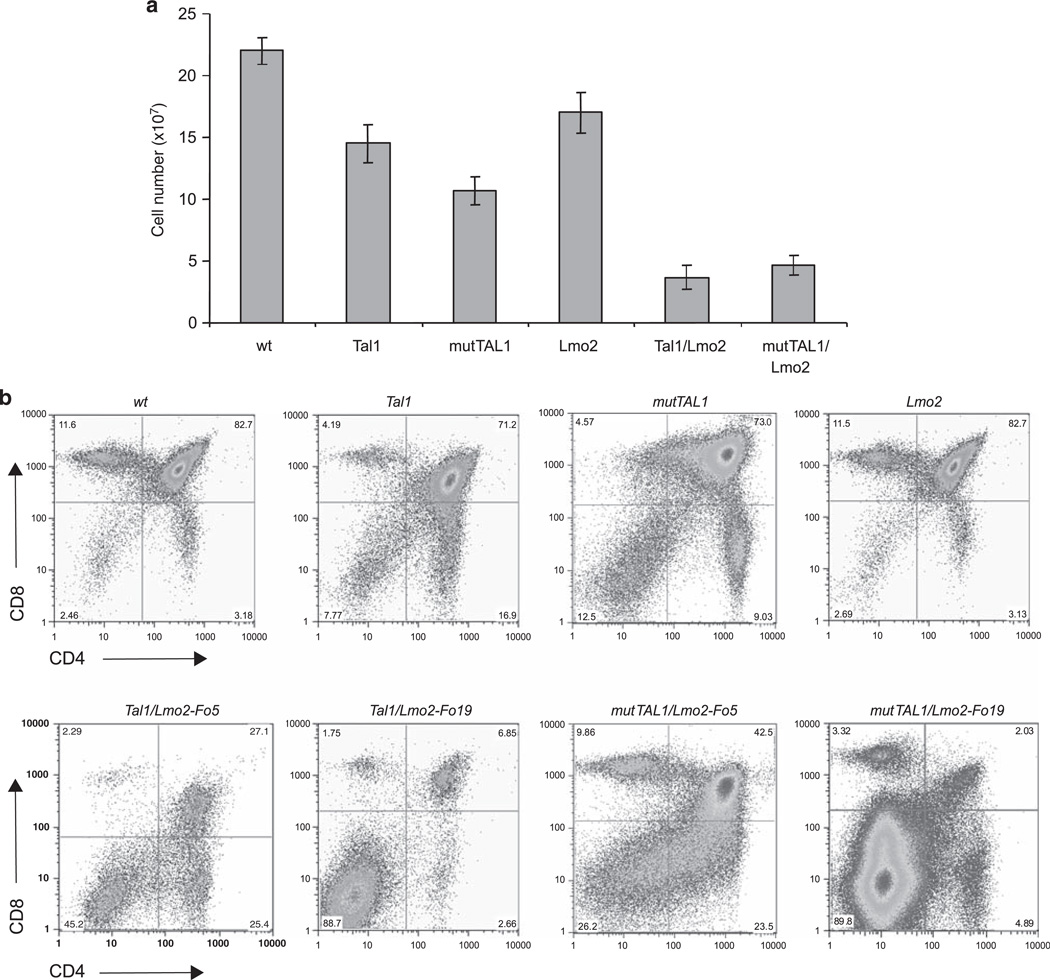
We have shown that thymic expression of Tal1 or the DNA-binding mutant results in a decrease in overall thymocyte cellularity because of fewer DP and CD8-positive thymocytes (Kelliher et al., 1996; O’Neil et al., 2001). Expression of Lmo2 under the control of the proximal Lck promoter appeared to have no detectable effects on DP thymocyte development (Figure 2b), however, Lmo2 expression decreased thymocyte cellularity and skewed DN development; although no increase in the absolute numbers of DN thymocytes was observed (Figures 2a and c). Consistent with published reports (Larson et al., 1996), co-expression of Tal1 and Lmo2 reduces thymic cellularity and blocks DP thymocyte development, resulting in increases in the relative and absolute number of DN thymocyte precursors in preleukemic Tal1/Lmo2 mice (Figures 2b and d). DP thymocyte development was consistently more severely affected in Tal1/Lmo2 mice derived from Lmo2 founder Fo19 compared with Fo5 (Figure 2b).
Figure 2.
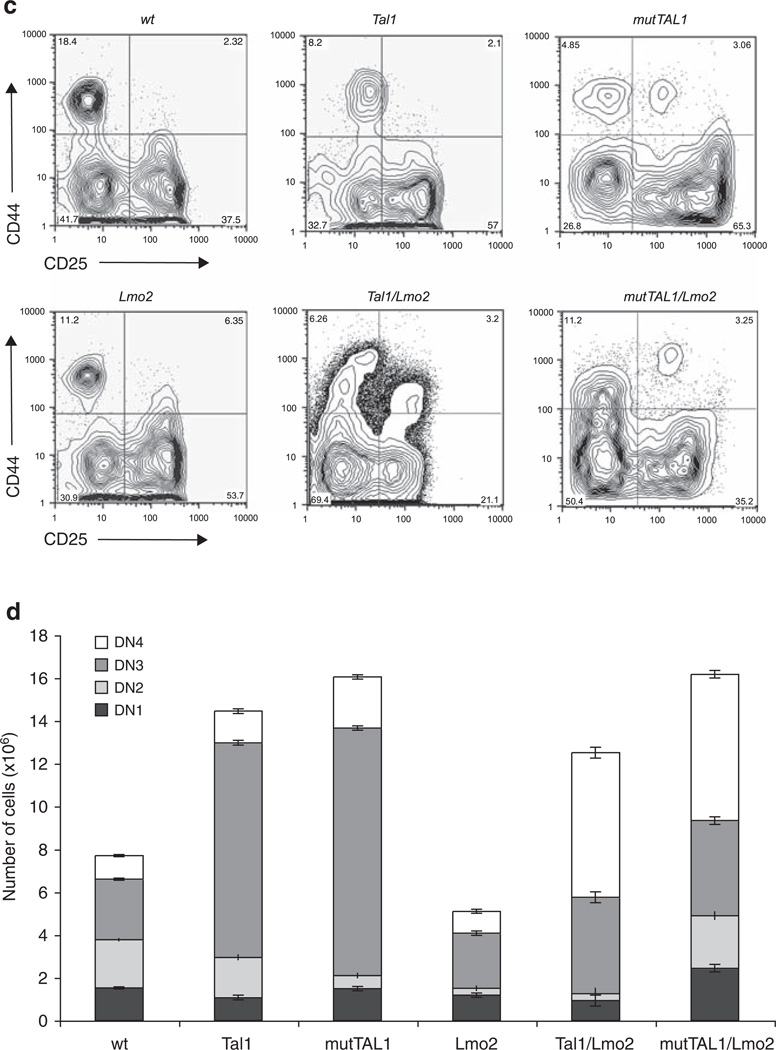
A DNA-binding mutant of TAL1 cooperates with Lmo2 to perturb thymocyte development. (a) Significant decreases in thymic cellularity are observed in Tal1/Lmo2 and mutTAL1/Lmo2 bitransgenic mice. Total thymocyte numbers from wild-type, Tal1, mutTAL1, Lmo2, Tal1/Lmo2 and mutTAL1/Lmo2 transgenic mice. When compared with wild type, Lmo2 expression causes a decrease in thymic cellularity (P = 8.0 × 10−6) that is exacerbated when either Tal1 or mut-TAL1 is co-expressed (P = 7.8×10−15 and P = 3.1×10−6, respectively). (b) Co-expression of Lmo2 with either Tal1 or mutTAL1 perturbs thymocyte development. Thymocytes from 4-week old, preleukemic mice were stained with CD4-PE and CD8-FITC and analyzed by flow cytometry. A total of 14 Lmo2, 19 Tal1/Lmo2 and 13 mutTAL1/Lmo2 preleukemic mice were analyzed. Representative profiles are shown. (c) DN thymocyte development is altered when Lmo2 is co-expressed with either Tal1 or mutTAL1. Thymocytes from 4-week old mice were stained with antibodies for the lineage markers (CD4, CD8, CD3, B220, Mac1, Gr1 and Terr119) and the lineage-negative cells were isolated and stained with CD44-APC and CD25-FITC. A total of 14 Lmo2, 19 Tal1/Lmo2 and 13 mutTAL1/Lmo2 preleukemic mice were examined. Representative profiles are shown. (d) Increase in the absolute numbers of DN3 and DN4 thymic progenitors in preleukemic Tal1 and mutTAL1 transgenic mice. Thymocytes from wild-type (n = 12), Tal1 (n = 7), MutTAL1 (n = 8), Lmo2 (n = 15), Tal1/Lmo2 (n = 11) and mutTAL1/Lmo2 (n = 12) mice were stained with CD4 and CD8 and lineage negative cells were stained with CD44 and CD25. Absolute numbers of DN1, DN2, DN3 and DN4 cells are indicated for each strain. A representative experiment is shown.
To determine whether the thymocyte developmental block reflects alterations in Tal1-mediated gene expression, we examined thymocyte development in mutTAL1/ Lmo2 pre-leukemic mice. We found thymocyte cellularity futher reduced in mutTAL1/Lmo2 mice compared with mice that express the TAL1 DNA-binding mutant only (Figures 2a). Overall cellularity was similarly reduced in all the Tal1/Lmo2 and mutTAL1/Lmo2 mice examined. Similar to the Tal1/Lmo2 mice, DP thymocyte development was consistently more severely affected in mutTAL1/Lmo2 mice derived from Fo19 compared with founder Fo5 (Figure 2b). Importantly, the DNA-binding TAL1 mutant cooperated with Lmo2 to induce similar effects on thymocyte development as wild-type TAL1 (Figure 2).
Expression of Lmo2 with Tal1 or mutTAL1 also altered DN thymocyte development, resulting in arrest at the DN3-DN4 precursor stage (Figure 2c and d). In contrast, an E2A or HEB deficiency results in an arrest of DN thymocyte development at the DN1 and DN3 stages, respectively (Bain et al., 1997; Greenbaum and Zhuang, 2002). These studies demonstrate that Lmo2 does not require the DNA-binding properties of TAL1 to perturb thymocyte development. The block in thymocyte development induced by either form of Tal1 is not complete, as DP and SP thymocytes are detected (Figure 2c). Moreover, immunophenotyping of Tal1/Lmo2 and mutTAL1/ Lmo2 tumors revealed that tumors from both bitransgenic lines were often heterogeneous, consisting of DN and DP and/or CD8 SP cells (Table 1). For example, although the primary Tal1/Lmo2 tumor 5338 is clonal, it contains DN, DP and CD8-SP cells, suggesting that differentiation proceeds at some level after leukemic transformation.
The expression of E47/HEB-regulated genes is decreased in both Tal1/Lmo2 and mutTAL1/Lmo2 mice
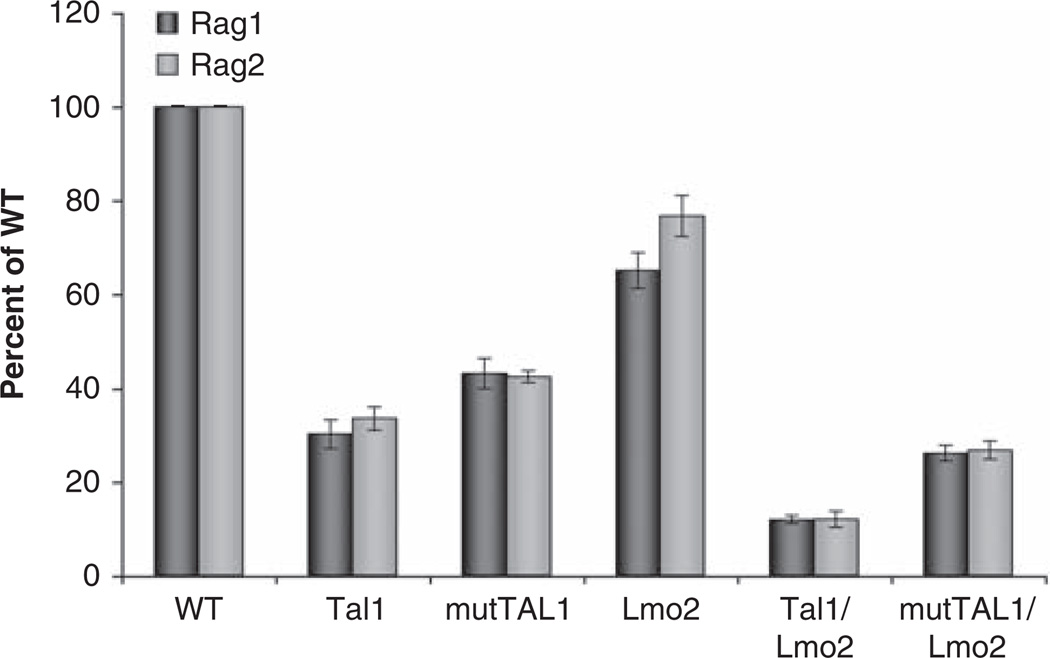
To understand how Lmo2 can cooperate with a DNA-binding mutant of TAL1 and interfere with thymocyte development, we examined the expression of E47/HEB target genes known to regulate thymocyte differentiation. Previously, we found the expression of PreTa, Rag1/2, CD3 CD4, T cell receptor α/β and CD5 repressed in the presence of the Tal1 oncogene (O’Neil et al., 2004). The expression of these thymocyte differentiation genes was further decreased in Tal1/E2A +/− or HEB +/− mice, indicating that Tal1 alters thymocyte development by interfering with the transcriptional activities of the E47/HEB heterodimer (O’Neil et al., 2004). Using real-time PCR, we quantified the expression of the immunoglobulin and T cell receptor gene recombinases Rag1 and Rag2 and the pre-Tα chain of the pre-TCR. We found the expression of Rag1 and Rag2 significantly decreased in Tal1 and mutTAL1 preleukemic thymocytes (Figure 3). Slight decrease in Rag1/2 expression were also detected in the preleukemic Lck-Lmo2 transgenic thymus, although no gross perturbations in the thymocyte developmental profile were observed. Co-expression of Tal1 or its DNA-binding mutant with Lmo2 resulted in consistent further decreases in Rag1 and Rag2 expression (Figure 3). Similarly, we found preTα expression further repressed in preleukemic Tal1/Lmo2 or mutTAL1/Lmo2 thymocytes than in thymocytes expressing Tal1, mutTAL1 or Lmo2 alone (data not shown). The reduced expression of these E47/HEB-regulated genes does not reflect changes in the relative percentage of DN versus DP stage thymocytes, as significant decrease in Rag1/2 and PreTα gene expression were observed in Tal1 and mutTAL1 animals, where the thymocyte developmental profile is only modestly altered (Figure 2b). These data reveal that in the presence of Tal1 and importantly, a DNA-binding mutant of TAL1, Lmo2 expression results in further reductions in E47/HEB transcriptional activity. These findings suggest that Lmo2 binding to Tal1 or mutTAL1/E47 or HEB heterodimers may stabilize the Tal1/E47 or HEB complex, resulting in more effective E47/HEB sequestration.
Figure 3.
Rag1 and Rag2 expression is further repressed in Tal1 and mut-TAL1 transgenic thymocytes when Lmo2 is co-expressed. Thymocytes were isolated from wild-type or preleukemic Tal1, mutTAL1, Lmo2, mutTAL1/Lmo2 or Tal/Lmo2 transgenic mice at 4–6 weeks of age. RNA was isolated and Rag1 and Rag2 expression levels quantified by qPCR. The copy number for the gene of interest was normalized to the copy number for β-actin. Data is represented as percent of wild-type control.
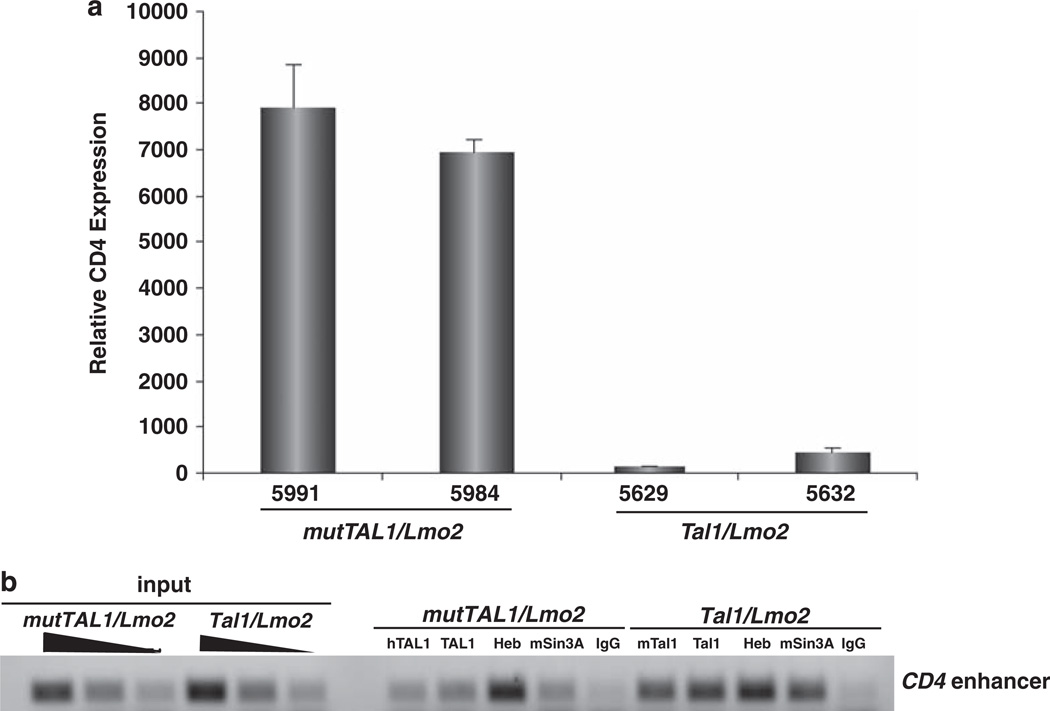
The TAL1 DNA-binding mutant is impaired in its ability to repress CD4 expression
Although we have demonstrated that mouse leukemic cells expressing a TAL1 DNA-binding mutant fail to bind E-box containing oligonucleotides (O’Neil et al., 2001), whether the TAL1 DNA-binding mutant binds native promoters or enhancers remains untested. Moreover, whether the DNA-binding mutant is recruited to regulatory regions through its incorporation into a mutTal1-E47(HEB)-Lmo2-Ldb1-Gata3 transcriptional complex is also unclear. We have shown that Tal1 actively represses CD4 expression in preleukemic mouse thymocytes by recruiting the msin3a corepressor (O’Neil et al., 2004). To determine whether mutant TAL1 binds the CD4 regulatory region and influences CD4 expression, we quantified CD4 messenger RNA levels in leukemic cell lines isolated from Tal1/Lmo2 and mutTAL1/Lmo2 mice. We found CD4 messenger RNA levels repressed on average 39-fold in Tal1/Lmo2 leukemic cells compared with those expressing the DNA-binding mutant (Figure 4a). These data suggest that the DNA-binding mutant may be impaired in its ability to stably bind E-box sequences and influence CD4 expression. Using chromatin immunoprecipitation, we analyzed Tal1 and mutTAL1 binding to the CD4 enhancer. Tal1, Heb and msin3a recruitment to the CD4 enhancer was observed in the Tal1/Lmo2 leukemic cell lines. Although Heb binding to the CD4 enhancer was detected in the mutTAL1/Lmo2 leukemic cells, decreased mutTAL1 binding was observed with both human TAL1 antibodies used for immunoprecipitation (Figure 4b). The decrease in mutTAL1 binding was accompanied by a reduction in msin3a recruitment, consistent with published studies demonstrating that Tal1 directly binds msin3a (Huang and Brandt, 2000). Thus, Tal1 and mutTAL1 bind and sequester E47 and HEB proteins, whereas active CD4 repression requires the DNA-binding properties of Tal1.
Figure 4.
The DNA-binding activity of TAL1 is required to stably bind and repress CD4. (a) The DNA-binding properties of TAL1 contribute to CD4 repression. RNA was isolated from Tal1/Lmo2 and mutTAL1/Lmo2 leukemic cell lines and CD4 expression levels quantified by qPCR. The copy number for CD4 was normalized to the copy number for β-actin. (b) The TAL1 DNA-binding mutant is impaired in its ability to bind the CD4 enhancer. Chromatin immunoprecipitation was performed on mutTAL1/Lmo2 and Tal1/Lmo2 leukemic cell lines using a rabbit IgG antibody or antibodies to HEB, mSin3a, mouse Tal1 (m258c), human TAL1 (Millipore, BTL73) or a TAL1 antibody that recognizes mouse and human TAL1 (Santa Cruz, C-21). Input-sheared DNA served as a positive control. PCR amplification was performed on the immunoprecipitated DNA with primers specific for the mouse CD4 enhancer region.
Collectively, these studies suggest that Lmo2 binding to Tal1 stabilizes the Tal1/E47 or HEB interaction. Consistent with this idea, the binding affinities for the Tal1/E47 heterodimer have been measured in vitro and shown to significantly increase in the presence of Lmo2 (KA of ~4 × 107 versus KA of ~1 × 108) (Ryan et al., 2008). Therefore, Lmo2 binding appears to increase the affinity of Tal1 for E47 or HEB and this interaction is greater than the affinity of Lmo2 for its LIM domain binding partner, Ldb1 (KA of 5 × 107) (Ryan et al., 2008). These estimates predict that E protein sequestration may be favored in a setting where Tal1 and Lmo2 are both expressed.
Our findings have therapeutic implications and suggest that interfering with Tal1/E47 or Tal1/Lmo2 binding may increase E47/HEB levels and release Tal1/E47 hetero-dimers from GATA3 or other transcriptional complexes. Disruption of these complexes may stimulate leukemic cell differentiation and/or induce apoptosis of leukemic cells. However, disrupting the Tal1/E47 or HEB protein interface with small molecules may not be feasible. In some instances, only a minor part of the protein dimer interfaces contribute to the affinity between proteins (Clackson and Wells, 1995; Kussie et al., 1996; Wells, 1996; Wells and de Vos, 1996). Targeting these regions may be sufficient to inhibit protein:protein interactions. Small molecules have in fact been identified that interfere with the Myc/Max bHLH/LZ interaction and these molecules have been shown to interfere with c-Myc-induced transformation in vitro (Berg et al, 2002). Alternatively, interfering with Lmo2 binding to Tal1 may be sufficient to induce apoptosis of T-ALL cells. Retroviral expression of an anti-Lmo2 single chain Fv antibody fragment or an Lmo2 aptamer have been shown to reduce leukemic growth in transplanted mice (Nam et al, 2008), suggesting that interfering with LMO2 protein interactions may be an effective therapeutic strategy for some relapsed T-ALL patients.
Materials and methods
Generation of transgenic mice
Murine Lmo2 complementary DNA was cloned into p1017, a plasmid containing the proximal lck promoter and the human growth hormone splice and poly adenylation addition sites. The plasmid DNA was sequenced and digested with SpeI for microinjection into the FVB/N pronuclei. Transgenic founders were identified by Southern blotting and mated with FVB/N mice. Proximal lck-Tal1 and mutTAL1 transgenic lines have been previously described (Kelliher et al., 1996; O’Neil et al., 2001).
Cell culture and flow cytometry
Mouse T-ALL tumors were minced into a single-cell suspension using frosted slides and cultured in RPMI with 10% fetal bovine serum, l% glutamine, penicillin/streptomycin and 50 mM b-mercaptoethanol at 37°C under 5% CO2. Media was replaced once/week and once the culture appeared confluent, cells were expanded to generate cell lines. Cytospin preparations of the cultures were stained with Wright/Giemsa to confirm their lymphoblastoid morphology. Mouse leukemic cell lines were immunophenotyped by staining with antibodies to hematopoietic lineage markers as described below. Thymi from preleukemic mice were gently disrupted with frosted glass slides in order to produce single cell suspensions. The thymocytes were washed with PBS and stained with fluorescent-labeled antibodies and subjected to flow cytometry at the fluorescence-activated cell sorter facility at the University of Massachusetts Medical Center. For double negative analysis, cells were stained with antibodies for the lineage antibodies, and the lineage-negative cells were stained with CD44-APC and CD25-Fitc. Antibodies used in flow cytometry included CD3-PE, CD4-PE, CD4-Cy5-PE, CD8-FITC, CD8-Cy5-PE, CD25-Fitc, CD44-APC and lineage markers (BD Pharmingen, San Diego, CA, USA). Data were analyzed using FlowJo software (Treestar, Inc., Ashland, OR, USA). For tumor immunophenotyping, primary tumors cells were stained with either PE or FITC-conjugated anti-mouse Ly-2 (CD8), L3T4 (CD4), CD25 and CD3 antibodies (BD Pharmingen), and analyzed by flow cytometry.
Quantitative PCR
RNA was extracted from primary thymocytes using Trizol. complementary DNA was synthesized using the Superscript firststrand synthesis system (Invitrogen, Carlsbad, CA, USA). Rag1, Rag2 and pre-Tα expression was assayed using primers as described in references (Hsu et al., 2003; Huang et al., 2003). To determine target-gene expression levels, complementary DNA was serially diluted and quantified using the SYBR green kit (Qiagen, Valencia, CA, USA) gene-specific primers and β-actinspecific primers using relative quantification analysis. The copy number for the gene of interest was normalized to the copy number for β-actin.
Chromatin immunoprecipitation
To determine whether mutTAL1 bound the regulatory regions of the mouse CD4 gene, chromatin immunoprecipitation was performed essentially as described in (Sharma et al., 2006). The following antibodies were used for immunoprecipitation: mouse Tal1 (generously provided by Richard Baer, Columbia University), human mutTAL1 (Millipore BTL73), TAL1 (Santa Cruz C-21, Santa Cruz, CA, USA), msin3a (Santa Cruz K-20), and HEB (Santa Cruz A-20). Rabbit IgG (Santa Cruz) was used as the negative control. The following primer sets were used to amplify the regulatory regions of the CD4 gene: Forward—5′-TTCTGAGCCCACC CTAAGATG-3′ and Reverse—5′-GTCTTTTTTCCAGAGC CCC-3′ .
Acknowledgements
This work is supported by an NIH/NCI CA096899 Grant to MK. Core resources supported by the Diabetes Endocrinology Research Center Grant DK32520 were also used.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Abraham N, Miceli MC, Parnes JR, Veillette A. Enhancement of T-cell responsiveness by the lymphocyte-specific tyrosine protein kinase p56lck. Nature. 1991;350:62–66. doi: 10.1038/350062a0. [DOI] [PubMed] [Google Scholar]
- Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, et al. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- Aplan PD, Jones CA, Chervinsky DS, Zhao X, Ellsworth M, Wu C, et al. An scl gene product lacking the transactivation domain induces bony abnormalities and cooperates with LMO1 to generate T-cell malignancies in transgenic mice. Embo J. 1997;16:2408–2419. doi: 10.1093/emboj/16.9.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, et al. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, et al. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci USA. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T, Foroni L, Kennedy M, Rabbitts TH. The rhombotin gene belongs to a class of transcriptional regulators with a potential novel protein dimerisation motif. Oncogene. 1990;5:1103–1105. [PubMed] [Google Scholar]
- Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Davenport J, Neale GA, Goorha R. Identification of genes potentially involved in LMO2-induced leukemogenesis. Leukemia. 2000;14:1986–1996. doi: 10.1038/sj.leu.2401913. [DOI] [PubMed] [Google Scholar]
- Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Fisch P, Boehm T, Lavenir I, Larson T, Arno J, Forster A, et al. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992;7:2389–2397. [PubMed] [Google Scholar]
- Garcia IS, Kaneko Y, Gonzalez-Sarmiento R, Campbell K, White L, Boehm T, et al. A study of chromosome 11p13 translocations involving TCR beta and TCR delta in human T cell leukaemia. Oncogene. 1991;6:577–582. [PubMed] [Google Scholar]
- Goardon N, Lambert JA, Rodriguez P, Nissaire P, Herblot S, Thibault P, et al. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. Embo J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum S, Zhuang Y. Regulation of early lymphocyte development by E2A family proteins. Semin Immunol. 2002;14:405–414. doi: 10.1016/s1044532302000751. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003a;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003b;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Wadman I, Tsan JT, Baer R. Positive and negative transcriptional control by the TAL1 helix-loop-helix protein. Proc Natl Acad Sci USA. 1994;91:5947–5951. doi: 10.1073/pnas.91.13.5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LY, Lauring J, Liang HE, Greenbaum S, Cado D, Zhuang Y, et al. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 2003;19:105–117. doi: 10.1016/s1074-7613(03)00181-x. [DOI] [PubMed] [Google Scholar]
- Huang EY, Gallegos AM, Richards SM, Lehar SM, Bevan MJ. Surface expression of Notch1 on thymocytes: correlation with the double-negative to double-positive transition. J Immunol. 2003;171:2296–2304. doi: 10.4049/jimmunol.171.5.2296. [DOI] [PubMed] [Google Scholar]
- Huang S, Brandt SJ. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol Cell Biol. 2000;20:2248–2259. doi: 10.1128/mcb.20.6.2248-2259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher MA, Seldin DC, Leder P. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIalpha. Embo J. 1996;15:5160–5166. [PMC free article] [PubMed] [Google Scholar]
- Krosl G, He G, Lefrancois M, Charron F, Romeo PH, Jolicoeur P, et al. Transcription factor SCL is required for c-kit expression and c-Kit function in hemopoietic cells. J Exp Med. 1998;188:439–450. doi: 10.1084/jem.188.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Lahlil R, Lecuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RC, Lavenir I, Larson TA, Baer R, Warren AJ, Wadman I, et al. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. Embo J. 1996;15:1021–1027. [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, et al. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- Nam CH, Lobato MN, Appert A, Drynan LF, Tanaka T, Rabbitts TH. An antibody inhibitor of the LMO2-protein complex blocks its normal and tumorigenic functions. Oncogene. 2008;27:4962–4968. doi: 10.1038/onc.2008.130. [DOI] [PubMed] [Google Scholar]
- O’Neil J, Billa M, Oikemus S, Kelliher M. The DNA binding activity of TAL-1 is not required to induce leukemia/lymphoma in mice. Oncogene. 2001;20:3897–3905. doi: 10.1038/sj.onc.1204519. [DOI] [PubMed] [Google Scholar]
- O’Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/ HEB. Cancer Cell. 2004;5:587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Ono Y, Fukuhara N, Yoshie O. Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or −2 and induces TALLA1, a highly specific tumor marker of T-ALL. J Biol Chem. 1997;272:4576–4581. doi: 10.1074/jbc.272.7.4576. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- Ryan DP, Duncan JL, Lee C, Kuchel PW, Matthews JM. Assembly of the oncogenic DNA-binding complex LMO2-Ldb1-TAL1-E12. Proteins. 2008;70:1461–1474. doi: 10.1002/prot.21638. [DOI] [PubMed] [Google Scholar]
- Schlaeger TM, Schuh A, Flitter S, Fisher A, Mikkola H, Orkin SH, et al. Decoding hematopoietic specificity in the helix-loop-helix domain of the transcription factor SCL/Tal-1. Mol Cell Biol. 2004;24:7491–7502. doi: 10.1128/MCB.24.17.7491-7502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valge-Archer VE, Osada H, Warren AJ, Forster A, Li J, Baer R, et al. The LIM protein RBTN2 and the basic helix-loop-helix protein TAL1 are present in a complex in erythroid cells. Proc Natl Acad Sci USA. 1994;91:8617–8621. doi: 10.1073/pnas.91.18.8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Venter D, Hahm K, Santamaria M, Sum EY, O’Reilly L, et al. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2001;98:14452–14457. doi: 10.1073/pnas.251547698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman I, Li J, Bash RO, Forster A, Osada H, Rabbitts TH, et al. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. Embo J. 1994;13:4831–4839. doi: 10.1002/j.1460-2075.1994.tb06809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. Embo J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA. Binding in the growth hormone receptor complex. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA, de Vos AM. Hematopoietic receptor complexes. Annu Rev Biochem. 1996;65:609–634. doi: 10.1146/annurev.bi.65.070196.003141. [DOI] [PubMed] [Google Scholar]
- Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/ Id1 double-knockout mice. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]