Abstract
We asked whether specific mesenchymal/epithelial (M/E) induction generates olfactory receptor neurons (ORNs), vomeronasal neurons (VRNs) and gonadotropin releasing hormone (GnRH) neurons—the major neuron classes associated with the olfactory epithelium (OE). To assess specificity of M/E-mediated neurogenesis, we compared the influence of frontonasal mesenchyme on frontonasal epithelium, which becomes the OE, with that of the forelimb bud. Despite differences in position, morphogenetic and cytogenic capacity, both mesenchymal tissues support neurogenesis, expression of several signaling molecules and neurogenic transcription factors in the frontonasal epithelium. Only frontonasal mesenchyme, however, supports OE-specific patterning and activity of a subset of signals and factors associated with OE differentiation. Moreover, only appropriate pairing of frontonasal epithelial and mesenchymal partners yields ORNs, VRNs, and GnRH neurons. Accordingly, the position and molecular identity of specialized frontonasal epithelia and mesenchyme early in gestation and subsequent inductive interactions, specifies the genesis and differentiation of peripheral chemosensory and neuroendocrine neurons.
Keywords: induction, olfactory, neurogenesis, limb bud
INTRODUCTION
The mesenchymal/epithelial (M/E) interactions that influence initial morphogenesis and differentiation of the olfactory epithelium (OE) in mammals are similar to those seen in the branchial arches, aortic arches and limb buds (LaMantia et al, 2000; Balmer & LaMantia, 2005; Bhattachyra & Bronner-Fraser, 2008). Induction at each site is accompanied by focal accumulation of mesenchymal cells in register with distinct epithelial placodal thickenings. Nevertheless, the genesis of peripheral chemosensory and neuroendocrine neurons from the frontonasal epithelium distinguishes the frontonasal mass from M/E other sites. Our previous results indicate that neurogenesis does not occur in the frontonasal epithelium in the absence of M/E interactions (LaMantia et al, 2000). Thus, we asked whether the capacity of frontonasal epithelium and mesenchyme to generate chemoreceptive olfactory receptor neurons (ORNs; Buck, 1996; Axel, 2005), phermone responsive vomeronasal neurons (VRNs; Dulac, 2000) and gonadotropin releasing hormone neurons (GnRN neurons; Wray et al, 1989; Schwanzel-Fukuda et al, 1989) that migrate from the OE to the hypothalamus to regulate reproduction reflects olfactory-specific frontonasal M/E inductive interactions.
Appropriate OE morphogenesis and cellular differentiation depend upon expression and activity of local signals that mediate M/E induction. These include: retinoic acid (RA; LaMantia et al, 1993; Anchan et al, 1997; Whitesides et al, 1998), sonic hedgehog (Shh; Balmer & LaMantia, 2004), bone morphogenetic proteins (especially Bmp4; Shou et al, 2000; LaMantia et al, 2000) and fibroblast growth factors (especially Fgf8; LaMantia et al, 2000; Bhasin et al, 2003; Kawauchi et al, 2005). These signals are also essential for M/E regulation of branchial arch, heart, and limb development. Mutation of genes that regulate each signaling pathway results in phenotypes at all sites, including the OE (reviewed by Balmer & LaMantia, 2005). Recent evidence suggests that the olfactory placodal ectoderm is specified within a broader cranial placodal domain quite early in development (Bhattacharyya & Bronner-Fraser, 2008). The nearly complete derivation of the frontonasal mesenchyme from the neural crest indicates that this mesenchyme may have distinct inductive capacity (Yoshida et al, 2008). Furthermore, embryological manipulation of epithelia and mesenchyme in the head suggests that local morphogenesis is largely determined by the mesenchyme (Richman & Tickle, 1989). Nevertheless, expression of key signals for OE differentiation, including Fgf8 and Bmp4, rely upon M/E interactions that parallel those at multiple non-axial sites of M/E apposition including the limb bud (Bhasin et al, 2003). Thus we asked whether frontonasal versus limb mesenchyme provides generic or specific support for expression and activity of shared signals as well as target transcription factors that facilitate musculo-skeletal and epidermal differentiation at each site or OE neurogenesis (Calof et al, 2002; Mangalpus et al, 2004; Balmer & LaMantia, 2005).
We devised an in vitro variation of a classical embryological experimental approach to exchange tissues from the embryonic mouse frontonasal mass and other sites of non-axial M/E induction and evaluate specificity or flexibility of OE morphogenesis, patterning, and neuronal differentiation. We focused on comparison of frontonasal and forelimb M/E interactions because—despite parallels during early development—they represent clearly distinct endpoints for morphogenesis and cellular differentiation. To fully assess neuronal identity in the OE, we determined molecular, cellular and physiological properties of neurons generated from frontonasal epithelium induced either by frontonasal or limb mesenchyme. We found that a significant amount of molecular, cellular, and neuronal differentiation in the OE can be supported by heterologous (i.e. limb) as well as homologous (i.e. frontonasal) M/E interactions. Nevertheless, only frontonasal M/E induction supports OE-characteristic patterning of a subset of local signaling molecules and transcription factors, as well as genesis and differentiation of ORNs, VRNs and GnRH neurons.
RESULTS
Frontonasal and forelimb mesenchyme support neuronal differentiation in the presumptive OE
At E9.0 in the mouse, there is little evidence of frontonasal epithelial thickening that defines the olfactory placode at slightly later stages, and there is no detectable neuronal differentiation (Figure 1A, upper box). Similarly, the forelimb bud is a slender, undifferentiated protuberance from the mid-trunk (Figure 1A, lower box). Nevertheless, each structure can be identified, microdissected and manipulated. We isolated frontonasal masses (Fn) and forelimb buds (Lb) from individual E9.0 embryos and separated the mesenchymal (M) and epithelial (E) compartments. These tissues were then recombined homologously (frontonasal epithelium with frontonasal mesenchyme: Fn:E/Fn:M; limb bud epithelium with limb bud mesenchyme: Lb:E/Lb:M) or heterologously (Fn:E/Lb:M; Lb:E/Fn:M), and grown for 48 hours in vitro, the time necessary for significant patterning and differentiation to occur in OE and limb explants based upon our previous observations (LaMantia et al, 2000; Bhasin et al, 2003).
Figure 1.
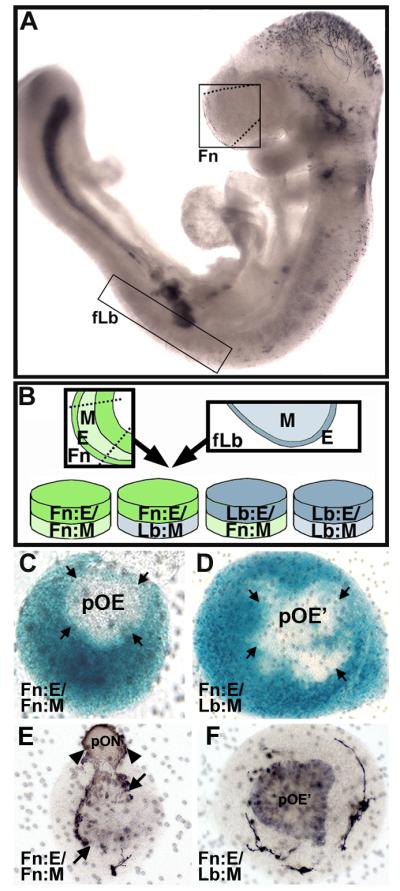
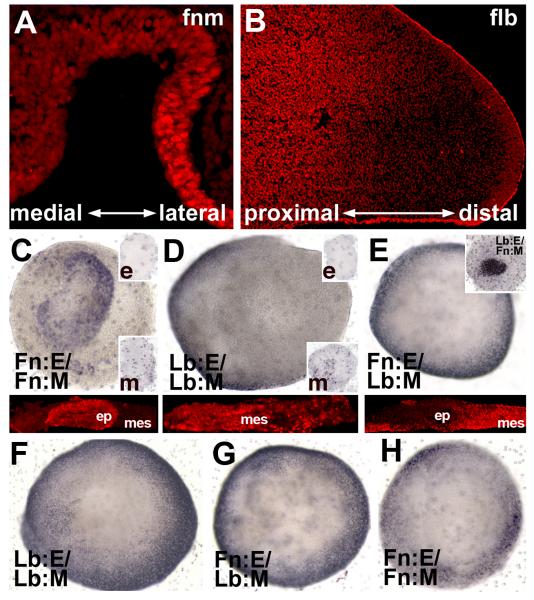
M/E induction supports neuronal differentiation. A) An E9.0 embryo, labeled for β-tubulin, a marker for early differentiating neurons, showing sites of initial neuronal differentiation including the hindbrain (hb), spinal cord (sc), telencephalon, and frontonasal region (Fn; box), but not the forelimb bud (Lb; box). B) The schematic shows in vitro “swapping” of frontonasal (Fn) and forelimb bud (Lb) epithelium (E) and mesenchyme (M). C,D) There is no cell migration between mesenchyme and presumed olfactory epithelium (pOE) in Fn:E/Fn:M and Fn:E/Lb:M explants. ROSA26/β-galactosidase (β-gal) labels mesenchyme blue, and wild type epithelium is unlabeled. E) In Fn:E/Fn:M explants of E9.0 tissue cultured for 48 hours, β-tubulin labeled cells are seen in the presumed medial OE (labeled cells left of arrows) and their processes (arrowheads) form a coherent nerve that corresponds, presumably, to the olfactory nerve in vivo (pON). F) In Fn:E/Lb:M explants, β-tubulin labels cells are seen throughout the entire placodal region (pOE), and their apparent processes surround the explant periphery.
Both Fn:E/Fn:M and Fn:E/Lb:M explants have an extensive epithelial placodal thickening by the 2nd day in vitro (Figure 1C,D). In both cases there is no mixing of epithelial and mesenchymal cells based upon recombination of M or E from indicator (ROSA26) and wild type embryos (Figure 1C,D). Neuronal differentiation occurs in the Fn:E of both Fn:E/Fn:M and Fn:E/Lb:M explants (Figure1E,F). In homologous Fn:E/Fn:M explants, however, neurons are concentrated in the presumed medial aspect of the explant and neurites form a coherent nerve as described previously (LaMantia et al, 2000). The position of these nascent OE neurons parallels that in the OE in vivo around E11.5, when the first presumed ORNs are seen in the medial OE (Whitesides & LaMantia, 1996; LaMantia et al, 2000). This organization is not seen when Fn:E is induced by Lb:M; instead neurons are seen throughout the placodal domain, with no apparent discontinuities in their distribution. Similar patterns are seen in homologous versus heterologous explants labeled for NCAM and GAP43 (data not shown), which also recognize early OE neurons (Calof & Chikaraishi, 1989; Verhaagen et al, 1989). Thus, Fn:M and Lb:M support placodal morphogenesis and neurogenesis in Fn:E; however, only Fn:M establishes the distribution of neurons and formation of an apparent nerve associated with OE differentiation at similar stages of development (E11.5) in vivo.
Limb mesenchyme has limited intrinsic capacity to support neurogenesis
These recombination experiments indicate that E9.0 Lb:M has previously unrecognized potential to support neurogenesis. We therefore asked whether this capacity reflects an intrinsic capacity of the Lb:M to support transient neural differentiation in Lb:E in vivo and in vitro, or a property acquired due to signals from the frontonasal epithelium. We first evaluated markers of neurogenesis in the apical ectodermal ridge (AER), the site of Fgf8 expression (Figure 2A) which resembles the initial OE placodal thickening of the frontonasal mass. β-tubulin, a marker of initial neuronal differentiation (Figure 2B), as well as Sox2 and NCAM (data not shown) are expressed transiently (from E9.5 to E11) in the AER, in register with Fgf8, similar to the registration of Fgf8 and neurogenic regions in the medial OE at similar stages (LaMantia et al, 2000; Kawauchi et al, 2005). This transient neurogenesis and its relationship with local Fgf8 expression is maintained in an AER-like structure in vitro when Lb:M is cultured with Lb:E (Figure 2C,D). Initial transient neurogenesis may be partially due to some intrinsic neurogenic capacity in the Lb:E; β-tubulin labeled cells are also seen in Lb:E/Fn:M explants (inset, Figure 2D). Neurogenesis in the limb epithelium, however, relys upon mesenchymal signals; we do not find neurons in Lb:E cultured in isolation (not shown). Thus, the initial capacity of Lb:M to support OE neuronal differentiation may reflect some intrinsic capacity for maintenance of Fgf8 expression and transient neurogenesis in the ectoderm of the AER.
Figure 2.
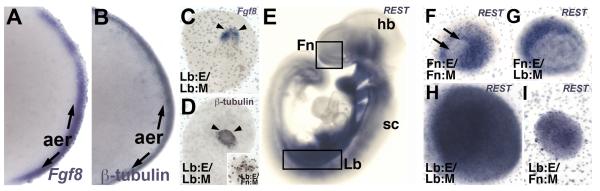
Intrinsic and induced neural inductive capacities of limb mesenchyme. A) Fgf8 and B) β-tubulin are expressed in the apical ectodermal ridge (aer, arrows) of the E 11.5 forelimb bud. C) Fgf8 and D) β-tubulin are maintained in a more limited aer-like region (arrowheads) in Lb:E/Lb:M explants. D) Inset: β-tubulin is expressed in Lb:E/Fn:M explants. E) By E10.5 (and thereafter), the neural gene silencing transcription factor REST is down-regulated at sites of neurogenesis throughout the CNS including the hindbrain (hb) and spinal cord (sc). There is diminished expression in the frontonasal mass (Fn, box), but not in the forelimb bud (Lb, box). F) In Fn:E/Fn:M explants, REST is down-regulated in the medial OE (arrows). G) In Fn:E/Lb:M explants, REST is down-regulated in the placodalized epithelium. H) In Lb:E/Lb:M and I) Lb:E/Fn:M explants, REST is maintained.
An additional hallmark of initial neuronal differentiation—including establishing a permissive transcriptional state for expression of neuronal voltage gated Na+ channels and other neurogenic genes—is local down-regulation of the neuronal gene silencing transcription factor REST (Chong et al, 1995; Schoenherr & Anderson, 1995). By E10.5-11.0 in the mouse, significant down-regulation of REST is seen throughout the central nervous system, in peripheral ganglia, and in the frontonasal epithelium that becomes the OE (Figure 2E, upper box). In contrast, there is no apparent down-regulation in the forelimb bud in vivo (Figure 2E, lower box). In Fn:E/Fn:M pairings, REST expression is substantially diminished in the presumed medial aspect of the explant, likely in register with neuronal differentiation (compare Figure 2F to Figure 1F). In Fn:E/Lb:M pairings, REST is down-regulated in a broader region of the placodal domain with no detectable pattern (Figure 2G). Such diminished expression is not seen when Fn:E is cultured without mesenchyme (data not shown). In Lb:E/Lb:M and Lb:E/Fn:M pairings, REST does not appear to be down-regulated in either epithelium or mesenchyme (Figure 2H,I). Thus, M/E interactions are necessary for significant down-regulation of REST, and this mesenchymal capacity apparently relies upon FnE signals that can modify heterologous mesenchyme like that from the limb bud.
Frontonasal M/E interactions support distinct inductive signaling
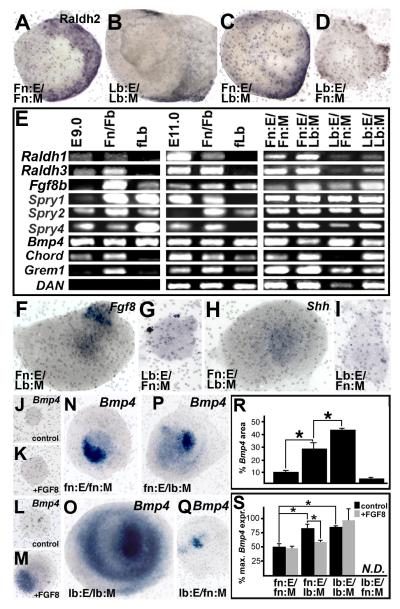
Shared inductive signals: RA, Fgf8, Bmp4, and Shh, are associated with morphogenetic axes that define M/E inductive mechanisms in the frontonasal mass and limb bud (reviewed by Balmer & LaMantia, 2005). Thus, we asked whether local M/E interactions establish and maintain region-specific expression and activity of these signals. The RA synthetic enzyme Raldh2, associated with local RA sources in the frontonasal mass and limb bud (reviewed by Rawson & LaMantia, 2006), is maintained in somewhat different patterns in Fn:E/FnM versus Lb:E/Lb:M explants (Figure 3A,B). When Fn:E is recombined with Lb:M, Raldh2 expression resembles that in the limb bud, rather than the frontonasal mass; Raldh2 expression is diminished in extent, and is more diffuse (Figure 3C,D). In the presence of Lb:E, Raldh2 expression in the Fn:M is greatly attenuated or absent (Figure 3D). Two additional RA synthetic enzymes, Raldh1 and Raldh3 are expressed robustly in the frontonasal mass (see also Bhasin et al, 2003), but hardly detected in the limb bud at E9 or E11 (Figure 3E). Expression of these two genes relies upon the presence of Fn:E; expression in Lb:E/Fn:M and Lb:E/Lb:M explants is diminished or absent, but maintained in Fn:E/Fn:M and Fn:E/Lb:M explants (Figure 3E). Similarly, homologous and heterologous explants maintain Fgf8 and Bmp4 expression, as well as local mediators of Fgf and Bmp signaling (Spry1, 2, and 4; Chord1, Grem, and Dan, respectively; Figure 3E). When Fn:E is cultured with Lb:M, Fgf8 remains expressed in the epithelium (Figure 3F); however Lb:E/Fn:M interactions do not support detectable expression (Figure 3G). Shh is maintained in Fn:E by Lb:M (Figure 3H), but not detected in Lb:E induced by Fn:M. Apparently, Fn:M is limited in its capacity to support Fgf8 and Shh expression beyond the context of Fn:E/Fn:M interactions. Despite modest variations, there are no absolute differences in the capacity of limb mesenchyme to establish or maintain expression of RA synthetic enzymes, Fgf8, Shh, Bmp4 or several related signals in the presence of Fn:E versus Lb:E.
Figure 3.
Distinct regulation of M/E inductive signals in frontonasal mass versus forelimb bud. A) In Fn:E/Fn:M explants, Raldh2, detected immunohistochemically, is seen throughout the presumed lateral frontonasal mesenchyme, as it is in vivo. B) In Lb:E/Lb:M, Raldh2 is limited to the presumed proximal mesenchyme, enhanced at the presumed dorsal and ventral margins (arrows). C) The pattern of Raldh2 in Fn:E/Lb:M explants resembles more closely that in Lb:E/Lb:M explants. D) Very little Raldh2 is detected in Lb:E/Fn:M explants. E) RT-PCR expression analysis of additional M/E signaling molecules and associated regulators in E9.0 and E11.0 embryos, as well as microdissected frontonasal mass/forebrain (Fn/Fb) and forelimb bud (fLB). Expression of the same molecules is evaluated in the four different M/E explant combinations. F) Epithelial expression of Fgf8, detected by in situ hybridization, is retained in Fn:E/Lb:M explants. G) Little specific labeling for Fgf8 is seen in Lb:E/Fn:M explants. H) Shh is maintained in the Fn:E in the presence of Lb:M. I) Shh is not detected in Lb:E/Fn:M explants. J) Bmp4 is not expressed in isolated frontonasal mesenchyme. K) Fgf8b does not restore Bmp4 expression in isolated frontonasal mesenchyme. L) Bmp4 is not expressed in isolated forelimb bud mesenchyme. M) Fgf8b restores Bmp4 expression in isolated limb bud mesenchyme. N) Frontonasal mesenchymal expression of Bmp4 in Fn:E/Fn:M explants. O) Bmp4 expression pattern in Lb:E/Lb:M explants. P) In Fn:E/Lb:M explants, Bmp4 expression domain appears larger and more diffuse than in Fn:E/Fn:M explants. Q) Minimal Bmp4 expression in Lb:E/Fn:M explants. R) Quantification of Bmp4 expression area. Asterisks indicate significant differences (p≤0.01, Mann-Whitney). S) qPCR measurement of Bmp4 mRNA in explants (n=3 pooled sets) and in response to exogenous Fgf8 (n=4 pooled sets). Brackets and asterisks indicate significant differences (Fn:E/Fn:M:FnE/Lb:M, p≤0.025; Fn:E/Fn:M:Lb:E/Lb:M, p≤0.005; Fn:E/Lb:M Fn:E/Lb:M+Fgf8b, p≤0.027, T-tests).
Multiple regulatory relationships are thought to exist between local M/E signals, and we asked whether such relationships might be specific to frontonasal M/E inductive interactions. We evaluated the capacity of Fgf8 to influence Bmp4 expression or activity, since this regulatory interaction is thought to modulate neural induction generally in neurogenic epithelia (Marchal et al, 2009), as well as digit formation in limb morphogenesis, perhaps via gremlin antagonism (Verheyden & Sun, 2008). In the frontonasal mass, Bmp4 is not expressed in mesenchyme, nor does Fgf8, normally available from the Fn:E, influence Bmp4 expression in Fn:M (Figure 3J, K; n=3/3 pairs analyzed). In the limb, however, Fgfs, including Fgf8, are known to regulate mesenchymal Bmp4 expression (Buckland et al, 1998). Accordingly, Fgf8 alone, independent of Lb:E, maintains Bmp4 in isolated Lb:M (Figure 3L, M; n=3/3 pairs analyzed). The patterning of Bmp4, as well as its local regulation by Fgf8, is specified by limb versus frontonasal M/E interactions. Patterns (Figure 3N-Q) and significant differences in area of Bmp4 expression in homologous and heterologous explants reflect limb versus frontonasal epithelial and mesenchymal identity (Figure 3R; p≤0.01, Mann-Whitney, for each comparison, n=at least 5 explants/combination), as does the regulatory influence of additional signals. When Lb:M is paired with Fn:E, Fgf8 diminishes rather than enhances or maintains Bmp4 expression (Figure 3S). Thus, frontonasal M/E interactions establish specific activity and expression levels of local signals including Fgf8 and Bmp4 during initial OE differentiation.
Regulation of OE-associated transcription factors by frontonasal M/E interaction
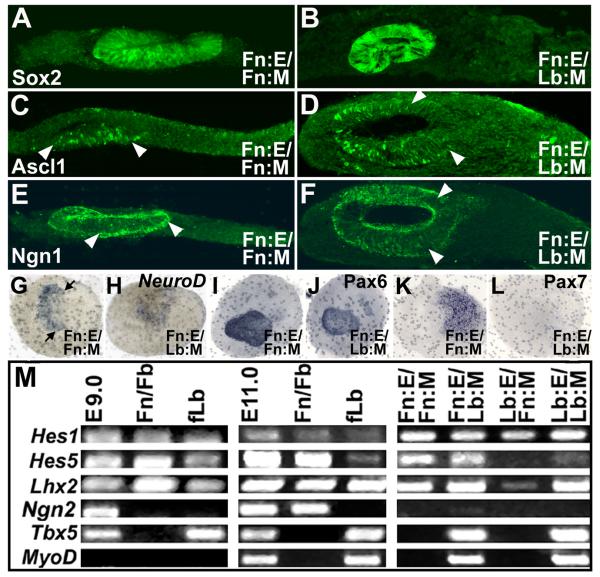
Neuronal differentiation in the OE relies upon specific transcriptional regulation (reviewed by Nicolay et al, 2006) as well as local signals. Thus, we asked whether multiple transcription factors associated with OE neurogenesis are specifically induced or maintained by frontonasal M/E interactions. Sox2 (Figure 4A,B), as well as Ascl1 (Figure 4C,D), Ngn1 (Figure 4E,F), and NeuroD (Figure 4G,H), are thought to be hierachically expressed in the OE (Cau et al, 1997; Kawauchi et al, 2005). Sox2, Ascl1 and Ngn1 are expressed only in the invaginated epithelium in homologous as well as heterologous M/E explants. Ascl1 and Ngn1 labeling in the heterologously induced epithelia seemed to expand (compare arrowheads in Figure 4C,D or 4E,F) suggesting that the pattern of these neurogenic bHLH genes associated with neurogenesis in the medial OE might be altered by heterologous induction (see also Figures 5, 6). Similarly, NeuroD, another neurogenic bHLH transcription factor associated with Ascl1 and Ngn1 in the developing OE (Cau et al, 1997) is patterned in the presumed medial aspect of whole Fn:E/Fn:M explants (Figure 4G, arrows), but seen throughout the entire placodal domain in Fn:E/Lb:M explants (Figure 4H). Pax6, which is associated with the placodal epithelium during initial morphogenesis and differentiating non-neural cells thereafter (Grindley et al, 1995; Bhattachyra & Bronner-Fraser, 2008) is maintained throughout the placodal domain of the Fn:E by either Fn:M or Lb:M (Figure 4I,J). In contrast, one well-established Fn:M selective transcription factor, Pax7 (Mansouri et al, 1998) is only expressed when Fn:E is apposed to Fn:M (Figure 4K,L; see also LaMantia et al, 2000). We evaluated M/E maintenance of expression of several other OE associated transcription factors. Hes1, which is widely expressed in vivo, is maintained non-specifically (Figure 4M). Hes5, thought to be regulated by Ascl1 and related bHLH genes in the OE (Cau et al, 2002) is maintained in Fn:E regardless of mesenchyme identity; however, Fn:M does not support Lb:E expression, nor is there substantial Hes5 in the fLB in vivo, or in recombined Lb:E/Lb:M explants. Lhx2, which is associated with OE neuronal differentiation (Kolterud et al, 2004), as well as limb patterning and morphogenesis (Tzchori et al, 2009), is maintained regardless of M/E combination. In contrast, Ngn2, a bHLH factor, excluded from the OE, absent from the limb buds, but expressed in the forebrain, is not anomalously induced by any frontonasal or limb M/E combination. Similarly, transcription factors seen primarily in the limb including Tbx5 and MyoD are seen only when limb mesenchyme, a site of mesoderm-derived myogenic precursors, is present—regardless of the epithelium. Thus, despite the maintenance of some aspects of Lb:M identity, the expression of several transcription factors associated with OE neurogenesis in the frontonasal epithelium can be supported by both Fn:M and Lb:M.
Figure 4.
Differential M/E regulation of OE-associated transcription factors. Expression of Sox2, Ascl1, and Ngn1, detected immunohistochemically in sectioned Fn:E/Fn:M (A, C, E) and Fn:E/Lb:M (B, D, F) explants. Arrowheads in (C),(D),(E), and (F) indicate differences in extent of labeled cells in homologous versus heterologous explants. G, H) NeuroD visualized using in situ hybridization in Fn:E/Fn:M and Fn:E/Lb:M explants. Arrows in (G) indicate the enhanced labeling in the presumed medial aspect of the explant. I, J) Pax6 is detected immunohistochemically in whole Fn:E/Fn:M and Fn:E/Lb:M explants. K,L) Pax7, labeled immunohistochemically, is detected in Fn:E/Fn:M explants, but not in Fn:E/Lb:M explants. M) RT-PCR detection of Notch related transcription factors Hes1 and Hes5, OE-selective Lhx2, forebrain-selective Ngn2, and limb-selective Tbx5 and MyoD in whole E9.0 and 11.0 embryos, dissected frontonasal mass/forebrain (Fn/Fb) and dissected forelimb bud (fLb) from E9.0 and 11.0 embryos.
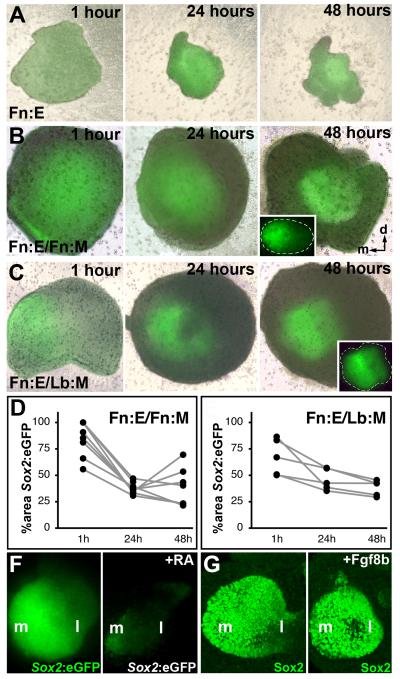
Figure 5: Patterning of Sox2 in Fn:E by Fn:M and Lb:M. Individual living explants from donor embryos carrying a Sox2:eGFP allele with or without +/+ mesenchyme imaged successively at 1, 24, and 48 hours after dissection. Fluorescence images for each explant have been superimposed upon with bright-field phase images. A) Isolated Fn:E has diffuse expression of Sox2:eGFP, that is maintained, with some increase in intensity and some local restriction, over the 48 hour culture period. B) Sox2:eGFP expression is progressively limited to a placodal domain and patterned in a presumed medial lateral gradient in Fn:E/Fn:M explants over 48 hours in vitro. A single explant is shown, imaged at 1, 24, and 48 hours after dissection. C) Sox2:eGFP expression is progressively limited to a placodal domain in Fn:E/Lb:M explants. Although present, the Sox2 gradient does not appear as distinct when Fn:E is induced by Lb:M. D) Sox2:eGFP signal area as a percentage of total explant area. There is an equivalent, linear, statistically significant decline in area of Sox2 signal in both explant pairings (p≤0.009, n=9, Fn:E/Fn:M; p<0.01, n=5, Fn:E/Lb:M; ANOVA). F) Retinoic Acid (RA) suppresses Sox2:eGFP expression in Fn:E/Fn:M explants. G) Fgf8b expands Sox2 expression (detected immunohistochemically) in Fn:E/Fn:M explants.
Figure 6.
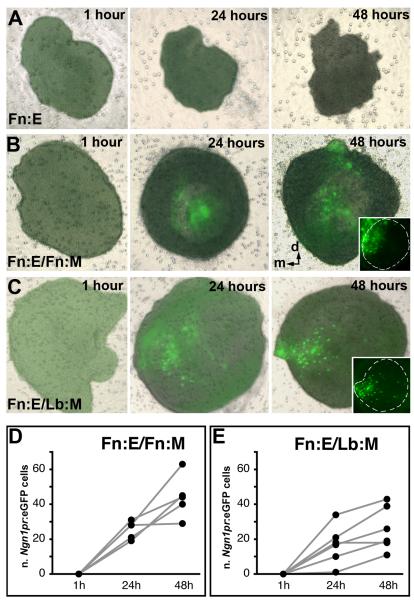
Induction and patterning of Ngn1 in Fn:E by Fn:M and Lb:M. Fb:E is dissected from Ngn1prTg:eGFP E9.0 embryos, and recombined with Fn:M or Lb:M from +/+ E9.0 embryos. Explants have been imaged at 1, 24, and 48 hours after dissection. A) A single Fn:E explant has been imaged at 1, 24 and 48 hours. Ngn1prTg:eGFP is not expressed in Fn:E in the absence of mesenchyme over the 48 hour culture period. B) A single Fn:E/Fn:M explant has been imaged at 1, 24, and 48 hours. Ngn1prTg:eGFP is induced and expressed in Fn:E cells concentrated in the apparent medial aspect of the explant over 48 hours in vitro. The inset at right shows the dense accumulation of Ngn1prTg:eGFP cells in the presumed medial aspect of the explant. C) A single Fn:E/Lb:M explant has been imaged at 1, 24, and 48 hours. Lb:M also induces Ngn1prTg:eGFP expression in cells within the Fn:E. The inset at right shows that the distribution of these cells, however, is more diffuse than in Fn:E/Fn:M explants. D) Numbers of Ngn1pr:eGFP cells increase significantly in Fn:E/Fn:M and Fn:E/Lb:M explants over time (p≤0.001, n=5, Fn:E/Fn:M; p ≤ 0.005, n=5, Fn:E/Lb:M; ANOVA); however, there are significantly more Ngn1pr:eGFP cells in Fn:E/Fn:M explants at 48 hours (p≤0.01;Mann-Whitney).
Induction and patterning in the nascent olfactory epithelium
Previous observations indicate that Sox2 is expressed in a lateral to medial gradient in the early OE (Ellis et al, 2004; Kawauchi et al, 2005). In parallel, OE-associated neurogenic bHLH genes are seen in the medial, but not lateral OE as it becomes distinct between E10 and E11 (Cau et al, 1997). Thus, we monitored differences in the capacity of different mesenchymes to induce or pattern these transcriptional regulators in living explants where the Fn:E was taken from either Sox2:eGFP knock-in allele (Ellis et al, 2004) or a Ngn1prTg:eGFP transgene (Nakada et al, 2004). Sox2 is modestly and diffusely expressed in the Fn:E at E9.0, based upon Sox2:eGFP signal in Fn:E after 1 hour in vitro (Figure 5A). We monitored subsequent patterning of Fn:E by Fn:M or Lb:M in the same explant over a 48 hour period in vitro. By 24 hours, there is significant placodal restriction regardless of mesenchymal identity. Some patterning is seen in isolated Fn:E, but it is not as consistent or distinct as that in recombined explants (compare Figure 5A with Figures 5B,C). The degree of placodal restriction in Fn:E/Fn:M and Fn:E/Lb:M explants is similar over 24 and 48 hours, based upon area occupied by Sox2:eGFP fluorescence measured in the same live explant (Figure 5D, E), with a linear, statistically significant decline in area of Sox2 signal in both explant pairings (p≤0.009, n=9, Fn:E/Fn:M; p≤0.01, n=5, Fn:E/Lb:M; ANOVA). The only difference was an apparently diminished gradient of Sox2:eGFP in the presumed medial-lateral axis (compare insets, Figure 5B,C). Thus, Fn:M and Lb:M support Sox2 placodal patterning in the Fn:E; however, the Fn:M may be more effective in patterning the presumed medial/lateral gradient.
We next asked whether Sox2 expression reflects the activity of RA, which may be differentially available from lateral Fn:M where the synthetic enzyme Raldh2 is expressed more robustly (see Figure 3A). We exposed Fn:E/Fn:M explants to exogenous RA (10−7M) for 48 hours and compared the level of Sox2:eGFP fluorescence. Exogenous RA diminishes Sox2:eGFP expression in Fn:E/Fn:M explants and eliminates the apparent lateral to medial gradient (Figure 5F; n=6/6). Fgf8 has apparently antagonistic effects on RA signaling in the OE (LaMantia et al, 2000; Bhasin et al, 2003), thus we asked whether Fgf8 enhances Sox2 expression. When Fgf8 concentrations are increased in Fn:E/Fn:M explants, Sox2 expression is consistently elevated across the entire placodal region (Figure 5G; n=4/6). Thus, differences in local availability of RA and Fgf8 may modulate the medial/lateral Sox2 gradient in the nascent OE.
We used a transgenic mouse line engineered to express GFP under the control of elements in the Ngn1 promoter (Ngn1prTg:eGFP; Nakada et al, 2004) to monitor the induction and patterning of Ngn1—a neurogenic bHLH gene that is part of a hierarchy of transcription factors associated with OE neurogenesis (Cau et al, 2002)—in living explants over time. Ngn1prTg:eGFP is not seen in the Fn:E at E9.0 (Figure 6A), nor is it detected subsequently in the isolated Fn:E. When cultured with either Fn:M or Lb:M within 24 hours there is significant expression of Ngn1 in individual cells (Figure 6B,C, middle). This expression becomes more pronounced by 48 hours (Figure 6B,C, right). There is a linear increase in the number of Ngn1prTg:eGFP cells in both homologous and heterologous pairings (p≤0.001, n=5, Fn:E/Fn:M; p ≤ 0.005, n=5, Fn:E/Lb:M; ANOVA). We noticed two distinctions, however, between Fn:E/Fn:M and Fn:E/Lb:M explants. First, Fn:E/Fn:M explants have more Ngn1prTg:eGFP cells (Figure 6D,E; p≤0.01;Mann-Whitney). Second, the distribution of the Ngn1prTg:eGFP cells is more diffuse in the placodal domain in Fn:E/Lb:M explants (compare insets in Figure 6F,G, right). Thus, although Fn:M and Lb:M induce Ngn1 expression in the same time frame, there are differences in the number and distribution of Ngn1-expressing cells.
Frontonasal-specific expression and induction of Meis1
Meis transcription factors, including Meis1, are known to be expressed in the OE as well as limb (Mercader et al, 1999; 2000; Toresson et al, 2000). Although not detected at E9.0 (not shown), Meis1 has a distinct compartmental distribution at each site by E10: epithelial in the OE, and mesenchymal in the Lb (Figure 7A,B). We asked whether the epithelial induction and expression of Meis1 in the OE depends upon frontonasal M/E interactions. An apparent lateral to medial expression gradient of Meis1 is maintained in the epithelium of Fn:E/Fn:M explants (Figure 7C, top and bottom panels). This patterning most likely reflects induction of Meis1, since we see no Meis1 in isolated Fn:E or Fn:M (Figure 7C, insets). In contrast, Lb:E/Lb:M explants maintain the limb pattern of Meis1: there is expression in the presumed proximal mesenchyme (Figure 7D top and bottom panels). This expression may not rely exclusively upon M/E induction, since some Meis1 expression is seen in isolated Lb:M, but not Lb:E (Figure 7D, insets). The Fn:E expression pattern of Meis1 is not maintained in the presence of Lb:M; instead, Meis1 expression shifts from the epithelium to the mesenchyme (Figure 7E top and bottom panels). In the presence of Fn:M, however, Lb:E expresses high levels of Meis1. Apparently, epithelial expression of Meis1—in Fn:E or other epithelia like Lb:E—is specifically induced and patterned by the Fn:M.
Figure 7.
Frontonasal M/E induction specifies epithelial expression of Meis1. A.) Meis1 is expressed in the lateral frontonasal epithelium (fnm, left), and B.) in the proximal forelimb mesenchyme (flb, right) at E10. C.) Fn:E/Fn:M interactions recapitulate in vivo Meis1 expression pattern. (Inset) Meis1 is not expressed in isolated Fn:E or Fn:M. (Below) A section through an Fn:E/Fn:M explant confirms expression in epithelium (ep), and exclusion from the mesenchyme (mes). D.) Lb:E/Lb:M interactions recapitulate in vivo Meis1 expression pattern. (Inset) Meis1 is not expressed in isolated Lb:E; however, some expression remains in isolated Lb:M. (Below) A section through an Lb:E/Lb:M explant confirms mesenchymal expression of Meis1. E.) In Fn:E/Lb:M explants Meis1 is excluded from the Fn:E and seen throughout the mesenchyme. (Below) is a section through an explant confirms Meis1 exclusion from the Fn:E and expression in the Lb:M. F) Elevated RA (10−7M) expands Meis1 expression throughout the mesenchyme in Lb:E/Lb:M explants. G) Elevated RA also expands Meis1 expression in the mesenchyme in Fn:E/Lb:M explants. H) Elevated RA shifts Meis1 expression to the mesenchyme and eliminates it in the epithelium in Fn:E/Fn:M explants.
Meis genes are thought to modulate RA and Fgf8 antagonism. Previous observations (Mercader et al, 2000) suggest that Meis genes in the proximal limb, regulated by locally produced RA (see Figure 3B), limit Fgf8 signaling to the distal limb mesenchyme. Accordingly, we asked whether Meis might contribute to similar antagonism in the OE by varying the levels of RA and Fgf8 and evaluating changes in Meis1 expression. As expected, exogenous RA expands Meis1 expression in Lb:E/Lb:M explants (Figure 7F; Mercader et al, 2000; n=4/4), while Fgf8 has no effect (data not shown). RA causes a similar expansion of Meis1 expression in Fn:E/Lb:M explants (Figure 7G; n=4/4), and Fgf8 has no detectable influence (data not shown). Finally, elevated RA’s influence on Meis1 expression in the frontonasal mass in some ways resembles that in the limb—it causes widespread mesenchymal expression; however, Meis1 expression in the Fn:E is lost (Figure 7H; n=7/7). As at other sites, Fgf8 has no apparent influence on Meis1 expression in Fn:E/Fn:M explants (not shown). Apparently, frontonasal M/E interactions establish a specific context for RA regulation of Meis1. Elevated RA concentrations disrupt this specific frontonasal signaling leading to the loss of epithelial expression of Meis1.
OE neuronal identity depends upon specific M/E interactions
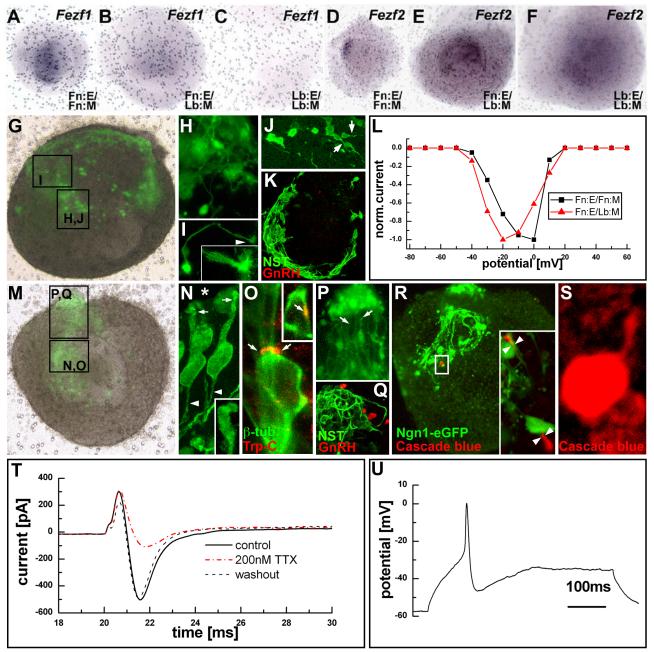
Specificity of pattern, rather than expression, of several signaling molecules and transcription factors in the frontonasal versus limb bud epithelium and mesenchyme raises a fundamental question: Are such quantitative distinctions sufficient to specify the three distinct classes of neurons generated from the nascent OE? Thus, we asked whether cells with fundamental molecular, cellular and physiological characteristics of ORNs, VRNs and GnRH neurons can be recognized only when Fn:E interacts with Fn:M. We first determined whether two transcription factors, Fezf1—associated with ORN differentiation (Hirata et al, 2006; Watanabe et al, 2009)—and Fezf2—associated with VRN differentiation (Hirata et al, 2004)—are regulated by specific frontonasal M/E interaction. In the presence of Fn:M, there is an apparent medial-lateral pattern of Fezf1 in the Fn:E (Figure 8A). This pattern is not seen when Fn:E is recombined with Lb:M; instead, Fezf1 is expressed minimally and ectopically in the mesenchyme (Figure 8B). In parallel with the limb bud in vivo (Hirata et al, 2006), we do not detect Fezf1 in Lb:E/Lb:M explants (Figure 8C). Fezf2 is also specifically patterned by Fn:E/Fn:M interactions, and seen in a limited region in the presumed medial aspect of the explant OE (Figure 8D) similar to the vomeronasal domain in vivo at E11.5. When Fn:E is induced by Lb:M, Fezf2, like Fezf1, shifts to the mesenchyme, and there is no substantial patterning (Figure 8E). This distribution of Fezf2 resembles that seen in Lb:E/Lb:M explants (Figure 8F). Thus, frontonasal M/E interactions specifically pattern two transcription factors, Fezf1 and Fezf2 associated with ORN and VRN differentiation.
Figure 8.
ORNs, VRNs and GnRH neurons generated only when lateral precursors are present. A) Fezf1, detected by in situ hybridization, is expressed in the presumed medial region in Fn:E/Fn:M explants. B) Fezf1 expression is diminished in Fn:E/Lb:M explants, and appears limited to the mesenchyme. C) Fezf1 is not expressed in Lb:E/Lb:M explants. D) Fezf2, detected by in situ hybridization, is expressed in a limited domain in Fn:E/Fn:M explants that may correspond to the presumptive vomeronasal organ in vivo. E) Fezf2 is expressed throughout the mesenchyme in Fn:E/Lb:M explants. F)Fezf2 is expressed widely in the mesenchyme in Lb:E/Lb:M explants. G) Diffuse neuronal differentiation and disorganized axon growth in heterologously induced Fn:E/Fn:M explant (live image, Ngn1pr:eGFP). H.) Transgene-labeled neurites form a loose plexus at the M/E interface. I) An isolated neurite just under the epithelial surface of an Fn:E/Lb:M explant ends in a lamellate growth cone (inset). J) Individual cells have single neurites (arrows), which extend, in isolation, for relatively long distances (not shown). K) GnRH neurons are not detected in the neuritic plexus in Fn:E/Lb:M explants. L) Placodal cells from Fn:E/Lb:M (red) and Fn:E/Fn:M (black) explants have slow, inward Ca++ currents with similar I/V relationships for normalized peak Ca++ currents. M) Fn:E/Fn:M explant imaged live showing Ngn1pr:eGFP labeled cells in the presumed medial OE (box N,O) and labeled processes in a coherent nerve and migratory mass (box P,Q). N) Ng1pr:eGFP labeled OE neurons have single apparent dendrites (arrows) oriented lumenally (asterisk) terminating in nascent dendritic knobs (arrows, inset). They also have single axons that exit the basal epithelium (arrowheads). O) A subset of Fn:E/Fn:M generated neurons (labeled in this explant for β-tubulin, green) express TrpC2 (red), the TrpC2 channel that identifies VRNs in vivo. P) Apparent ORN axons (arrows) and additional Ngn1pr:eGFP labeled cells form a coherent nerve and “migratory mass” in vitro. Q) GnRH neurons (red) are seen at the terminus of the nascent olfactory nerve/migratory mass in vitro (processes labeled for neuron-specific tubulin, NST;green). R) A single cell among the Ngn1:eGFP labeled population in a homologous explant filled with cascade blue (red) during physiological recording. (Inset) Another cascade blue filled cell, imaged live, has a distinct cell body (asterisk), single axon (arrowheads) and growth cone (gc). S) The cascade blue labeled cell in (R) has a single apical process (asterisk). T) Transient inward currents in homologously induced cells are voltage-activated Na+ currents, based on TTX sensitivity. 200 nM TTX diminishes this current (voltage steps from −80 to −20 mV) 77%, with recovery after washout. U) Current injection in Fn:E/Fn:M explant cell with large inward currents elicits an action potential.
To securely determine whether ORNs, VRNs and GnRH neurons are generated only by frontonasal M/E interactions, we characterized the cellular, molecular and physiological properties of neurons induced in the Fn:E by Fn:M versus Lb:M. Neurons in Fn:E/Lb:M explants have neurites that neither form one coherent, well-fasiculated nerve, nor enter the underlying mesenchyme (Figure 8G, H). Instead, these neurites have tortuous trajectories and end in lamellate, highly branched growth cones (Figure 8I). The cell bodies have only a single neurite—there is no suggestion of dendritic versus axonal processes (Figure 8J,K). Finally, although these cells have voltage activated Ca++ conductances found widely in neurons (Figure 8L), they lack other excitable properties seen in ORNs at this embryonic stage (see below). In contrast, when Fn:E is induced by Fn:M, neurons in the presumed OE, labeled by the Ngn1pr:eGFP transgene, are clustered together in the apparent medial region, and give rise to a single well-fasiculated nerve (Figure 8M-K). These cells have morphological hallmarks of ORNs and VRNs in vivo at E11.5: a single dendrite ends in a nascent dendritic knob, and a single axon extends toward the basal lamina to exit into the mesenchyme (Figure 8N). A subset of these cells can be labeled on their surfaces with antisera against the TrpC2 channel (Figure 8O), which is seen primarily in VRNs (Stowers et al, 2002). The fasciculated nerve is populated with migratory cells (Figure 8P), as is the case in vivo (where the combination of axons and migrating cells is referred to as the migratory mass). In vivo, some of the cells in migratory mass at this time are GnRH neurons en route through the mesenchyme, to the basal forebrain toward their final target in the hypothalamus (Schwanzel-Fukuda and Pfaff, 1988; Wray, 1988). Similarly, a subset of cells within the presumed migratory mass in Fn:E/Fn:M explants can be labeled for GnRH (Figure 8Q; 17/35 explants); this is not the case for Fn:E/Lb:M explants (see Figure 8K; 2/12 explants had one faintly labeled cell with no relationship to the placodal domain or neurites).
Finally, neurons within the presumed OE in Fn:E/Fn:M explants have age-appropriate physiological characteristics: voltage activated TTX-sensitive Na+ channels (n=33/63 cells recorded; Figure 8T). We did not find similar Na+ conductances in Fn:E/Lb:M explants (very small, <2pA, inward currents seen in 3/28 cells). In the Fn:E/FnM explants, the Na+ channels apparently have sufficient conductance to support the generation of action potentials (action potentials seen in 10/19 cells tested Figure 8U) which is unique to ORNs compared to other special sensory receptor cells (i.e. photoreceptors, auditory, and vestibular hair cells). We did not detect action potentials in cells with Ca++ conductances found in Fn:E/Lb:M explants (n=0/28 cells recorded). Thus, despite a significant amount of shared inductive capacity, only Fn:M is capable of inducing the differentiation of cells with age-appropriate properties of ORNs, VRNs, and GnRH neurons in the Fn:E.
DISCUSSION
Non-axial M/E induction seen in the frontonasal mass and limbs is apparently a singular mechanism modified in the anterior-posterior axis to facilitate distinct differentiation programs. Mesenchymal cells with disparate origins share the capacity to induce or maintain several inductive signals and transcription factors seen normally in the frontonasal epithelium, and to support initial, generic, neurogenesis in the OE. Nevertheless, only frontonasal mesenchyme facilitates appropriate expression or activity of a subset of OE-associated signals and transcription factors including Raldh2, Fgf8, Bmp4, Meis1, Fezf1 and Fezf2, as well as acquisition of key cellular, molecular, and physiological properties of the three major neuron classes derived from the nascent OE: ORNs, VRNs, and GnRH neurons (Figure 9). Thus, frontonasal M/E interactions are specifically adapted from a generic non-axial M/E inductive mechanism either to define OE neural progenitors or limit newly post-mitotic OE neurons to chemosensory or neuroendocrine identity and function.
Figure 9.
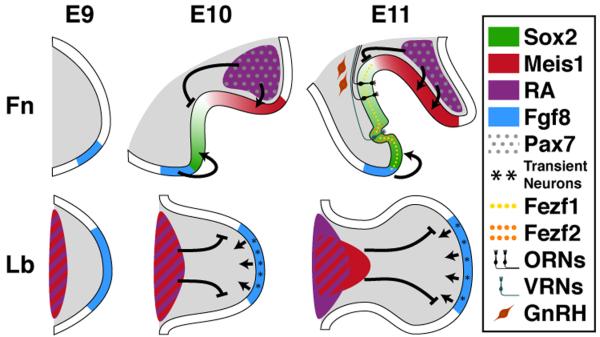
Parallel and divergent mechanisms of M/E induction in the frontonasal mass and limb. The top three panels show the mesenchymal and epithelial compartments of the frontonasal mass as they differentiate into the nascent olfactory epithelium (OE). The bottom three panels show the mesenchymal and epithelial compartments of the forelimb bud. Key distinctions in the local availability of RA (purple), which is in register with uniquely Pax7 expressing mesenchyme in the frontonasal mass (gray dots), expression of Meis1 (red), graded Sox2 expression, local Fezf1 and Fezf2 expression (yellow and orange dots), and ORN (black), VRN (teal), and GnRH (maroon) neuron differentiation between E9 and E11.5 are shown. Arrows and lines indicate signaling interactions established by our experiments. With the exception of Pax7 Fezf1, several factors found in the frontonasal mass are also expressed in the limb bud; however, as shown, their compartmental distribution is distinct.
A novel approach for studying ORN differentiation in vitro
We took advantage of the accessibility of M/E co-cultures, often used to evaluate initial induction and patterning (Vainio et al, 1993; Neubeusser et al, 1997; Dassule & McMahon, 1998; LaMantia et al, 2000; Bhasin et al, 2003) to study the initial specification of peripheral chemosensory and neuroendocrine neuron differentiation in the nascent OE. Removal or relocation of tissues has been an essential experimental approach for understanding specificity of induction and differentiation in accessible embryos for nearly a century (reviewed by Saunders et al, 1976; Hamburger, 1988; LeDourain et al, 1997; Fernandez-Teran et al, 1999; Landmesser, 2001); however, even the most sophisticated intrauterine imaging and manipulation (e.g. Gaiano et al, 1999) is not likely to facilitate consistent, successful extirpation or translocation of frontonasal tissue, which give rise to the OE, in intact mammalian embryos. Our in vitro approach circumvents these limitations, and we found that it is possible to assess molecular, cellular and functional differentiation of ORNs, VRNs, and GnRH neurons in vitro, using M/E explant cultures. Critical aspects of signaling and patterning are preserved in our explants, and definitive molecular, cellular and physiological properties are recapitulated.
Of the 33 signaling molecules, transcription factors and differentiation markers we analyzed, all were detected in our in vitro explants in parallel with their in vivo expression. Indeed, the degree of ORN differentiation seen in frontonasal M/E explants is far greater than that reported for odontogenic cells in branchial arches, or cartilage, muscle and bone in limb buds in vitro (reviewed by Fernandez-Teran et al, 1999; Hay, 2005; Modino & Sharpe, 2005). By varying M/E interactions specifically and reproducibly alter signaling and transcriptional regulation, we elicit tissue specific (i.e. M or E) loss or gain of expression (and presumably function) of transcription factors like Meis1, Fezf1 and Fezf2 that may be critical for ORN VRN, or GnRH neurogenesis or differentiation. It is unlikely that such mechanistic experiments on local induction could be easily accomplished using current genetic methods. Thus, our in vitro approach permits analysis of induction critical for molecular, cellular and functional differentiation of peripheral chemosensory and neuroendocrine neurons.
Parallel and divergent M/E identities influence OE induction
We found that mesenchyme and epithelium from the frontonasal mass versus limb bud has significant shared capacity for induction and differentiation, suggesting parallel mechanisms for M/E induction at diverse non-axial sites (Figure 9). Our data indicates that non-axial mesenchyme in the frontonasal mass and forelimb between E9 and E11 is either inductively similar or remarkably plastic despite distinct anterior-posterior (A-P) position and divergent levels of derivation from neural crest (Lumsden, 1988; Richman & Tickle, 1989; Serbedzija et al, 1992; Osumi-Yamashita et al, 1994; Prince & Lumsden 1994; Yoshida et al, 2008). When recombined with Fn:E, limb mesenchyme supports parallel expression and patterning of local signals and transcription factors associated with OE neurogenesis. In fact, limb mesenchyme transiently supports some aspects of neurogenesis or differentiation in the limb epithelium itself. Thus, a significant amount of parallel capacity for induction and differentiation is established quite early during development and retained by epithelia and mesenchyme at divergent sites of non-axial M/E interaction, independent of additional anterior posterior specification and differences in mesodermal versus neural crest origin.
Mesenchyme and epithelia at M/E sites like the frontonasal mass and limb, once established, seem to retain much of their identity. Thus, despite the presence of Fn:E, Lb:M uniquely expresses Tbx5 and MyoD, maintains its own pattern of Raldh2 and Meis1, and does not express Pax7. These distinctions may help explain—or at least provide evidence for—differences in the capacity of Lb:M versus Fn:M to support generic neurogenesis versus ORN, VRN and GnRH neuronal differntiation. The available data, however, does not resolve the contribution of these genes. MyoD, coupled with Myf5, seems necessary for genesis of skeletal muscle from mesodermal precursors in the trunk and limbs, but not from neural crest precursors in the head (reviewed by Arnold & Braun, 1996). There are no reported cranial or nasal phenotypes in Tbx5 mutants, which lack forelimbs (Rallis et al, 2003). Raldh2 constitutive loss of function is lethal prior to extensive OE morphogenesis (Niederreither et al, 1999). Meis1 null embryos have no identifiable OE phenotypes, perhaps due to functional redundancy (Hisa et al, 2004), and the OE in Pax7 mutants is apparently normal (Mansouri et al, 1998). Thus, while expression of these genes suggests distinct identities of frontonasal versus limb mesenchyme, additional factors are likely to be critical for specific inductive capacities, including that of limb mesenchyme for primarily musculo-skeletal development, and frontonasal mesenchyme for OE neuronal differentiation.
The epithelium at each M/E site possesses some identity that establishes signaling and differentiation capacity in adjacent mesenchyme (see also Richman & Tickle, 1989; 1992). Fn:E alone induces expression of Pax7 in Fn:M (see also LaMantia et al, 2000). This suggests that cranial neural crest, which expresses Pax7 at high levels (Mansouri et al, 1998) relies upon signals from the Fn:E to maintain some aspects of its identity (see also Bhattachyra & Bronner-Fraser, 2009). Fgf8 from the frontonasal versus limb epithelium has distinct effects on expression of Bmp4 in adjacent mesenchyme: In the limb, epithelial Fgf8 promotes mesenchymal Bmp4 expression and patterning, while in the frontonasal mass, epithelial Fgf8 has no influence on Bmp4. Fn:E apparently regulates this lack of Fgf8 responsiveness, since in the presence of Fn:E, Fgf8 regulation of Bmp4 is diminished in Lb:M. These differences may underlie frontonasal specific modulation of Bmp4 levels that influence bHLH gene activity and survival of OE neurogenic precursors or newly generated neurons. Lower concentrations of Bmp4 maintain Ascl1, downstream bHLH gene expression, and facilitate OE neurogenesis (Shou et al, 1999; 2000; LaMantia et al, 2000). A distinct context of Fgf8 signaling established by the Fn:E may maintain appropriately low Bmp4 levels and thus facilitate subsequent neurogenesis. Indeed, we find more Ngn1prTg:eGFP expressing cells in Fn:E/Fn:M than Fn:E/Lb:M explants, in parallel with differing Bmp concentrations. Thus, epithelia may establish or maintain distinct inductive signaling from adjacent mesenchyme—including Fn:E signals that support Fn:M’s ability to specify appropriate numbers or identity of ORNs, VRNs, and GnRH neurons.
Inductive Specificity and OE neuronal differentiation
We found that OE neurons acquire characteristic molecular and cellular properties only when induced by frontonasal M/E interactions (Figure 9). Appropriate expression levels and patterns of two transcription factors associated with ORN and VRN differentiation, Fezf1 and Fezf2 (Hirata et al, 2004, 2006; Watanabe et al, 2009), are only supported by Fn:M. Similarly, ORNs and VRNs, like photoreceptors and hair cells, acquire a distinctive polarized morphology—including a single apical process and nascent cilia, which eventually will serve as the site for signal transduction—central for their function (reviewed by Barald & Kelley, 2004; Malicki, 2004) only in the context of frontonasal M/E interaction. The acquisition of key OE cellular features is apparently independent of specific expression of Ascl1 or other bHLH factors including Ngn1 and NeuroD (Guillemot et al, 1993; Cau et al, 2002), as well as Pax6 (Grindley et al, 1995; Davis & Reed, 1996; Anchan et al, 1997). Lb:M induces or maintains these factors in Fn:E without eliciting differentiation of ORNs, VRNs, or GnRH neurons. Fn:M also provides a uniquely supportive territory for fasciculation of pioneering ORN/VRN axons, as well as their coalescence with placode-derived cells into a migratory mass. Apparently, Fn:M has the capacity to provide either appropriate signals, or local adhesion cues including semaphorins (Schwarting et al, 2000), and other adhesion molecules (Whitesides & LaMantia, 1996) necessary for olfactory nerve formation. Clearly, frontonasal M/E interactions establish key OE cellular features that are correlated with odorant transduction, neuronal migration, and relay of signals to the CNS. Nevertheless, the capacity of these interactions to influence odorant/pheromone receptor expression and signal transduction in fully functional ORNs or VRNs, or appropriate targeting and differentiation of GnRH neurons in the hypothalamus, remains to be determined.
M/E induction specifies OE neuron physiological characteristics
Frontonasal M/E interactions induce essential functional properties of the earliest ORNs and VRNs—differential expression of TrpC2 channels that distinguish VRNs (Stowers et al. 2002), transient inward Na+ currents and the ability to generate action potentials (Figure 9). This represents the first report of acquisition of such physiological properties in neurons generated in vitro from undifferentiated embryonic epithelial precursors. The TTX sensitivity of the Na+ currents is similar to that in other immature neurons (Yoshida, 1994). Accordingly, it is unlikely that these neurons are terminally differentiated ORNs or VRNs. Acquisition of additional functional properties, including expression of odorant receptors, cyclic nucleotide gated channels, and ORN-associated G-proteins may depend upon systemic or target derived signals unavailable in our explant cultures (in vivo these genes appear around E15). Nevertheless, among special peripheral sensory neurons (photoreceptors, hair cells, ORNs and VRNs), TTX sensitive Na+ currents and the parallel generation of action potentials is seen only in ORNs and VRNs. Our results show clearly that frontonasal M/E induction accounts for this characteristic feature of peripheral chemosensory receptor neurons.
Consistent with some shared neuronal inductive capacity of mesenchyme, voltage regulated Ca++ and K+ currents are established in the presence of limb mesenchyme. These currents may influence general aspects of neuronal maturation including axon outgrowth and growth cone motility (Spitzer and Ribera, 1998); however, they are not sufficient to establish appropriate ORN/VRN morphology, dendritic differentiation, axonal growth or excitable properties. The dissociation of developing Na+ currents and action potentials from Ca++ and K+ currents suggests that integrated physiological properties in some nascent peripheral neurons are not acquired through a single linear developmental mechanism. Apparently, a subset of essential physiological properties of ORNs or VRNs are either intrinsic to ectodermal/epithelial precursors from an early time (we do not see Ca++ currents in mesenchyme; data not shown), or supported by general M/E interactions while others—including expression of TrpC2 and TTX-sensitive Na+ channels as well as the ability to relay signals to the CNS via action potentials—depend upon specific frontonasal M/E induction.
Transcriptional Codes and Specific Induction of ORNs, VRNs, and GnRH neurons
We found that M/E mediated expression in the Fn:E of Ascl1, Ngn1, and NeuroD, associated with a proposed sequential transcription code for ORNs (Guillemot et al, 1993; Cau et al, 1997; 2002; Calof et al, 2002; Manglapus et al, 2004) is not sufficient to establish key cytological, molecular and functional ORN properties. Mesenchyme from heterologous sites induces or maintains these bHLH factors in frontonasal epithelial cells with neuronal properties; nevertheless, such heterologously induced neurons do not acquire ORN characteristics. Our data does not preclude a general role for Ascl1, Ngn1 and NeuroD in neuronal maturation or expansion of ORN numbers, despite an apparently negligible contribution to ORN identity. The high degree of differentiation in an apparently small number of early-generated ORNs in Ascl1−/− mice (Guillemot et al, 1993; Cau et al, 1997), accords well with this conclusion. It is possible that the bHLH factors influence transit amplifying neurogenic precursor populations in concert with local signals, including Fgfs and TGFb family members (Kawauchi et al, 2005). Nevertheless, transcriptional regulation critical for ORN identity requires specific frontonasal M/E interactions that precede or super-cede induction and activity of Ascl1, Ngn1, and NeuroD.
Our current results suggest that maintenance of Sox2 gradients as well as induction of Meis1 in the OE is specified by frontonasal M/E interactions. This suggests that graded Sox2 or local Meis1 function may contribute to the establishment of OE specific progenitors, or the differentiation of ORNs, VRNs, or GnRH neurons. Sox2 is the primary SoxB1 factor expressed in the OE (Ellis et al, 2004). The obligate function of Sox2 for progression of retinal progenitors to ganglion cell fates suggests that Sox2 may contribute to OE neuronal differentiation (Tarranova et al, 2006). The known function of Meis1—establishing a transcriptional context, perhaps via interaction with Pbx factors, that favors particular cell fates (reviewed by Sagerstrom, 2004)—also suggests a contribution to OE lineages, perhaps by modulation of precursor capacity or identity. Indeed, recent evidence suggests that Meis1 influences retinal ganglion cell differentiation (Heine et al, 2008). RA regulates Meis1 in a variety of contexts (Mercader et al, 2000, 2005; Qin et al, 2002) and our results indicate that RA signaling can differentially influence Meis1 expression in the OE versus limb. It remains to be determined if RA-dependent mesenchymal induction of Meis1-expressing cells in the OE is a singular necessity for ORN genesis. Our recent evidence (Tucker et al, 2010, submitted) suggests that the Meis1 cells in the OE cells are slowly-dividing, self-renewing precursors capable of giving rise to multiple classes of OE neurons.
Common Tools, different uses: specificity in ORN induction
ORN specification via quantitative and positional variation of shared molecules raises a central question: are there any unique molecular determinants? The number of signals, transcription factors, and differentiation markers shared by olfactory primordia and limbs—in this report alone, we show common expression and partially parallel regulation of over 30 genes—is a striking feature of these apparently divergent M/E inductive sites. Given this high degree of similarity, one can fairly ask how do distinctive cell types, including ORNs, VRNs, and GnRH neurons acquire unique features? Our results do not preclude the existence of novel signaling molecules that induce peripheral chemosensory and neuroendocrine neurons. Nevertheless, our data demonstrate a distinct molecular mechanism for specificity: quantitative differences in expression, localization, and regulation of common determinants by distinct tissue compartments. This strategy may be used throughout the embryo, as well as in the adult peripheral and central olfactory pathway where stem cells retain the ability to generate different classes of neurons despite many molecular similarities (Alvarez-Buylla and Lim, 2004; Muotri & Gage, 2006). If this is the case, ex vivo generation of cytologically and functionally distinct neurons from multipotent stem cells is likely to be even more challenging than currently expected.
EXPERIMENTAL PROCEDURES
Animals
Embryos were harvested from timed-pregnant mothers maintained by the Department of Laboratory Animal Medicine at the University of North Carolina at Chapel Hill or the Animal Care Facility of the Monell Chemical Senses Center. Rosa26, Sox2:eGFP (Ellis et al, 2004) and Ngn1prTg:eGFP (gift of J.E. Johnson; Nakada et al, 2004) indicators were bred into CF-1 females from homozygous (Rosa26, Ngn1) or heterozygous (Sox2) males. Mother mice were sacrificed by rapid cervical dislocation, and embryos immediately dissected for culture or molecular analysis. The Institutional Animal Care and Use Committees (IACUC) of the University of North Carolina at Chapel Hill or the Monell Chemical Senses Center approved all procedures.
Explant Cultures
E9.0 frontonasal masses and forelimb buds and were dissected and layers separated as described previously (LaMantia et al, 2000; Bhasin et al, 2003). For homologous explants (Fn:E/Fn:M; Lb:E/Lb:M), layers were immediately reapposed; for heterologous explants (Fn:E/Lb:M; Lb:E/Fn:M), layers were exchanged. All explants were cultured under 95% air/5%CO2 at 37°C for 48 hours, with fresh medium after 24 hours. For Rosa26, Sox2 and Ngn1 indicator lines, epithelium was from a transgenic donor (confirmed by β-gal or fluorescence) and mesenchyme from a +/+ donor from a parallel litter. RA (10−7M) and Fgf8 (100ng/ml) were added to media at the outset of the culture period. Except where noted otherwise, we confirmed consistency of all markers analyzed in at least three sets of explants from embryos from different litters.
Live imaging
At 1, 24, and 48 hours, individual Sox2:eGFP and Ngn1prTg:eGFP explants were removed briefly from the incubator and transferred to a Leica DMIRB inverted fluorescent microscope with a SPOT-RT color digital camera. Each explant was illuminated briefly with low-level incandescent light to obtain a bright field image, then briefly for fluorescence imaging, and returned immediately to the incubator. Consistent detection thresholds were set to quantify GFP area or cell numbers in fluorescence images from each time point. Brightfield and fluorescent images were superimposed using Adobe Photoshop to simultaneously visualize explant boundaries and transgene expression.
Immunohistochemistry and in situ hybridization
Whole explants were fixed and prepared for antibody labeling or in situ hybridization as described previously (LaMantia et al, 2000; Bhasin et al, 2003). In addition, subsets were fixed in 2.5% paraformaldehyde/4% sucrose in 0.1M NaPO4 buffer (pH 7.4), cryoprotected removed from the filter in a drop of agar/sucrose, positioned in an embedding mold, surrounded by additional agar, immersed in cryoembedding media, and frozen in liquid nitrogen-cooled 2-Methylbutane for cryosectioning. These blocks were sectioned at 15μm, and sections immunostained as described above. Images for illustration and quantification were obtained using a Leica DMR epifluorescence microscope or a Zeiss LSM510 multiphoton laser confocal microscope.
RT-PCR and qPCR
mRNA from whole E9.0 and 11.0 rapidly dissected embryo parts pooled from an entire litter or combined sets of 6 explants of a particular type (e.g. Fn:E/Fn:M) was extracted using Trizol (Gibco), DNAse treated, and reverse transcribed using standard protocols. cDNA concentrations were standardized using competitive quantitative PCR for β-actin. Primers were designed with PrimerDesign software (supplemental material) and validated by sequencing amplicons from E11.5 whole embryo cDNAs. 3-5 separate pooled sets of embryos or explant samples were probed for each gene. Quantitative PCR was carried out using Taqman qPCR (ABI) with primers designed using ABI Primer Express software.
Physiological Recording
Explants were transferred from filters to collagen-coated glass coverslips, and then mounted in a flow-through recording chamber on an inverted microscope. Perforated patch recordings were performed with gramicidin as the ionophore (Lischka et al., 1999) using borosilicate pipettes (Kimax 51). Pipettes were filled with (concentration in mM) KCl 36, K-aspartate 110, CaCl2 1, MgCl2 1, HEPES 10. pH was adjusted with 1M KOH to 7.2, and solutions were approximately 300 mOsmol. The bath solution was NaCl 145, KCl 5, CaCl2 1, MgCl2 1, Na-pyruvate 1, HEPES 20, approx. 320 mOsmol, pH adjusted with 1M NaOH to 7.2. Only cells exhibiting stable outward currents were analyzed. In a subset of cells with stable inward currents, Cascade blue (Molecular Probes, Eugene OR) included in the pipette, diffused into the cell during recording. These explants were imaged using a Leica TCS SP2 Spectral Confocal Microscope.
ACKNOWLEDGEMENTS
The National Institute of Child Health and Human Development supported this project (HD029178 to A-S.L/N.R.). We thank Thomas Sugimoto, Clifford Heindel, Yonqin Wu, and Robert Peterson for expert technical assistance during various phases of this project. In situ hybridization and confocal microscopy were performed in the Expression Localization and Microscopy Cores of the UNC Neuroscience Center supported by National Institute of Neurological Diseases and Stroke (NS031768), as well as the Monell Center Confocal Microscopy facility supported by National Science Foundation (DBI-0216310).
Grant Sponsors: National Institutes of Health, National Science Foundation
Grant Numbers: NIH grant HD29178 to ASL as well as NS031768 to the UNC Neuroscience Center for Core Facilities, and NSF Grant DBI-0216310 to the Monell Chemical Senses Center Imaging Core Facility.
REFERNCES
- Anchan RM, Drake DP, Haines CF, Gerwe EA, LaMantia A-S. Disruption of local retinoid-mediated gene expression accompanies abnormal development in the mammalian olfactory pathway. J. Comp. Neurol. 1997;379:171–184. [PubMed] [Google Scholar]
- Arnold HH, Braun T. Targeted inactivation of myogenic factor genes reveals their during mouse myogenesis: a review. Int. J. Dev. Biol. 1996;40:345–353. [PubMed] [Google Scholar]
- Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41:683–686. doi: 10.1016/s0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Axel R. Scents and sensibility: a molecular logic of olfactory perception (Nobel lecture) Agnew Chem. Int. Ed. Engl. 2005;44:6110–6127. doi: 10.1002/anie.200501726. [DOI] [PubMed] [Google Scholar]
- Balmer CW, LaMantia A-S. Loss of Gli3 and Shh function disrupts olfactory axon trajectories. J. Comp. Neurol. 2004;472:292–307. doi: 10.1002/cne.20053. [DOI] [PubMed] [Google Scholar]
- Balmer CW, LaMantia A-S. Noses and neurons: Induction, morphogenesis and neuronal differentiation in the peripheral olfactory pathway. Dev. Dyn. 2005;234:464–481. doi: 10.1002/dvdy.20582. [DOI] [PubMed] [Google Scholar]
- Barald KF, Kelley MW. From placode to polarization: new tunes in inner ear development. Development. 2004;131:4119–4130. doi: 10.1242/dev.01339. [DOI] [PubMed] [Google Scholar]
- Bhasin N, Maynard TM, Gallagher P, LaMantia A-S. Mesenchymal/Epithelial Interactions Regulate Retinoid Signaling in the Olfactory Placode. Develop.Biol. 2003;261:82–98. doi: 10.1016/s0012-1606(03)00295-1. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Bronner-Fraser M. Competence, specification and commitment to an olfactory placode fate. Development. 2008;135:4165–4177. doi: 10.1242/dev.026633. [DOI] [PubMed] [Google Scholar]
- Buck LB. Information coding in the vertebrate olfactory system. Ann. Rev. Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- Buckland RA, Collinson JM, Graham E, Davidson DR, Hill RE. Antagonistic effects of FGF4 on BMP induction of apoptosis and chondrogenesis in the chick limb bud. Mech. Dev. 1998;71:143–150. doi: 10.1016/s0925-4773(98)00008-2. [DOI] [PubMed] [Google Scholar]
- Calof AL, Chikaraishi DM. Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron. 1989;3:115–127. doi: 10.1016/0896-6273(89)90120-7. [DOI] [PubMed] [Google Scholar]
- Calof AL, Bonnin A, Crocker C, Kawauchi S, Murray RC, Shou J, Wu HH. Progenitor cells of the olfactory receptor neuron lineage. Microsc. Res. Tech. 2002;58:176–188. doi: 10.1002/jemt.10147. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–80. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Dassule HR, McMahon AP. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev Biol. 1998;202:215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J. Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHamer MK, Guevara JL, Hannon K, Olwin BB, Calof AL. Genesis of olfactory receptor neurons in vitro: regulation of progenitor cell divisions by fibroblast growth factors. Neuron. 1994;13:1083–1097. doi: 10.1016/0896-6273(94)90047-7. [DOI] [PubMed] [Google Scholar]
- Dulac C. Sensory coding of pheromone signals in mammals. Curr. Opin. Neurobiol. 2000;10:511–518. doi: 10.1016/s0959-4388(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Fernandez-Teran M, Piedra ME, Ros MA, Fallon JF. The recombinant limb as a model for the study of limb patterning, and its application to muscle development. Cell Tissue Res. 1999;296:121–129. doi: 10.1007/s004410051273. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Kohtz JD, Turnbull DH, Fishell G. A method for rapid gain-of-function studies in the mouse embryonic nervous system. Nat Neurosci. 1999;2:779–780. doi: 10.1038/12186. [DOI] [PubMed] [Google Scholar]
- Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Hamburger V. The Heritage of Experimental Embryology: Hans Spemann and the Organizer. Oxford University Press; New York/Oxford: 1988. [Google Scholar]
- Hay ED. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev. Dyn. 2005;233:706–720. doi: 10.1002/dvdy.20345. [DOI] [PubMed] [Google Scholar]
- Heine P, Dohle E, Bumsted-O’brien K, Engelkamp D, Schulte D. Evidence for an evolutionary conserved role of homothorax/Meis1/2 during vertebrate retina development. Development. 2008;135:805–811. doi: 10.1242/dev.012088. [DOI] [PubMed] [Google Scholar]
- Hirata T, Suda Y, Nakao K, Narimatsu M, Hirano T, Hibi M. Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev. Dyn. 2004;230:546–556. doi: 10.1002/dvdy.20068. [DOI] [PubMed] [Google Scholar]
- Hirata T, Nakazawa M, Yoshihara S, Miyachi H, Kitamura K, Yoshihara Y, Hibi M. Zinc-finger gene Fez in the olfactory sensory neurons regulates development of the olfactory bulb non-cell-autonomously. Development. 2006;133:1433–1443. doi: 10.1242/dev.02329. [DOI] [PubMed] [Google Scholar]
- Hisa T, Spence SE, Rachel RA, Fujita M, Nakamura T, Ward JM, Devor-Henneman DE, Saiki Y, Kutsuna H, Tessarollo L, Jenkins NA, Copeland NG. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- Kolterud A, Alenius M, Carlsson L, Bohm S. The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development. 2004;131:5319–5326. doi: 10.1242/dev.01416. [DOI] [PubMed] [Google Scholar]
- LaMantia A-S, Colbert MC, Linney E. Retinoic acid induction and regional differentiation prefigure olfactory pathway formation in the mammalian forebrain. Neuron. 1993;10:1035–1048. doi: 10.1016/0896-6273(93)90052-s. [DOI] [PubMed] [Google Scholar]
- LaMantia A-S, Bhasin N, Rhodes K, Heemskerk J. Mesenchymal/ epithelial induction mediates olfactory pathway formation. Neuron. 2000;28:411–425. doi: 10.1016/s0896-6273(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Landmesser LT. The acquisition of motoneuron subtype identity and motor circuit formation. Int. J. Dev. Neurosci. 2001;19:175–182. doi: 10.1016/s0736-5748(00)00090-3. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Catala M, Batini C. Embryonic neural chimeras in the study of vertebrate brain and head development. Int. Rev. Cytol. 1997;175:241–309. doi: 10.1016/s0074-7696(08)62128-2. [DOI] [PubMed] [Google Scholar]
- Lischka FW, Teeter JH, Restrepo D. Odorants suppress a voltage-activated K+ conductance in rat olfactory neurons. J. Neurophysiol. 1999;82:226–236. doi: 10.1152/jn.1999.82.1.226. [DOI] [PubMed] [Google Scholar]
- Lumsden AG. Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development. 1988;103(Suppl):155–169. doi: 10.1242/dev.103.Supplement.155. [DOI] [PubMed] [Google Scholar]
- Malicki J. Cell fate decisions and patterning in the vertebrate retina: the importance of timing, asymmetry, polarity and waves. Curr. Opin. Neurobiol. 2004;14:15–21. doi: 10.1016/j.conb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Manglapus GL, Youngentob SL, Schwob JE. Expression patterns of basic helix-loop-helix transcription factors define subsets of olfactory progenitor cells. J. Comp. Neurol. 2004;479:216–233. doi: 10.1002/cne.20316. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Stoykova A, Torres M, Gruss P. Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development. 1996;122:831–838. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- Marchal L, Luxardi G, Thomé V, Kodjabachian L. BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17437–17442. doi: 10.1073/pnas.0906352106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N, Leonardo E, Azpiazu N, Serrano A, Morata G, Martinez C, Torres M. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402:425–429. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- Mercader N, Leonardo E, Piedra ME, Martinez AC, Ros MA, Torres M. Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development. 2000;127:3961–3970. doi: 10.1242/dev.127.18.3961. [DOI] [PubMed] [Google Scholar]
- Mercader N, Tanaka EM, Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132:4131–4142. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- Modino C, Sharpe PT. Tissue engineering of teeth using adult stem cells. Arch Oral Biol. 2005;50:255–258. doi: 10.1016/j.archoralbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Nakada Y, Parab P, Simmons A, Omer-Abdalla A, Johnson JE. Separable enhancer sequences regulate the expression of the neural bHLH transcription factor neurogenin 1. Dev. Biol. 2004;271:479–487. doi: 10.1016/j.ydbio.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Neubuser A, Peters H, Balling R, Martin GR. Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Nicolay DJ, Doucette JR, Nazarali AJ. Transcriptional regulation of neurogenesis in the olfactory epithelium. Cell Mol. Neurobiol. 2006;26:803–821. doi: 10.1007/s10571-006-9058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi-Yamashita N, Ninomiya Y, Doi H, Eto K. The contribution of both forebrain and midbrain crest cells to the mesenchyme in the frontonasal mass of mouse embryos. Dev. Biol. 1994;164:409–419. doi: 10.1006/dbio.1994.1211. [DOI] [PubMed] [Google Scholar]
- Prince V, Lumsden A. Hoxa-2 expression in normal and transposed rhombomeres: independent regulation in the neural tube and neural crest. Development. 1994;120:911–923. doi: 10.1242/dev.120.4.911. [DOI] [PubMed] [Google Scholar]
- Qin P, Cimildoro R, Kochhar DM, Soprano KJ, Soprano DR. PBX, MEIS, and IGF-I are potential mediators of retinoic acid-induced proximodistal limb reduction defects. Teratology. 2002;66:224–234. doi: 10.1002/tera.10082. [DOI] [PubMed] [Google Scholar]
- Rajendra S, Lynch JW, Barry PH. An analysis of Na+ currents in rat olfactory receptor neurons. Pflugers Arch. 1992;420:342–346. doi: 10.1007/BF00374468. [DOI] [PubMed] [Google Scholar]
- Rallis C, Bruneau BG, Del Buono J, Seidman CE, Seidman JG, Nissim S, Tabin CJ, Logan MP. Tbx5 is required for forelimb formation and continued growth. Development. 2003;130:2741–2751. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- Rawson NE, LaMantia A-S. Once and again: retinoic acid signaling in the developing and regenerating olfactory pathway. J. Neurobiol. 2006;66:653–676. doi: 10.1002/neu.20236. [DOI] [PubMed] [Google Scholar]
- Richman JM, Tickle C. Epithelia are interchangeable between facial primordia of chick embryos and morphogenesis is controlled by the mesenchyme. Dev. Biol. 1989;136:201–210. doi: 10.1016/0012-1606(89)90142-5. [DOI] [PubMed] [Google Scholar]
- Sagerstrom CG. PbX marks the spot. Dev. Cell. 2004;6:737–738. doi: 10.1016/j.devcel.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Saunders JW, Jr, Gasseling MT, Errick JE. Inductive activity and enduring cellular constitution of a supernumerary apical ectodermal ridge grafted to the limb bud of the chick embryo. Dev. Biol. 1976;50:16–25. doi: 10.1016/0012-1606(76)90063-4. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- Schwarting GA, Kostek C, Ahmad N, Dibble C, Pays L, Pushel AW. Semaphorin 3A is required for guidance of olfactory axons in mice. J. Neurosci. 2000;20:7691–7697. doi: 10.1523/JNEUROSCI.20-20-07691.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development. 1992;116:297–307. doi: 10.1242/dev.116.2.297. [DOI] [PubMed] [Google Scholar]
- Shou J, Rim PC, Calof AL. BMPs inhibit neurogenesis by a mechanism involving degradation of a transcription factor. Nat. Neurosci. 1999;2:339–345. doi: 10.1038/7251. [DOI] [PubMed] [Google Scholar]
- Shou J, Murray RC, Rim PC, Calof AL. Opposing effects of bone morphogenetic proteins on neuron production and survival in the olfactory receptor neuron lineage. Development. 2000;127:5403–5413. doi: 10.1242/dev.127.24.5403. [DOI] [PubMed] [Google Scholar]
- Spitzer NC, Ribera AB. Development of electrical excitability in embryonic neurons: mechanisms and roles. J. Neurobiol. 1998;37:190–197. [PubMed] [Google Scholar]
- Stowers L, Holy TE, Meister M, Dulac C, Koentges G. Loss of sex discrimination and male-male aggression in mice deficient for TRP2. Science. 2002;295:1493–1500. doi: 10.1126/science.1069259. [DOI] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toresson H, Parmar M, Campbell K. Expression of Meis and Pbx genes and their protein products in the developing telencephalon: implications for regional differentiation. Mech. Dev. 2000;94:183–187. doi: 10.1016/s0925-4773(00)00324-5. [DOI] [PubMed] [Google Scholar]
- Trombley PQ, Westbrook GL. Voltage-gated currents in identified rat olfactory receptor neurons. J Neurosci. 1991;11:435–444. doi: 10.1523/JNEUROSCI.11-02-00435.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzchori I, Day TF, Carolan PJ, Zhao Y, Wassif CA, Li L, Lewandoski M, Gorivodsky M, Love PE, Porter FD, Westphal H, Yang Y. LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development. 2009;136:1375–1385. doi: 10.1242/dev.026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- Verhaagen J, Oestreicher AB, Gispen WH, Margolis FL. The expression of the growth associated protein B50/GAP43 in the olfactory system of neonatal and adult rats. J. Neurosci. 1989;9:683–681. doi: 10.1523/JNEUROSCI.09-02-00683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Inoue K, Okuyaman-Yamamoto A, Nakai N, Nakatani J, Nibu K, Sato N, Iiboshi Y, Yusa K, Kondoh G, Takeda J, Terashima T, Takumi T. Fezf1 is required for penetration of the basal lamina by olfactory axons to promote olfactory development. J. Comp. Neurol. 2009;515:565–585. doi: 10.1002/cne.22074. [DOI] [PubMed] [Google Scholar]
- Whitsides JG, LaMantia A-S. Differential adhesion and the initial assembly of the mammalian olfactory nerve. J. Comp. Neurol. 1996;373:240–254. doi: 10.1002/(SICI)1096-9861(19960916)373:2<240::AID-CNE7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Whitesides JG, Hall M, Anchan R, LaMantia A-S. Retinoid signaling distinguishes a subpopulation of olfactory receptor neurons in the developing adult mouse. J. Comp. Neurol. 1998;394:445–461. [PubMed] [Google Scholar]
- Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc. Natl. Acad. Sci. USA. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S. Tetrodotoxin-resistant sodium channels. Cell Mol. Neurobiol. 1994;14:227–244. doi: 10.1007/BF02088322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech. Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]