Abstract
During retroviral maturation, the CA protein oligomerizes to form a closed capsid that surrounds the viral genome. We have previously identified a series of deleterious surface mutations within human immunodeficiency virus type 1 (HIV-1) CA that alter infectivity, replication, and assembly in vivo. For this study, 27 recombinant CA proteins harboring 34 different mutations were tested for the ability to assemble into helical cylinders in vitro. These cylinders are composed of CA hexamers and are structural models for the mature viral capsid. Mutations that diminished CA assembly clustered within helices 1 and 2 in the N-terminal domain of CA and within the crystallographically defined dimer interface in the CA C-terminal domain. These mutations demonstrate the importance of these regions for CA cylinder production and, by analogy, mature capsid assembly. One CA mutant (R18A) assembled into cylinders, cones, and spheres. We suggest that these capsid shapes occur because the R18A mutation alters the frequency at which pentamers are incorporated into the hexagonal lattice. The fact that a single CA protein can simultaneously form all three known retroviral capsid morphologies supports the idea that these structures are organized on similar lattices and differ only in the distribution of 12 pentamers that allow them to close. In further support of this model, we demonstrate that the considerable morphological variation seen for conical HIV-1 capsids can be recapitulated in idealized capsid models by altering the distribution of pentamers.
Retroviruses initially assemble as noninfectious spherical particles that are organized by the viral structural protein Gag (17, 23). Upon budding, these immature virions rearrange dramatically to form mature infectious viral particles (31). Viral maturation is triggered by the proteolytic cleavage of Gag into three new proteins, namely, MA, CA, and NC. As the virus matures, MA remains associated with the plasma membrane, NC associates with the dimeric RNA genome, and CA condenses to form a closed capsid shell that surrounds the NC-RNA complex.
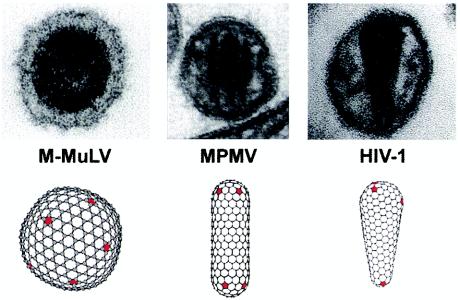
Although the immature capsids of all retroviruses are spherical, the shapes of mature retroviral capsids vary significantly between genera (see Fig. 1) (51). Specifically, the capsids of retroviruses from most genera are roughly spherical, whereas the capsids of beta- and type D retroviruses are cylindrical (e.g., Mason-Pfizer monkey virus [M-PMV]) and the capsids of lentiviruses are conical (e.g., human immunodeficiency virus [HIV]) (4, 55). Despite these large variations in capsid morphology, however, the tertiary structures of all retroviral CA proteins are similar (9), suggesting that the different capsid morphologies may be created by the utilization of common underlying principles (21, 35).
FIG. 1.
Variation in the mature capsid morphologies of retroviruses from different genera. EM images and idealized computer models of the mature capsids of a gammaretrovirus (Moloney murine leukemia virus [M-MuLV]), a betaretrovirus (M-PMV), and a lentivirus (HIV-1) are shown. Note that in all cases, the structures are assembled from hexagonal lattices and are allowed to close by the introduction of 12 pentameric defects (red).
Retroviral CA proteins contain two structural domains, the N-terminal assembly domain (NTD) and the C-terminal dimerization domain (CTD). The NTD is a tapered domain composed of seven α-helices and an N-terminal β-hairpin (5, 9, 10, 12, 22, 26, 28, 29, 40, 47). Biochemical and genetic analyses have demonstrated that this domain is especially important for mature capsid assembly and for subsequent early steps in the replication cycle (3, 6, 13, 43, 52, 53). Nevertheless, several NTD mutants have been identified that are also defective in immature capsid assembly, suggesting a role for the domain in this process as well (36, 43, 53).
The CTD is a roughly spherical domain composed of four α-helices and a 310 helix (5, 9, 19, 26, 28, 29, 56). Unlike the NTD, most deleterious mutations in the CTD impair the ability of Gag to assemble into immature particles (13, 43, 52, 53). The HIV type 1 (HIV-1) CA CTD dimerizes in solution and is also crystallized as a dimer (19, 44, 56). Mutational analyses have revealed that this crystallographic CTD dimer interface is important for infectivity and assembly (18, 52, 53, 56). In spite of their structural similarities, however, not all retroviral CA proteins form stable dimers in solution. One example is the equine infectious anemia virus (EIAV) CA protein, which is monomeric in solution but has been crystallized as a dimer. Interestingly, the two CTDs in the EIAV CA dimer associate via the same surface as the HIV CA CTD dimer, but with different orientations (26).
At high salt concentrations, HIV-1 CA assembles into hollow cylinders (16, 21, 25, 35). Cryo-electron microscopic reconstructions have revealed that the cylinders are composed of helical arrays of CA hexamers (35). Similar CA hexamers were also observed in Fourier-filtered images of cylindrical cores isolated from HIV-1 virions, indicating that CA forms similar assemblies in vitro and in vivo (8). Moreover, electron diffraction patterns from cylindrical and conical viral capsids share common reflections, indicating that both cones and cylinders utilize the same extended CA lattice, and therefore, that the cylinders formed in vitro are reasonable models for mature capsid assembly (8). The unusual shape of the CA NTD allowed it to be fit into the reconstructed electron density, resulting in a pseudoatomic structural model for the CA lattice (35). In this model, the hexameric rings are formed by six NTDs and are linked to adjacent rings via CTD dimers. Helices 1 and 2 of each NTD appear to form a 12-helix bundle that holds the hexamer together. However, this structure must still be considered a working model owing to the moderate resolution of the reconstruction (∼20 Å). Indeed, two other groups have recently proposed variations on this basic model (32, 38).
Based upon the hexameric lattice observed in the CA cylinder reconstructions, together with geometric analyses of synthetic HIV-1 cores, we have proposed that retroviral capsids are organized on hexameric lattices and that the incorporation of 12 CA pentamers allows each lattice to close. The relative positioning of the pentamers in the hexameric lattice defines the overall shape of the mature capsid (21) (Fig. 1). In support of this model, CA proteins from Rous sarcoma virus (39) and Moloney murine leukemia virus (20) have also been shown to form hexameric lattices that are similar to those formed by the HIV-1 CA protein.
The ambiguities inherent in fitting crystal structures to moderate-resolution density maps make it essential to test putative CA-CA interfaces by using complementary approaches. Members of our laboratory recently reported an extensive alanine-scanning mutational analysis of HIV-1 CA functioning in the context of a cultured virus (53). That study defined four surfaces that are important for HIV-1 assembly, infectivity, and replication. However, the complexity of viral replication made it difficult to determine precisely how and when these different surfaces function. For this study, we used a simple CA assembly assay to define which residues are important for the formation of the mature viral capsid structure. Specifically, purified mutant CA proteins that harbored the detrimental mutations defined in previous genetic assays (53) were tested for cylinder formation. We also performed computer modeling studies of retroviral capsids to explain the considerable size and shape variation observed for authentic HIV-1 capsids.
MATERIALS AND METHODS
Plasmid construction.
HIV-1NL4-3 CA mutants previously tested in viral replication assays (53) were transferred from the proviral R9ΔApaI plasmid to the pET11a expression vector by PCR amplification and standard cloning techniques. The PCR primers were designed to introduce an NdeI site and a start codon at the 5′ end and a BamHI site and a stop codon at the 3′ end of the CA gene. The four mutants presented in this paper that were not previously built into proviral constructs (M39A, R18A, N21A, and Δ87-97) were made by using PCR-based megaprimer mutagenesis (42). The complete open reading frame of every CA mutant was confirmed by DNA sequencing.
Protein expression and purification.
All CA proteins were expressed and purified as previously described for the wild-type protein (52). Purified CA proteins were dialyzed overnight against 20 mM Tris (pH 8.0)-5 mM β-mercaptoethanol. After dialysis, the mutant proteins were concentrated to ∼30 mg/ml, divided into aliquots, flash frozen in liquid N2, and stored at −70°C prior to analysis. The expected mass of each mutant protein was confirmed by electrospray mass spectrometry (except for E75A/E76A, which precipitated when dialyzed into water).
Assembly and EM analyses of recombinant CA proteins.
Frozen aliquots of each CA protein were thawed and requantitated prior to use. CA assembly reaction mixtures were made in 20-μl volumes (16 h at 4°C; 50 mM Tris [pH 8.0], 1 M NaCl, 5 mM β-mercaptoethanol). For electron microscopy (EM) analyses, carbon-coated grids were placed on 10-μl aliquots of each assembly reaction for 90 s, washed with 3 to 4 drops of 0.1 M KCl, stained with 3 or 4 drops of 4% uranyl acetate, and air dried. Samples were analyzed on Hitachi 7100 and Philips Tecnai 12 transmission electron microscopes at magnifications between ×2,500 and ×50,000. Assembly reactions were analyzed at three different protein concentrations (15, 10, and 5 mg/ml) for each CA mutant. Each reaction was repeated at least three times with proteins from at least two separate purifications.
Isolation and EM analysis of HIV-1 cores.
Mature virus cores were isolated as previously described (30). In brief, the virus was produced by transfection of 293T cells with the R9 proviral HIV-1 expression construct. After 48 h, the supernatant was harvested, filtered through a 0.45-μm-pore-size filter, and concentrated through a 20% sucrose cushion. Subsequently, the viral envelope was stripped and the resulting viral cores were purified by sedimenting the concentrated viral sample through a 30 to 70% sucrose cushion overlaid with layers of a 1% Triton X-100 solution in 15% sucrose and a 7.5% sucrose barrier cushion. After sedimentation, fractions containing viral cores were combined and concentrated. For EM analyses, viral cores were applied to glow-discharged carbon-coated grids, briefly washed with buffer, and stained with 1% uranyl acetate. Samples were examined in a Philips CM12 transmission electron microscope.
Molecular modeling.
Models for the different retroviral capsid shapes were constructed manually from hexameric and pentameric rings composed of sp2 hybridized carbon atoms (21) by using Chem3D software (CambridgeSoft Corporation). Different shapes were explored by inserting pentamers at different positions in the hexagonal lattice. After crude models had been constructed manually, the bond lengths and geometries of the hexameric and pentameric rings were regularized by energy minimization programs within the Chem3D software. The minimization protocol was demonstrated in histograms, which showed narrow ranges of bond lengths in the structure.
RESULTS AND DISCUSSION
Protein purification, solubility, and assembly.
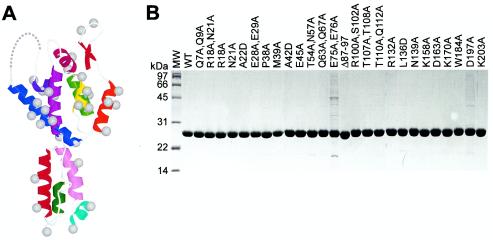
An ensemble of 27 mutant HIV-1NL4-3 CA proteins was tested for in vitro assembly (Table 1). The ensemble included 18 of the 21 mutations that reduced viral infectivity at least 25-fold in a previous mutagenesis study (53). The remaining three mutations were D51A and M185A, whose in vitro assembly was described previously (52), and P224A, which resides in an unstructured C-terminal region of CA that can be deleted without blocking assembly in vitro (data not shown). The ensemble also included five control mutations that were distributed throughout the protein and that reduced infectivity <25-fold. We also included one mutant (Δ87-97), which was created by replacing CA residues 87 to 97 with two glycine residues, to test the importance of the cyclophilin A (CypA) binding loop for CA assembly. Twenty-six of the 27 mutant CAs were soluble in Escherichia coli and were purified to high levels of homogeneity (Fig. 2B). An exception was CA mutant E75A/E76A, which was less soluble than the other proteins and therefore did not purify as well (Fig. 2B).
TABLE 1.
List of CA mutants and their assembly phenotypes in vitro and in vivo
| Mutation(s) | Locationa | In vitro cylinder formationb | In vivoc assembly
|
||
|---|---|---|---|---|---|
| Presence of conical capsid | Particle production | Infectivityd (n-fold less than wild type) | |||
| WT | ++ | ++ | ++ | 1.0 | |
| Q7A/Q9A | NTD β-hairpin | ++ | ++ | ++ | 10 ± 3.5 |
| R18A/N21A | NTD helix 1 | Altered | + (multiple capsids) | ++ | 0 |
| R18A | NTD helix 1 | Altered | ND | ND | ND |
| N21A | NTD helix 1 | ++ | ND | ND | ND |
| A22D | NTD helix 1 | + | − | ++ | 0 |
| E28A/E29A | NTD helix 1 | + | − | ++ | 0 |
| P38A | NTD helix 2 | ++ | ++ | ++ | 33 ± 16 |
| M39Ae | NTD helix 2 | − | ND | ND | ND |
| A42D | NTD helix 2 | − | − | ++ | 0 |
| E45A | NTD helix 2 | ++ | ++ | ++ | 29 ± 3.2 |
| T54A/N57A | NTD helix 3 | ++ | + | ++ | 80 ± 9 |
| Q63A/Q67A | NTD helix 4 | ++ | ++ | ++ | 34 ± 14 |
| K70A | NTD helix 4 | + | + (few conical capsids) | ++ | 21 ± 8 |
| E75A/E76A | NTD helix 4 | ++ | − | + | 0 |
| Δ87-97 | NTD CypA loop | Enhanced | ND | ND | ND |
| R100A/S102A | NTD helix 5 | ++ | − | + | 0 |
| T107A/T108A | NTD helices 5 and 6 | ++ | − | + | 0 |
| T110A/Q112A | NTD helix 6 | ++ | − | + | 0 |
| R132A | NTD helix 7 | ++ | ++ | ++ | 6.2 ± 0.7 |
| L136D | NTD helix 7 | ++ | + | ++ | 91 ± 13 |
| N139A | NTD helix 7 | ++ | ++ | ++ | 1.2 ± 0.2 |
| K158A | CTD turn (MHR) | Enhanced | − | − | 0 |
| D163A | CTD helix 8 (MHR) | ++ | ++ | ++ | 15 ± 7 |
| K170A | CTD helix 8 (MHR) | ++ | ++ | ++ | 0 |
| W184A | CTD helix 9 | − | − | + | 0 |
| D197A | CTD helix 10 | ++ | − | − | 0 |
| K203A | CTD helix 10 | + | ++ | ++ | 0 |
Position in the secondary structure of HIV-1 CA (see Fig. 1).
++, similar to WT CA at all protein concentrations tested; +, cylinders were formed at 15 mg/ml but were severely attenuated at lower protein concentrations; −, severely attenuated at all protein concentrations tested; enhanced, significantly longer cylinders than WT CA; altered, assembled efficiently into spheres, cones, and short capped cylinders.
In vivo data were previously reported in reference 53. ND, no data.
Mutations that reduced infectivity >100-fold are listed as having an infectivity of 0.
An alternative mutation in the same residue (M39D) blocked viral capsid assembly and infectivity (53).
FIG. 2.
Mutant CA proteins. (A) Locations of the 35 mutations employed in this study mapped onto the structure of the monomeric HIV-1 CA protein. The amino-terminal β-hairpin and helices 1 to 11 are red, orange, yellow, green, blue, indigo, magenta, violet, dark red, light purple, cyan, and green, respectively. Gray balls show positions of the Cβ atoms. (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of 26 of 27 purified mutant CA proteins. Note the following: (i) the E75A/E76A mutant was less soluble than the WT and was therefore more difficult to purify, (ii) the E28A/E29A mutant was slightly less soluble than the WT but could still be purified to homogeneity, and (iii) the K70A mutant was made late in the study and was therefore not included in this panel. MW, molecular size marker.
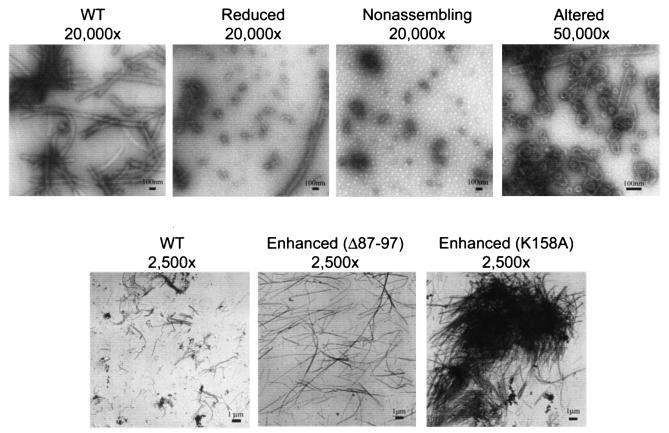
Assembly competence was assayed by incubating each CA protein in a high salt concentration at three different protein concentrations (15, 10, and 5 mg/ml) and by analyzing cylinder formation in transmission electron micrographs (TEM) of negatively stained samples. An analysis of the assembly characteristics of the different CA proteins revealed five distinct phenotypes, which we termed normal, enhanced, reduced, nonassembling, and altered (Table 1). Mutant CA proteins that assembled with efficiencies similar to that of the wild type (WT) at all protein concentrations tested were defined as normal (++; 16 of 27), while mutants which failed to form cylinders, even at 15 mg/ml, were defined as nonassembling (−; 3 of 27). Mutant proteins with an intermediate phenotype (i.e., cylinder formation was observed at 15 mg/ml but was severely attenuated at the two lower concentrations) were classified as reduced (+; 4 of 27), and mutants that produced longer cylinders at higher efficiencies than WT CA were defined as enhanced (2 of 27). Mutant proteins that did not form long cylinders but instead assembled efficiently into spheres, cones, and short capped cylinders were classified as altered (2 of 27) (Table 1). Examples of all five of these phenotypes are provided in Fig. 3.
FIG. 3.
Negatively stained EM images illustrating the five different assembly phenotypes observed for HIV-1 CA proteins (WT, wild-type CA; reduced, A22D CA; nonassembling, A42D CA; altered, R18A/N21A CA). The images in the top panels show assembly reactions performed at a protein concentration of 15 mg/ml, whereas the images in the lower panels show assembly reactions performed at a protein concentration of 5 mg/ml. Note that lower protein concentrations increased the lengths of cylinders for all CA proteins tested.
Mutations in the NTD of CA.
Twenty proteins with mutations in the NTD of CA were initially analyzed. Although most of these mutations adversely affected HIV-1 replication, only four significantly reduced or blocked cylinder assembly in vitro (Fig. 4A and Table 1). Notably, a series of mutations in helices 5, 6, and 7 that diminished HIV-1 replication in culture did not affect CA cylinder formation in vitro. These results are consistent with the idea that this region of CA is important for Gag assembly and particle production but does not play a direct role in the assembly of the mature viral capsid (53). In contrast, all four of the CA NTD mutations in our initial screen that exhibited detrimental effects on CA assembly in vitro mapped to helix 1 or 2. Helix 2 harbored the only two nonassembling mutations in the NTD (M39A and A42D), and helix 1 harbored two mutations that reduced, but did not eliminate, CA assembly (A22D and E28A/E29A).
FIG. 4.
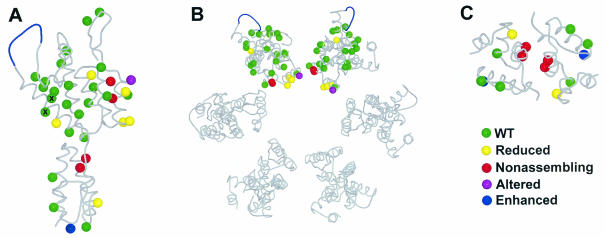
Locations and phenotypes of the different CA mutants described in this study. (A) Structure of the HIV-1 CA protein. (B) Structural model of the hexameric rings formed by the CA NTD. (C) Crystallographic model of the dimer formed by the CA CTD. Color code: green, WT assembly phenotype; red, nonassembling phenotype; yellow, reduced phenotype; purple, altered phenotype; blue, enhanced phenotype. The E75A/E76A mutant protein (denoted by ×) was less soluble than the WT.
Helix 1 also harbored one mutant with an altered CA assembly phenotype (R18A/N21A). This mutant CA protein assembled very efficiently, but instead of forming long cylinders it produced large numbers of small spheres, of ∼30 to 50 nm in diameter, as well as cones, spirals, and short capped cylinders (Fig. 3). To test which of the two residue changes in the double mutant were responsible for this phenotype, we also created the R18A and N21A mutations separately. CA proteins harboring only the R18A mutation were altered in assembly, demonstrating that this residue was responsible for the phenotype (data not shown).
Outside of those in helices 1 and 2, the only other mutation in the CA NTD that affected cylinder formation was the deletion mutation (Δ87-97), which enhanced cylinder production. This mutation removes the extended loop located between N-terminal helices 4 and 5 which acts to package cyclophilin A, a cellular prolyl isomerase, into the virion (36, 48).
Implications for pseudoatomic models of the CA hexamer.
There is general agreement that the hexameric rings observed in reconstructed CA assemblies are formed by six copies of the CA NTD (35, 38). It was previously proposed that the CA NTD can be fit into the reconstructed density from helical CA cylinders such that the six NTD molecules associate via interactions between helices 1 and 2 at the interior of the hexamer (Fig. 4B) (35). The results of our mutational analyses agree well with this model, as all of the detrimental CA NTD assembly mutations observed in our studies mapped to helix 1 or 2. Helices 1 and 2 are also protected from deuterium exchange upon CA cylinder assembly, which again suggests that they likely make important contacts in the assembled CA hexamer (32). Note, however, that not all mutations in helices 1 and 2 reduced CA cylinder assembly. In particular, the N21A (helix 1) and P38A (helix 2) mutant proteins, which are both adjacent in the primary sequence to detrimental mutations, assembled with normal efficiencies. This result may demonstrate the specificity of the CA assembly interface(s), and it emphasizes the importance of utilizing extensive mutagenesis screens when attempting to define protein-protein interaction surfaces.
Two groups have recently proposed variations on the CA hexamer model shown in Fig. 4B, and it is therefore of interest to consider these alternative models in light of our mutational analyses. In one alternative model for the CA hexamer, the NTDs are rotated ∼180° about axes that are parallel to the six-fold axis of the hexamer (38). As a result, helices 1, 2, and 7 from each CA NTD subunit face outward, and the cyclophilin A binding loops face toward the center of the ring (where they would presumably make intersubunit contacts that would help stabilize the hexamer). Several of our observations appear to be inconsistent with this model. Firstly, we have now shown that CA can form helical cylinders even when the entire cyclophilin binding loop is deleted (Fig. 2). Secondly, it is not obvious how this model can be reconciled with the importance of helices 1 and 2 for CA assembly. One possibility, suggested by Mayo et al., is that helix 2 might have an interhexamer interaction with helix 7 from a subunit of a different hexamer (38). To support this idea, they noted that an intermolecular four-helix bundle formed between helices 2 and 7 of adjacent CA subunits was observed in the crystal lattice of EIAV CA (26). However, in the crystallographic EIAV CA interaction, the two copies of helix 7 pack in an antiparallel orientation, whereas in the interaction proposed for HIV CA they would pack in a parallel orientation (which would therefore require a very different interaction). In any event, mutations in helix 7 generally do not block capsid assembly in vivo (53) or CA assembly in vitro (this work), and CA helix 7 is not protected from deuterium exchange in helical assembles of CA (32).
A second recent study has proposed that the NTD makes intermolecular contacts with the CTD of an adjacent molecule within the assembled lattice of CA hexamers. This proposal is based upon data from chemical cross-linking and deuterium exchange protection (32) and is consistent with assembly inhibitor and second-site suppressor studies (7, 33, 46). Specifically, Lanman et al. have shown that upon cylinder formation, the C-terminal end of helix 3 and the N-terminal end of helix 4 are protected from solvent exchange, and that Lys-70 (helix 4, in the NTD) of one CA subunit can be efficiently cross-linked to Lys-182 (helix 9, in the CTD) in a second subunit (32). In contrast, none of our original mutations in this region adversely affected cylinder formation (T54A/N57A [helix 3], Q63A/Q67A [helix 4], and E75A/E76A [helix 4]). Our initial experiments did not test the effects of the K70A mutation because this mutation did not reduce infectivity below our cutoff threshold (21-fold versus the threshold of 25-fold). To help with our testing of the validity of the NTD-CTD interface, however, we subsequently prepared the CA K70A mutant (53) and found that it exhibited an intermediate assembly phenotype. Specifically, the mutation reduced CA assembly in the cylinder formation assay, but short cylinders were still formed at CA concentrations of 15 mg/ml (data not shown). Similarly, the K70A mutation reduced, but did not eliminate, conical capsid formation in vivo (53). Thus, the phenotype of the K70A mutation is consistent with the involvement of this residue in a protein-protein interface, although mutations in this region of CA were generally not as detrimental as mutations elsewhere in the protein.
Implications for retroviral capsid morphology.
As described above, we have proposed that all retroviral capsids may be organized on hexameric lattices, with the positions of pentameric “defects” defining the overall capsid morphology. The assembly phenotype of R18A CA is consistent with this unifying model for capsid assembly, since this single protein can simultaneously form all of the known retroviral capsid structures (spheres, cones, and capped cylinders). Moreover, many of the structures formed by R18A CA resemble authentic retroviral capsids in both shape and size (Fig. 2).
One possible explanation for the phenotype of CA R18A is that the different capsid structures arise from an increased frequency of pentamer incorporation into the assembling hexameric lattice. Precedents for this idea are provided by a number of mutant icosahedral viruses that exhibit similar phenotypes (11, 14, 27, 37, 50). For example, point mutations in the structural protein of the T=3 icosahedral ilarvirus cause the random insertion of pentamers into the expanding lattice, and the resulting capsids exhibit highly variable diameters, similar to the spheres produced by the R18A mutant protein (50). Similarly, point mutations in the VP1 protein of polyomavirus cause the assembly of spirals (34, 45), which are also seen in R18A CA assembly reactions (not shown). Spirals are created when pentamers are improperly incorporated into an assembling hexagonal lattice, preventing closure of the structure (14, 27, 37). Finally, a single point mutant in the coat protein of alfalfa mosaic virus can result in a complete shift from spheres to cylinders in in vitro assembly reactions (11).
Importance of the cyclophilin A binding loop.
Previous studies have demonstrated that the addition of recombinant cyclophilin A to assembly reactions increases cylinder length (i.e., gives an enhanced phenotype) (24). Moreover, two point mutations in the cyclophilin A binding loop, G94D and A92E, which were originally discovered as cyclophilin-independent viral escape mutations (1, 57), can also enhance CA cylinder assembly in vitro (35). Taken together, these observations are consistent with the idea that the cyclophilin A binding loop actually inhibits CA cylinder assembly, suggesting that at least in vitro, cyclophilin A may increase the efficiency of CA assembly by suppressing off-pathway CA aggregation caused by the large, hydrophobic cyclophilin A binding loop (24). Off-pathway aggregation can alternatively be suppressed by mutations that increase loop hydrophilicity and charge (e.g., G94D and A92E) or by deletion of the entire loop. Interestingly, HIV-1 CA is the only retroviral CA protein known to use an exposed loop to bind cyclophilin A, and this loop is larger in HIV-1 than in other retroviruses (9). Conceivably, the selective advantage(s) of binding cyclophilin A (49) comes at the cost of reduced CA assembly efficiency.
Mutations in the CTD of CA.
The assembly phenotypes of six different CA proteins harboring mutations in the CTD were also analyzed. All of these mutations reduced the infectivity of the cultured virus, but only three of the six mutant proteins (W184A, K203A, and K158A) affected cylinder assembly in vitro (Fig. 4A).
Two of the mutations that diminished cylinder assembly are located within the crystallographic CTD dimer interface (W184A and K203A). The W184A mutation caused a nonassembling phenotype and is located in the hydrophobic core of the crystallographic CA dimer (Fig. 4C). This phenotype is similar to that of another hydrophobic dimer interface mutation (M185A) which also blocks cylinder formation in vitro (52). The K203A mutation was less severe but still clearly had a reduced assembly phenotype. K203 makes a salt bridge with D152 across the dimer interface. These results confirm the importance of the crystallographic dimer interface for cylinder assembly and support the idea that the CA CTD serves to make dimeric linkages between adjacent CA hexamers.
A second surface located at the “bottom” of the CTD is also important for particle production and viral infectivity (46). However, the two mutations within this surface that were tested (K158A and D197A) did not diminish CA cylinder assembly, consistent with the idea that this surface functions during immature particle formation. Indeed, the K158A mutation actually appeared to enhance CA cylinder assembly. However, unlike the long discrete cylinders produced by the Δ87-97 CA protein, the long cylinders produced by K158A aggregated into very large clumps (Fig. 3). We can provide no obvious structural explanation for this phenotype, but we speculate that the replacement of a charged lysine surface residue with an alanine may create a more hydrophobic surface that contributes to CA aggregation.
Molecular modeling.
EM analyses of thin-sectioned mature virions and isolated HIV capsids have revealed significant variations in capsid size and shape (2, 8, 30, 31, 54). In an attempt to understand whether a fullerene cone model can account for the observed variation in authentic viral cores, we used a molecular modeling program to build three different idealized models for the HIV cone.
From Euler's theorem, only five conical hexagonal lattices are allowed, which correspond to cones with mean angles of 19.2°, 38.9°, 60°, 83.6°, and 112.9° (for reviews, see references 15 and 41). Structures corresponding to the narrowest of the allowed cone angles predominate in both synthetic and actual HIV-1 capsids (2, 8, 21, 54). Therefore, the three different idealized models for HIV-1 capsids were built with conical hexagonal lattices with 19.2° cone angles. The cones were closed by incorporating seven pentamers into the wide end and five pentamers into the narrow end, and the precise positions of the pentamers were varied between the different models (21). The models were built with 1,000 to 1,500 CA molecules/capsid (assuming each hexamer represents six CA subunits and each pentamer represents five CA subunits), which was judged to be a reasonable approximation for the viral core, as recent reports have suggested that 4,000 to 5,000 copies of Gag are present in each virion (V. Vogt, personal communication), and multiple conical capsids are often seen in single virions (8).
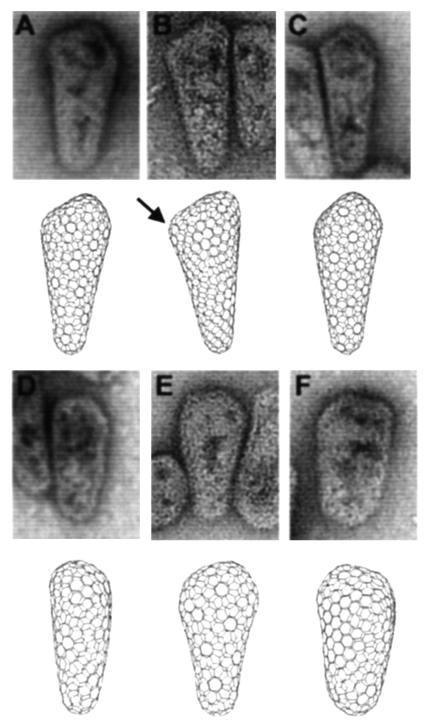
Six different views of these three idealized cones are shown in Fig. 5, and these images illustrate how the apparent shape of the capsid can be altered by changing the pentamer placements and viewing orientations. This is because (i) the inherent asymmetry of the lattice means that cones have different shapes when viewed from different orientations and (ii) introducing pentamers into different positions in the hexameric lattice causes one cone to differ from another. As shown in Fig. 5, EM images of bona fide HIV-1 capsids also often show significant variations in capsid shape (e.g., rounded caps versus pointed caps with abrupt edges and angles, differences in apparent cone angles, etc.) (54). Similar variations were also apparent in our computer models when they were viewed from different angles or constructed with different pentamer distributions. For example, clustering of the pentameric defects within the lattice increases the local declination and causes the capsid to appear more pointed. This principle is nicely illustrated by the model in Fig. 5B, in which an acute cap angle (marked by an arrow) arose because two pentamers were placed in adjacent positions. Conversely, more dispersion of the pentameric defects resulted in rounder caps. Similarly, the observed cone morphology and apparent cone angle can also vary dramatically depending on the orientation from which a single model is viewed. This can be seen by comparing the models in Fig. 5D and E, which show two different views of the same cone. Overall, we conclude that the size and shape variations seen for authentic HIV-1 capsids can be mimicked in idealized cones, providing strong support for the idea that lentiviral capsids are assembled on the principles of fullerene cones (21).
FIG. 5.
Comparisons of bona fide HIV-1 capsids with idealized models. Upper panels show negatively stained EM images of HIV-1 cores purified from detergent-treated virions. Lower panels show computer-generated images of idealized conical capsids assembled from mixtures of hexamers and pentamers. The location of a single pentamer in the cone shown in panel B is highlighted by an arrow to illustrate how pentameric defects give rise to sites of declination in the hexagonal lattice. The six images represent different views of three different computer-generated conical capsids, each of which has five pentamers capping the narrow end and seven pentamers capping the wide end.
Conclusions.
Our studies have defined two CA surfaces that are important for the in vitro assembly of CA cylinders and, by extension, the mature HIV-1 capsid. Specifically, we have shown that helices 1 and 2 in the NTD and the crystallographic CTD dimer interface are required for efficient CA assembly. Our data also allow for the possibility that there is a third CA-CA interface between the NTD and the CTD (32), as a single mutation (K70A) in helix 4 also diminished CA assembly. These data are generally in good agreement with deuterium exchange mapping experiments, which implicate these same surfaces in homo-oligomeric interactions in the CA cylinders (32). Mutations in these interfaces also reduced viral infectivity, supporting the idea that in vitro CA cylinder assembly accurately mimics some (though clearly not all) aspects of mature capsid assembly (53).
We have also demonstrated that a single point mutation, R18A, allows the HIV-1 CA protein to assemble into cylinders, spheres, and cones in the same reaction mixture. As these structures match the three known retroviral capsid morphologies, this observation provides support for the idea that all retroviral capsids can be generated by using similar underlying lattice structures.
Finally, we have used molecular modeling experiments to demonstrate that computer models built with hexamer-pentamer lattices can mimic the considerable shape variation observed for actual HIV-1 capsids, which emphasizes the fact that although these objects are assembled on hexagonal nets using repeating protein-protein interfaces, they are nevertheless fundamentally asymmetric structures.
Acknowledgments
We thank Alex Kotov and Paula Flicker for HIV-1 core preparation and imaging, Donovan Anderson for preparing and testing assembly of the CA K70A mutant protein, and Nancy Chandler and Kurt Albertine for electron microscopy support in Utah.
This work was funded by an NIH grant to W.I.S. and an NIH predoctoral training grant fellowship to B.K.G.-P.
REFERENCES
- 1.Aberham, C., S. Weber, and W. Phares. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 70:3536-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola, M. A., A. Ohagen, and H. G. Göttlinger. 2000. Isolation of human immunodeficiency virus type 1 cores: retention of Vpr in the absence of p6Gag. J. Virol. 74:6198-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2002. GenBank. Nucleic Acids Res. 30:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowzard, J. B., J. W. Wills, and R. C. Craven. 2001. Second-site suppressors of Rous sarcoma virus Ca mutations: evidence for interdomain interactions. J. Virol. 75:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs, J. A., T. Wilk, R. Welker, H. G. Krausslich, and S. D. Fuller. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campos-Olivas, R., J. L. Newman, and M. F. Summers. 2000. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol. 296:633-649. [DOI] [PubMed] [Google Scholar]
- 10.Campos-Olivas, R., and M. F. Summers. 1999. Backbone dynamics of the N-terminal domain of the HIV-1 capsid protein and comparison with the G94D mutant conferring cyclosporin resistance/dependence. Biochemistry 38:10262-10271. [DOI] [PubMed] [Google Scholar]
- 11.Choi, J., and L. S. Loesch-Fries. 1999. Effect of C-terminal mutations of alfalfa mosaic virus coat protein on dimer formation and assembly in vitro. Virology 260:182-189. [DOI] [PubMed] [Google Scholar]
- 12.Cornilescu, C. C., F. Bouamr, X. Yao, C. Carter, and N. Tjandra. 2001. Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein. J. Mol. Biol. 306:783-797. [DOI] [PubMed] [Google Scholar]
- 13.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earnshaw, W., and J. King. 1978. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J. Mol. Biol. 126:721-747. [DOI] [PubMed] [Google Scholar]
- 15.Ebbesen, T. 1998. Cones and tubes: geometry in the chemistry of carbon. Acct. Chem. Res. 31:558-566. [Google Scholar]
- 16.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 18.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 19.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 20.Ganser, B. K., A. Cheng, W. I. Sundquist, and M. Yeager. 2003. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J. 22:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 22.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 23.Gottlinger, H. G. 2001. The HIV-1 assembly machine. AIDS 15:S13-S20. [DOI] [PubMed] [Google Scholar]
- 24.Grattinger, M., H. Hohenberg, D. Thomas, T. Wilk, B. Muller, and H. G. Krausslich. 1999. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology 257:247-260. [DOI] [PubMed] [Google Scholar]
- 25.Groß, I., H. Hohenberg, and H. G. Krausslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 26.Jin, Z., L. Jin, D. L. Peterson, and C. L. Lawson. 1999. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J. Mol. Biol. 286:83-93. [DOI] [PubMed] [Google Scholar]
- 27.Kemp, C. L., A. F. Howatson, and L. Siminovitch. 1968. Electron microscopy studies of mutants of lambda bacteriophage. I. General description and quantitation of viral products. Virology 36:490-502. [DOI] [PubMed] [Google Scholar]
- 28.Khorasanizadeh, S., R. Campos-Olivas, and M. F. Summers. 1999. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 291:491-505. [DOI] [PubMed] [Google Scholar]
- 29.Kingston, R. L., T. Fitzon-Ostendorp, E. Z. Eisenmesser, G. W. Schatz, V. M. Vogt, C. B. Post, and M. G. Rossmann. 2000. Structure and self-association of the Rous sarcoma virus capsid protein. Struct. Fold Des. 8:617-628. [DOI] [PubMed] [Google Scholar]
- 30.Kotov, A., J. Zhou, P. Flicker, and C. Aiken. 1999. Association of Nef with the human immunodeficiency virus type 1 core. J. Virol. 73:8824-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krausslich, H. G. (ed.). 1996. Morphogenesis and maturation of retroviruses. Springer-Verlag, Berlin, Germany.
- 32.Lanman, J., T. T. Lam, S. Barnes, M. Sakalian, M. R. Emmett, A. G. Marshall, and P. E. Prevelige. 2003. Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J. Mol. Biol. 325:759-772. [DOI] [PubMed] [Google Scholar]
- 33.Lanman, J., J. Sexton, M. Sakalian, and P. E. Prevelige, Jr. 2002. Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 76:6900-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, M., M. K. Lyon, and R. L. Garcea. 1995. In vitro phosphorylation of the polyomavirus major capsid protein VP1 on serine 66 by casein kinase II. J. Biol. Chem. 270:26006-26011. [DOI] [PubMed] [Google Scholar]
- 35.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 36.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 37.Marvik, O. J., T. Dokland, R. H. Nokling, E. Jacobsen, T. Larsen, and B. H. Lindqvist. 1995. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J. Mol. Biol. 251:59-75. [DOI] [PubMed] [Google Scholar]
- 38.Mayo, K., D. Huseby, J. McDermott, B. Arvidson, L. Finlay, and E. Barklis. 2003. Retrovirus capsid protein assembly arrangements. J. Mol. Biol. 325:225-237. [DOI] [PubMed] [Google Scholar]
- 39.Mayo, K., M. L. Vana, J. McDermott, D. Huseby, J. Leis, and E. Barklis. 2002. Analysis of Rous sarcoma virus capsid protein variants assembled on lipid monolayers. J. Mol. Biol. 316:667-678. [DOI] [PubMed] [Google Scholar]
- 40.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 41.Moody, M. F. 1999. Geometry of phage head construction. J. Mol. Biol. 293:401-433. [DOI] [PubMed] [Google Scholar]
- 42.Picard, V., E. Ersdal-Badju, A. Lu, and S. C. Bock. 1994. A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 22:2587-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose, S., P. Hensley, D. J. O'Shannessy, J. Culp, C. Debouck, and I. Chaiken. 1992. Characterization of HIV-1 p24 self-association using analytical affinity chromatography. Proteins 13:112-119. [DOI] [PubMed] [Google Scholar]
- 45.Salunke, D. M., D. L. Caspar, and R. L. Garcea. 1989. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys. J. 56:887-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, C., E. Loeliger, I. Kinde, S. Kyere, K. Mayo, E. Barklis, Y. Sun, M. Huang, and M. F. Summers. 2003. Antiviral inhibition of the HIV-1 capsid protein. J. Mol. Biol. 327:1013-1020. [DOI] [PubMed] [Google Scholar]
- 47.Tang, C., Y. Ndassa, and M. F. Summers. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537-543. [DOI] [PubMed] [Google Scholar]
- 48.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 49.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 50.van Vloten-Doting, L., R. I. Francki, R. W. Fulton, J. M. Kaper, and L. C. Lane. 1981. Tricornaviridae—a proposed family of plant viruses with tripartite, single-stranded RNA genomes. Intervirology 15:198-203. [DOI] [PubMed] [Google Scholar]
- 51.Vogt, V. M. 1997. Retroviral virions and genomes, p. 27-70. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Press, Plainview, N.Y. [PubMed]
- 52.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Welker, R., H. Hohenberg, U. Tessmer, C. Huckhagel, and H. G. Krausslich. 2000. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 74:1168-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wheeler, D. L., C. Chappey, A. E. Lash, D. D. Leipe, T. L. Madden, G. D. Schuler, T. A. Tatusova, and B. A. Rapp. 2000. Database resources of the National Center for Bio/Technology Information. Nucleic Acids Res. 28:10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worthylake, D. K., H. Wang, S. Yoo, W. I. Sundquist, and C. P. Hill. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr. D 55:85-92. [DOI] [PubMed] [Google Scholar]
- 57.Yin, L., D. Braaten, and J. Luban. 1998. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J. Virol. 72:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]