Abstract
Developing cells acquire mature fates in part by selective (i.e. qualitatively different) expression of a few cell-specific genes. However, all cells share the same basic repertoire of molecular and sub-cellular building blocks. Therefore, cells must also specialize according to quantitative differences in cell-specific distributions of those common molecular resources. Here we propose the novel hypothesis that evolutionarily-conserved transcription factors called scaling factors (SFs) regulate quantitative differences among mature cell types. SFs: 1) are induced during late stages of cell maturation; 2) are dedicated to specific sub-cellular domains; and, thus, 3) allow cells to emphasize specific sub-cellular features. We identify candidate SFs and discuss one in detail: MIST1 (BHLHA15, vertebrates)/ DIMM (CG8667, Drosophila); professional secretory cells use this SF to scale up regulated secretion. Because cells use SFs to develop their mature properties and also to adapt them to ever-changing environmental conditions, SF aberrations likely contribute to diseases of adult onset.
Keywords: differentiation, DIMM, MIST1, sub-cellular domains, transcription factor
Introduction
During embryonic development individual cells read their positions within the body axes to acquire patterning information that determines their mature cell fates. These cell-extrinsic signals then trigger cascades of fate determining transcription factors (TFs) [1] thereby ultimately resolve single cell fates (Fig. 1). Studies of fate determination tend to mark the end of this process by the first appearance of so-called ‘terminal differentiation’ markers. However, it is important to bear in mind that the initial onset of cell-specific gene expression does not indicate that all the important developmental work is done. For example, a mature pancreatic β cell produces 3-10 times more insulin than an immature Neurogenin3+ precursor β cell present in late-stage mouse embryos [2]. Thus development involves important late-stage quantitative aspects of cell maturation as well as early-stage qualitative specification. Quantitative maturation may continue over long periods of time and may also underlie cellular adaptation and physiological regulation. Here we address mechanisms cells use to orchestrate quantitative specialization and thus bridge the periods of cell development and of cellular function.
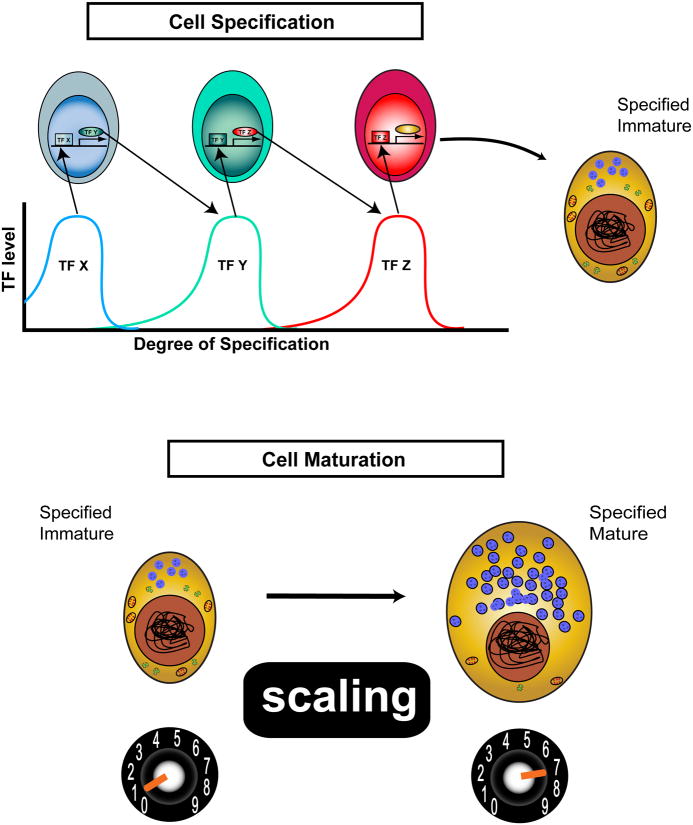
Figure 1.
Cell development is more than just cell specification - cells must mature by Scaling up certain properties. During development, cell fates are first determined by the transient, sequential expression of transcription factors (TFs). In this fictive example, a 3-step linear pathway restricts a cell to produce a specific but immature “Gold” cell – Cell Specification, upper. However, a truly mature cell has scaled its specific features to optimally perform its physiological functions. The transition from immature to mature Gold cell differentiation involves a set of poorly understood processes that we term “Scaling” – Cell Maturation, lower.
We focus on how maturing and adult cells scale subcellular resources (such as organelles) to become specialists in certain physiological functions or to adapt to changing daily conditions. In short, we propose that to become specialists, cells maximize efficiency by greatly enhancing certain subcellular features while suppressing others. We further suppose that the process is controlled by dedicated transcriptional regulators, or scaling factors (SFs). SFs are transcription factors and co-factors that promote the maturation of different cell types – after cell fate is determined – by organizing and amplifying specific sub-cellular domains. The discussion starts with a prototypical SF family: DIMM/MIST1, then extends to consider two other, non-related SFs. We close by examining how the concept of sub-cellular scaling factors can help frame questions at the interface of cell development and cell physiology.
MIST1/DIMM are prototypes for a category of TFs we call scaling factors
The biology of the DIMM/MIST1 family of TFs – organizers of cell specialization
MIST1 (aka BHLHA15) is a bHLH transcription factor expressed in diverse secretory lineages that, in all cases studied so far, are long-lived, high capacity secretors of proteins. For example, its expression has been characterized in pancreatic and salivary gland acinar cells and in gastric chief (zymogenic) cells secreting digestive enzymes, in lacrimal glands secreting tear proteins, in epithelial cells of lactating mammary glands secreting milk proteins, in intestinal Paneth cells secreting anti-microbial agents, and in plasma cells secreting antibodies[3-8]. MIST1-expressing cells are thus not related by lineage, only by functionality. MIST1 expression commences only during terminal differentiation of a cell and is sustained thereafter throughout its lifespan [8-10]. With few exceptions [8], cells that normally would express MIST1 still form in its absence, but they are structurally and functionally abnormal.
In pancreatic and gastric digestive enzyme secreting cells, where MIST1 physiology has been most explored, MIST1 is required for basal polarization of nuclei, and for formation/maintenance of large, apical digestive-enzyme containing granules [3, 5, 6, 9](Fig. 2, top). Thus, Mist1−/− gastric chief cells and pancreatic acinar cells show a dramatic reduction in apical cytoplasm with apically oriented nuclei and small secretory granules and dramatically reduced secretion of digestive enzymes in response to secretagogues [11]. Under standard laboratory conditions, MIST1 deficient mice are viable with normal lifespan and reproductive capacity; however, as mice age, the long-lived acinar cells of the pancreas begin to degenerate in the absence of MIST1, undergoing acinar to ductal metaplasia by 9-10 months [5]. Similarly, Mist1−/− acinar cells are also more sensitive to acute, chemically induced pancreatitis [12]. Thus, the inability of specialized secretory cells to scale up regulated secretion in the absence of MIST1 makes them less able to adapt over the lifetime of the animal to their extracellular environment.
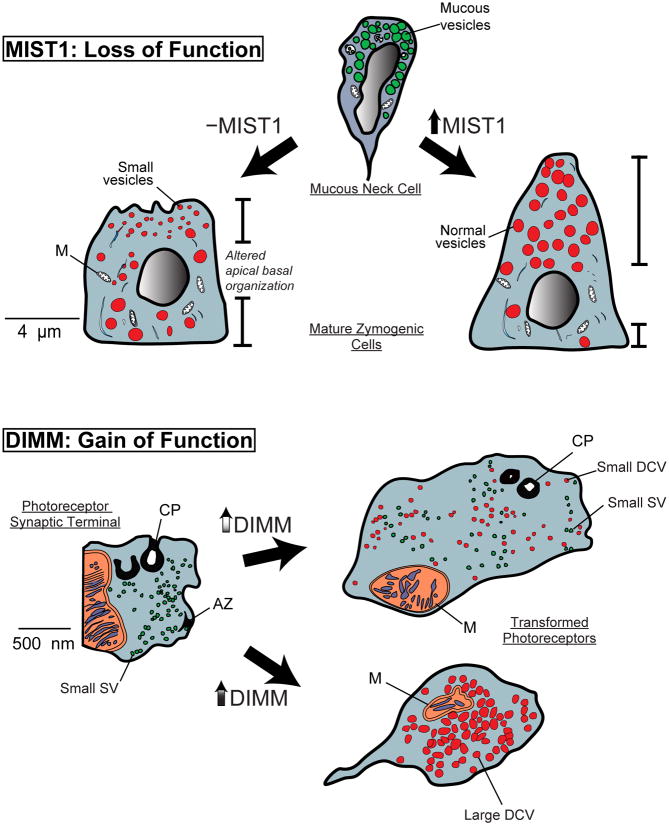
Figure 2.
Genetic analyses of Mist1/DIMM functions reveal their scaling factor properties.
Top: A schematic of results observed in a MIST1 loss of function model [3]. During normal zymogenic chief cell development, the chief cell begins to express MIST1 as it arises from its mucous granule-containing neck cell precursor. MIST1 is required for the expansion of apical cytoplasm and formation/maintenance of large, zymogenic vesicles. In its absence, cell apices are stunted, vesicles are smaller, and nuclei are nearer the cell apex.
Bottom: A schematic of results observed in a DIMM gain of function model [17].
Drosophila photoreceptors project axons that terminate within the brain, and store the fast neurotransmitter histamine within small clear synaptic vesicles (SSV). SSVs are released from T-bar type active zones (AZ) and retrieved by endocytosis at glia invaginations – capitate projections (CP). Following mis-expression of DIMM at moderate (top) or high (bottom) levels), the properties of the photoreceptor are transformed. Low-level DIMM mis-expression results in accumulation of small dense-core vesicles (Small DCV) in addition to the SSV. Hi-level DIMM mis-expression results in loss of SSVs, AZs and CPs, and the heavy accumulation of large DCVs. With co-mis-expression of a neuropeptide precursor transgene, the large DCVs process and store mature neuropeptide.
The DIMM transcription factor is a basic helix-loop-helix protein of the Atonal class normally restricted to diverse peptidergic neurosecretory neurons and endocrine cells in Drosophila [13]. DIMM is associated with secretion of most of the ∼35 families of insect neuropeptides. In the larval central nervous system, approximately 3% of neurons are DIMM-positive but they are not related by lineage, position, axonal projection or specific peptide expression [14, 15]. With loss of dimm function, neurosecretory neurons survive, express antecedent developmental markers of cell fate, but fail to display their normal accumulation of large amounts of secretory peptides or biosynthetic enzymes (the cells are “dimmed”) [15]. DIMM expression commences only after cells undergo terminal divisions and for most DIMM cells, it is sustained thereafter throughout the cell's lifespan [14, 15]. Cells that normally would express DIMM still form in its absence, but they are structurally and functionally abnormal. Hence, the DIMM loss of function phenotype argues for its role in cell structure and function and not in the specification of cell fate or in the control of cell survival.
DIMM over-expression in photoreceptors, a cell lineage that does not normally express DIMM, is sufficient to establish a neuroendocrine-type secretory apparatus[16]. Ectopic DIMM over-expression leads to accumulation of large (∼60 nm) dense core vesicles (LDCVs), similar to those seen in normal DIMM expressing cells, and does so at the expense of small clear vesicles and pre-synaptic secretory machinery [17] (Fig. 2, bottom). The scaling aspect of DIMM actions is evident in this suppressive activity: small clear synaptic vesicles are either retained or completely lost depending on the level of DIMM mis-expression. Additionally, DIMM misexpression causes loss of other components of fast neurotransmission within photoreceptors such as active zones, as well as a loss of its principal differentiated feature the rhabdomere, where rhodopsin normally is accumulated in high density. Interestingly, rhodopsin expression remains unchanged, only its normal trafficking to the rhabdomeric membranes. Thus DIMM misexpression has major consequences on the details of sub-cellular domain organization, and not so much on the essential features of cell fate determination.
With combined co-misexpression of a pro-neuropeptide transgene, photoreceptors accumulate the ectopic neuropeptide within the large dense core vesicles and biochemically process the pro-form of the neuropeptide to its active form. Hence, the DIMM transcription factor ramps up two cardinal features of the classic regulated secretory pathway – (i) packaging within a dedicated sub-cellular organelle (the LDCV) and (ii) processing by dedicated biosynthetic enzymes resident within the trans-Golgi and in secretory granules. Thus, DIMM is normally dedicated to scaling up regulated secretion (and scaling down other types of secretion) within that subset of peptidergic cells in Drosophila that have a strong “professional” dedication to the secretion of bioactive peptides.
The essential properties of DIMM and MIST1 suggest a new category of transcription factor activity that underlies sub-cellular scaling
DIMM and MIST1 are sequence orthologues, with high similarity throughout the bHLH region of the molecules. The fundamental similarities in molecular genetic studies of DIMM and MIST1 suggest that they regulate transcription of genes that perform a shared, evolutionarily-conserved purpose. But what kind of transcription factors are they? Unlike other transcription factors that are part of developmental cascades, MIST1/DIMM expression occurs predominantly in mature cells and persists well after development of those cells. Furthermore, MIST1/DIMM do not seem to affect cell specification (the qualitative features of cell development), because loss of mature, lineage-specific markers is not a phenotype seen following loss of MIST1/DIMM function, and there is no evidence of paralogous transcription factors in vertebrates or flies. Thus, the lack of cell specification phenotype is not likely to be due to redundancy. Rather, MIST1/DIMM loss of function causes cellular defects that are quantitative: they are essentially limited to a reduced capacity for regulated secretion of polypeptides; MIST1/DIMM gain of function is sufficient to re-direct cell resources to promote that subcellular process.
Here, we argue that MIST1/DIMM are prototypes for a new class of TFs that we term scaling factor TFs. These have critical distinguishing characteristics that we outline below.
They control entire sub-cellular domains
First and foremost, their target genes will dictate expansion of a specific subcellular domain or resource (Fig. 3). For MIST1/DIMM, this subcellular resource is the secretory vesicle trafficking apparatus, the extended domain that permits regulated peptide secretion. Note, however, that certain direct targets of scaling factors may not at first glance appear to fit the definition of a sub-cellular domain component, thus belying the original scaling factor definition. However, such apparent contradictions may in truth reflect a lack of understanding for how such proteins actually operate.
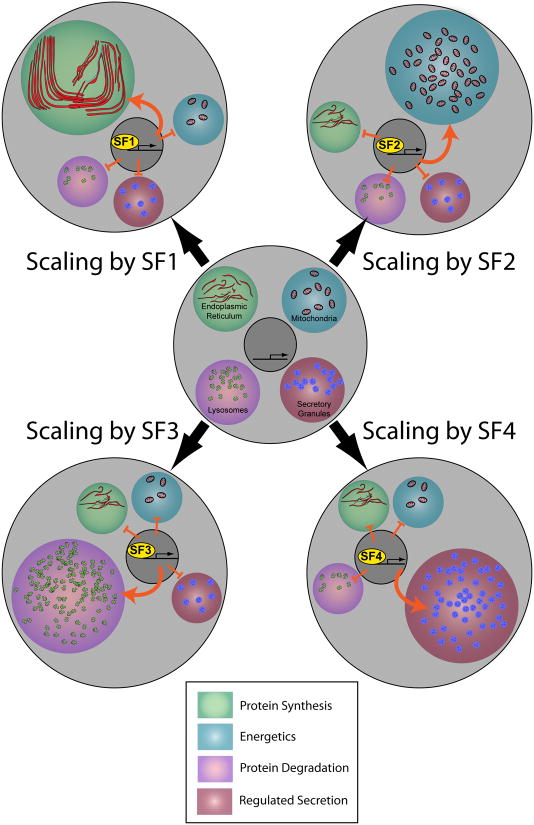
Figure 3.
Scaling factors function by scaling specific sub-cellular domains. In the illustration shown at center, an immature cell devotes resources equally to each of four sub-cellular domains – domains involved in protein synthesis (endoplasmic reticulum), energetics (mitochondria), protein degradation (lysosomes) and (regulated secretion) secretory granules. We propose the concept of scaling factors (SFs), whose expression is induced during terminal cell fate specification. SFs regulate change the distribution of a cell's resources as indicated by the four examples SF1-SF4. This figure highlights several features of SFs that are discussed in the text: namely, (i) that SFs are not required to generate particular sub-cellular domains, only to scale them appropriately; (ii) that each is devoted to a certain sub-cellular domain, and (iii) that SFs scale up single sub-cellular domains while repressing others.
For example, we know that regulated secretion is profoundly decreased by loss of MIST1/DIMM, but MIST1/DIMM's effects on the secretory subcellular domain will not be exerted only by modulating expression of secretory vesicle-associated genes. Known MIST1 targets include Connexin32 (GJB1) [18] and SPCA2 (Atp2c2) [19], which mediate either cell-cell or intracellular transport of the calcium signal that regulates release of secretory vesicles; and p21 (CDKN1A) [20]which inhibits mitosis to allow development of an elaborate secretory vesicle apparatus.
Furthermore in the case of DIMM, its identified direct targets include genes that encode bone fide secretory granule proteins like PHM and cyt-b561-1, but also proteins like CAT-4, a putative arginine transporter [21]. Such a transporter protein has never previously been implicated in secretory pathway regulation, yet down-regulation of CAT-4 RNA severely abrogates DIMM's ability to support a functional secretory pathway [21]. Thus a general point is that the study of Scaling Factors like DIMM and MIST1 provides a basis to create testable hypotheses, and thereby potentially implicate critical components of subcellular domains that were not or could not be otherwise identified.
They control quantitative features
DIMM/MIST1 scale up the secretory apparatus, but they do not appear to be required for its establishment. Consistent with this scaling function MIST1 and DIMM are not exclusive regulators of their respective gene targets; indeed many of their target genes are still expressed at low levels even in their absence. Therefore the second critical characteristic of SFs is that they amplify expression of their target genes, rather than gate them - OFF to ON - in binary fashion [6, 15, 22, 23].
They persist for the life of the cells
MIST1/DIMM expression continues throughout cell lifespan. It follows that, because SFs are used by cells to maintain preferential, high levels of specific subcellular organelles, they must express the TFs themselves at persistent, high levels to maintain, in turn, high levels of all the constantly turning over protein components SFs not only initiate a program of sub-cellular differentiation, but also contribute to the maintenance of cell specialization throughout a cell's lifespan (Fig. 4).
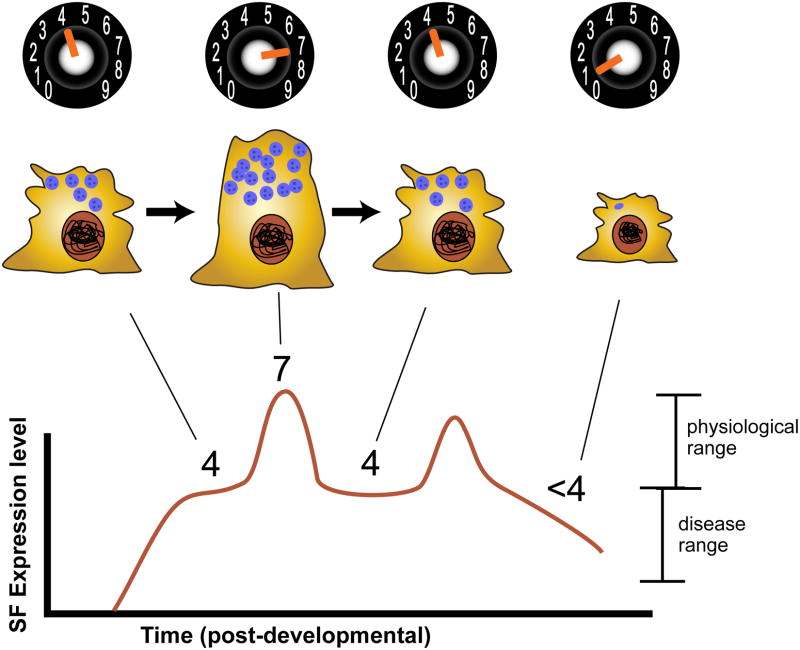
Figure 4.
Scaling factors are persistently expressed and mediate cellular adaptive responses. SF levels fluctuate throughout a cell's lifespan, with higher levels generating greater specialization – in this example, more secretory granules. As long as SF levels remain within a certain range (in this example, 4-7), the cell functions normally; however, SF expression may fall outside the normal range, leading, in the depicted example, to a lack of balance among the cell's different sub-cellular domains and ultimately to cellular pathophysiology.
Do other SFs bridge the stages of development and cell function?
Next we discuss two other TFs PGC-1alpha and TFEB because their functional properties suggest a fundamental similarity with those of MIST1/DIMM. They amplify distinct sub-cellular compartments and they persist throughout the life of a cell.
PGC-1 α controls mitochondrial functions
Certain tissues (like cardiac tissues and brown fat) require a greater commitment of cellular resources to ATP production than most. This specialization is in part executed by the actions of a transcriptional coactivator, the peroxisome proliferator-activated receptors gamma (PPARγ) coactivator-1 alpha (PGC1α)[24, 25]. In cardiac myocytes, its actions govern cell maturation, not cell fate determination. In these cells, PGC1α arises weeks after cells are born; and its expression persists thereafter [26]. It is responsible for the stereotyped switch in fuel these myocytes display from glucose in the fetal period to fatty acids following birth [25]. Remarkably, mis-expression of PGC1α in cell culture promotes mitochondrial biogenesis at the expense of other sub-cellular organelles such as sarcomeres[26, 27], and cardiac specific over-expression leads to increased mitochondrial mass in the heart in vivo [28]. PGC1α executes its pro-mitochondrial program by promoting activation of many key mitochondrial genes[29]. Thus, PGC1α conforms well to the SF category because it amplifies cellular differentiation, it is persistent in its expression and it exerts higher order control over a key sub-cellular domain.
TFEB controls lysosomal functions
Because the requirements for protein degradation vary among cells, Ballabio and colleagues postulated the presence of a cellular program that cells could use to scale lysosomal biogenesis and activity according to physiological needs. Analyzing ∼100 known lysosomal genes, they uncovered a common cis regulatory element which eventually led to the identification of a bHLH-zipper transcription factor –TFEB. This factor appears highly dedicated to regulating the lysosomal sub-cellular domain. It directly regulates a network of ∼300 target genes, whose protein products are lysosomal structural components, or mediate protein import to the lysosome, or protect against leakage of proteases from it. Significantly, TFEB over-expression enhances cellular clearance and lysosomal biogenesis, indicating it is sufficient to produce a coherent and coordinated program of lysosomal operations. TFEB splice variants display specific patterns of tissue expression which supports the possibility that it might be most important for cell lineages that require high lysosomal activity [30]. These observations argue that the actions of TFEB conform to some of the proposed SF cardinal properties; it directly amplifies the essential genes to promote cell differentiation and does so by scaling the structural components of a specific sub-cellular domain.
The SF concept suggests new avenues of investigation
We now use the new SF framework to suggest new experimental lines of investigation at the interface of cell development and cell physiology.
Are all sub-cellular domains regulated by specific SFs?
We have indicated subcellular domains that are quantitatively regulated by transcriptional regulation (e.g. mitochondria, lysosomes, LDCVs). But are other major subcellular domains also regulated by dedicated SFs? There are many organelles or structures that may also display such regulation – peroxisomes, the primary cilium, Golgi apparatus, pre- and post-synaptic domains, to name a few. Identifying cell types that, to accomplish their physiological functions, exaggerate display of particular organelles or domains may be a useful first step in discovering new SFs and new modes of cellular organization.
Do SFs scale down certain sub-cellular domains while scaling up others?
The SF hypothesis emphasizes patterns of developmental gene regulation that are grouped by sub-cellular domains (Fig. 3). One important consideration is the extent to which up-regulation of specific sub-cellular domains during adaptive responses may come at the expense (i.e. repression) of other domains. When DIMM is mis-expressed in non-secretory cells, those normally histaminergic neurons gain peptide secretory properties, but they lose hallmark features of fast neurotransmission (e.g. normal histamine levels, histamine decarboxylase expression, small clear synaptic vesicles, and the pre-synaptic machinery needed for exocytosis and endocytosis of synaptic vesicles) [17]). Likewise, when PGC1α up-regulates mitochondrial cell number in cardiac tissues, it does so at the expense of sarcomeres [26]. Thus SF actions involve compensation among sub-cellular domains: promotion of one likely involves repression of others (Fig. 3).
A corollary to this question is how different SFs might operate at the same time in the same cell? For example, can PGC1α and TFEB be increased at the same time and increase both mitochondriogenesis and lysosomogenesis, or are there restrictions to how many SFs can operate concurrently? Both in vitro and in vivo molecular genetic experiments could be designed to help address this unexplored question.
If SFs are good for some cells, are they good for all?
We have focused on a few specific cell types that appear to have “professional” dedication to certain subcellular function (e.g. lysosomal clearance). It is reasonable therefore to ask: do the ideas we put forward here apply to just a few cell types, or to the majority? On the one hand, professional dedication may be the lifestyle of just a small number of cell types within the body. Alternatively, we might begin to understand cellular physiology more broadly if professional dedication describes the physiology of many or most cell types. According to the latter hypothesis, cell diversification reflects a cell-type specific emphasis of certain cellular features (sub-cellular domains) and a de-emphasis of others. We submit that SFs are perfectly positioned to mediate cell diversification and adaptation in many different cell types.
Do SFs contribute to cellular adaptation?
Because SFs persist and because they scale cellular attributes, they offer a mechanism for adaptation to extracellular challenges (Fig. 4). SF regulation of specific sub-cellular domains lies within a dynamic quantitative range. Importantly, the baseline for this dynamic range is not zero; in molecular terms this means that SF target genes are also regulated at moderate levels by other TFs. For example, the regulation of lysosomal biosynthetic genes is also not exclusive to TFEB, but TFEB is an especially potent regulator and does so in concert with regulation of many other lysosomal biosynthetic genes [31]. Thus, we speculate that SFs work together with other more general regulators to help a mature cell tune its cellular response after the period of its overt differentiation from other lineages.
Are malfunctioning SFs a cause of disease? Can they be harnessed to promote regeneration?
Finally, what happens to cells when the capacity to scale sub-cellular domains is exceeded or lost (Fig. 4)? We speculate that studying SFs may help us better understand the etiology of diverse adult diseases, because new therapies can be designed based on the identification of genetic control circuits that normally scale critical sub-cellular domains. Understanding SFs may also help us generate de novo specific cellular phenotypes to replace precisely those cell populations that are lost in different disease states. For example, there is currently great interest in generating newly differentiated β islet cells from embryonic or induced pluripotent stem cells to address certain diabetic syndromes. If success is scored only by the initial appearance of terminal differentiation features (i.e. insulin expression), these efforts may fall short [32]. Given the quantitative maturation of specialized properties that islet cells display after their specification [2], there may be considerable value in assuring the involvement of those SFs that govern the specialization of β cell secretory systems.
Conclusions
We have presented the scaling factor hypothesis concerning mechanisms of cellular organization. The central premise is that different sub-cellular domains of a cell – each dedicated to different cellular functions – are governed by dedicated transcriptional control. Such scaling control is exerted by transcription factors first at the end of cell development to help specialize the cell beyond its initial specification. Regulation of scale then extends throughout the period of a cell's maturity and functionality: continued ability to scale different sub-cellular domains thereby representing a key feature underlying cellular adaptations to changing environmental conditions. The hypothesis raises several intriguing questions. For example, what signaling pathways do cells use to detect changing conditions and modify SF expression? What homeostatic functions limit SF actions within physiological ranges? How extensive is the set of gene targets SFs control? In addition to addressing normal cell functions, the SF hypothesis also raises possibilities for defining new experimental approaches to study cellular disease states. Their potency and coordinating actions make them plausible agents by which to better understand and treat disease states.
Acknowledgments
We thank Adam Joyce and Dr, Jim Skeath for their insights and critique of the manuscript. We especially thank Dr. Joe Corbo for reading, discussion and encouragement. Funding for work in our labs on this problem comes from the National Institutes of Health: R01 DK-079798-01,02,03 (JCM) and R01 NINDS-21749 (PHT).
Abbreviations
- SF
scaling factor
- TF
transcription factor
Literature Cited
- 1.Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–24. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 2.Jiang FX, Mehta M, Morahan G. Quantification of insulin gene expression during development of pancreatic islet cells. Pancreas. 2010;39:201–8. doi: 10.1097/MPA.0b013e3181bab68f. [DOI] [PubMed] [Google Scholar]
- 3.Ramsey VG, Doherty JM, Chen CC, Stappenbeck TS, et al. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development. 2007;134:211–22. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Johansson C, Tran T, Bettencourt R, et al. Identification of a basic helix-loop-helix transcription factor expressed in mammary gland alveolar cells and required for maintenance of the differentiated state. Mol Endocrinol. 2006;20:2187–98. doi: 10.1210/me.2005-0214. [DOI] [PubMed] [Google Scholar]
- 5.Pin CL, Rukstalis JM, Johnson C, Konieczny SF. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J Cell Biol. 2001;155:519–30. doi: 10.1083/jcb.200105060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson CL, Kowalik AS, Rajakumar N, Pin CL. Mist1 is necessary for the establishment of granule organization in serous exocrine cells of the gastrointestinal tract. Mech Dev. 2004;121:261–72. doi: 10.1016/j.mod.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Capoccia BJ, Lennerz JK, Bredemeyer AJ, Klco JM, et al. Transcription factor MIST1 in terminal differentiation of mouse and human plasma cells. Physiol Genomics. 2011;43:174–86. doi: 10.1152/physiolgenomics.00084.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bredemeyer AJ, Geahlen JH, Weis VG, Huh WJ, et al. The gastric epithelial progenitor cell niche and differentiation of the zymogenic (chief) cell lineage. Dev Biol. 2009;325:211–24. doi: 10.1016/j.ydbio.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huh WJ, Esen E, Geahlen JH, Bredemeyer AJ, et al. XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology. 2010;139:2038–49. doi: 10.1053/j.gastro.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo X, Shin DM, Wang X, Konieczny SF, et al. Aberrant localization of intracellular organelles, Ca2+ signaling, and exocytosis in Mist1 null mice. J Biol Chem. 2005;280:12668–75. doi: 10.1074/jbc.M411973200. [DOI] [PubMed] [Google Scholar]
- 12.Kowalik AS, Johnson CL, Chadi SA, Weston JY, et al. Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein-induced pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1123–32. doi: 10.1152/ajpgi.00512.2006. [DOI] [PubMed] [Google Scholar]
- 13.Park D, Taghert PH. Peptidergic neurosecretory cells in insects: organization and control by the bHLH protein DIMMED. Gen Comp Endocrinol. 2009;162:2–7. doi: 10.1016/j.ygcen.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Park D, Veenstra JA, Park JH, Taghert PH. Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS One. 2008;3:e1896. doi: 10.1371/journal.pone.0001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewes RS, Park D, Gauthier SA, Schaefer AM, et al. The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development. 2003;130:1771–81. doi: 10.1242/dev.00404. [DOI] [PubMed] [Google Scholar]
- 16.Allan DW, Park D, St Pierre SE, Taghert PH, et al. Regulators acting in combinatorial codes also act independently in single differentiating neurons. Neuron. 2005;45:689–700. doi: 10.1016/j.neuron.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 17.Hamanaka Y, Park D, Yin P, Annangudi SP, et al. Transcriptional orchestration of the regulated secretory pathway in neurons by the bHLH protein DIMM. Curr Biol. 2010;20:9–18. doi: 10.1016/j.cub.2009.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rukstalis JM, Kowalik A, Zhu L, Lidington D, et al. Exocrine specific expression of Connexin32 is dependent on the basic helix-loop-helix transcription factor Mist1. J Cell Sci. 2003;116:3315–25. doi: 10.1242/jcs.00631. [DOI] [PubMed] [Google Scholar]
- 19.Garside VC, Kowalik AS, Johnson CL, DiRenzo D, et al. MIST1 regulates the pancreatic acinar cell expression of Atp2c2, the gene encoding secretory pathway calcium ATPase 2. Exp Cell Res. 2010;316:2859–70. doi: 10.1016/j.yexcr.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia D, Sun Y, Konieczny SF. Mist1 regulates pancreatic acinar cell proliferation through p21 CIP1/WAF1. Gastroenterology. 2008;135:1687–97. doi: 10.1053/j.gastro.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park D, Hadzic T, Yin P, Rusch J, et al. Molecular Organization of Drosophila Neuroendocrine Cells by Dimmed. Curr Biol. 2011;18:1515–24. doi: 10.1016/j.cub.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian X, Jin RU, Bredemeyer AJ, Oates EJ, et al. RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol Cell Biol. 2010;30:1269–84. doi: 10.1128/MCB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park D, Shafer OT, Shepherd SP, Suh H, et al. The Drosophila basic helix-loop-helix protein DIMMED directly activates PHM, a gene encoding a neuropeptide-amidating enzyme. Mol Cell Biol. 2008;28:410–21. doi: 10.1128/MCB.01104-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–70. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol. 2002;29:339–45. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 26.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, et al. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–56. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z, Puigserver P, Andersson U, Zhang C, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 28.Russell LK, Mansfield CM, Lehman JJ, Kovacs A, et al. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–33. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 29.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–38. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 30.Kuiper RP, Schepens M, Thijssen J, Schoenmakers EF, et al. Regulation of the MiTF/TFE bHLH-LZ transcription factors through restricted spatial expression and alternative splicing of functional domains. Nucleic Acids Res. 2004;32:2315–22. doi: 10.1093/nar/gkh571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sardiello M, Palmieri M, di Ronza A, Medina DL, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–7. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 32.Borowiak M, Melton DA. How to make beta cells? Curr Opin Cell Biol. 2009;21:727–32. doi: 10.1016/j.ceb.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]