Abstract
Mutation rates of human immunodeficiency virus type 1 (HIV-1) genomes have been estimated using purified reverse transcriptase or single-round infection system. Since small sequences were used as templates, the overall mutation rates could only be extrapolated and the biological significance of mutations is unknown. For direct estimation of HIV-1 mutation rates and understanding of the potential biological influences of mutations, we obtained 19 complete or nearly full-length proviral genomes from single-round-infected adherent cells of lymphocytes by using a lambda phage library method and a long-range PCR technique. Analysis of 160,000 bp of sequences showed that the overall mutation rate of HIV-1 genomes was 5.4 × 10−5 per base per replication cycle. On average, 1.1 mutations (range, 0 to 3) were generated in each viral genome during one infection cycle. Inspection of the mutations in the HIV-1 genome revealed that all site mutations within protein-coding regions were nonsynonymous mutations. Among all mutations, half were deleterious (premature stop codon and deletions) and would result in defective genomes. By applying the same system to an HIV-1 genome with a G262A mutation in the thumb region of the reverse transcriptase, a significant increase was observed in deletion and insertion mutation rates but no increase in the overall mutation rate in viral genomes was found.
Genetic variation among human immunodeficiency virus (HIV) isolates is extensive. The highly error-prone reverse transcriptase (RT) is the primary reason for the high levels of genetic variation in HIV (6, 16, 29, 32, 41-43). High yields of virus production and a short viral-replication cycle also contribute to the mutation frequency in the HIV genome (14, 39, 46). On the basis of genetic variation and positions in the phylogenetic tree, HIV type 1 (HIV-1) has been divided into three groups (M, N, and O). Among them, group M viruses are most prevalent and responsible for the global AIDS epidemic. They have been further divided into 9 subtypes and 15 circulating recombinant forms (22; http://hiv-web.lanl.gov/content/hiv-db/CRFs/CRFs.html). The variation in amino acid sequences in the envelope gene among those subtypes can be as high as 30%. This high genetic variability has become a major obstacle to the development of a broadly protective AIDS vaccine and has caused difficulties in gene-based viral detection and viral load measurement. In addition, the variation in amino acid sequences within an infected individual can be as high as 20% (44). This high level of genetic variation is one of the main reasons for the rapid emergence of resistant viral strains during antiretroviral therapy and immune escape mutants in infected individuals. Therefore, it is imperative to study and understand mechanisms of viral mutation and to determine how mutations affect viral biology, drug resistance, and vaccine development.
Mutation rates in HIV-1 genomes have been studied in two systems. One is an in vitro system in which purified RT was used for primer extension of short RNA or DNA templates (2-4, 17, 19, 41, 42). Many factors, including the purity, source, and concentration of RT, the deoxynucleoside triphosphate pool, and other components in the reaction, influence the RT mutation rate in this system. The other system is a single-round viral-replication method (21, 28-30, 33, 38). The mutation rate is studied by determining cDNA sequences after reverse transcription in live cells and is generally not affected by the factors that affect the previously described system. Since these two systems are very different, a wide range of mutation rates has been reported. On average, reported mutation rates are high with purified HIV-1 RT (3× 10−4 to 6 × 10−4) and 10- to 20-fold lower with the single-round infection system (3.4 × 10−5 × 10−5). All previous studies used small DNA or RNA templates, and the overall mutation rate for the complete viral genome could only be extrapolated (2-4, 17, 19, 21, 28-30, 33, 38, 41, 42).
When RT fidelity is studied using partial viral or nonviral gene fragments, it is difficult to fully understand how these mutations affect the biological function of viral genes. To more accurately estimate the viral mutation rate, we have used either the lambda phage library method or the long-range PCR technique in a single-round infection system to obtain and analyze complete and near-full-length HIV-1 genomes. We sequenced a total of 160,000 bp from 19 viral genomes and calculated the mutation rate to be 5.4 × 10−5 per base per replication cycle, which is equivalent to 1.1 mutations in each genome per infection cycle. Inspection of mutations showed that all site mutations in the protein-coding regions were nonsynonymous mutations and that half of the mutations were deleterious. This study is the first to directly examine the mutation rate of the HIV-1 genome by analyzing complete and near-full-length viral genomes.
MATERIALS AND METHODS
Selection of Gpt-resistant cell clones.
The selection of guanosine phosphoribosyl transferase (Gpt) gene-resistant cells was carried out as previously reported (8). Briefly, the R7/3-gpt clone (the env gene was deleted and replaced with simian virus 40-gpt) was cotransfected with an amphotropic murine leukemia virus-env expression vector into 293T cells. HeLa cells (MAGI) were infected with the pseudovirions and selected for Gpt-resistant colonies in selection medium (Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 20 mM HEPES, 14 μg of hypoxanthine/ml, 250 μg of xanthine/ml, and 50 μg of mycophenolic acid/ml). A total of 5,000 colonies were selected from a 100-mm-diameter petri dish and expanded in a T75 flask for 10 days.
Cloning and sequencing of complete HIV-1 genomes from Gpt-resistant cells.
Genomic DNA was extracted from expanded Gpt-resistant cells through the use of a Qiagen blood and tissue kit (Qiagen Inc., Valencia, Calif.) and digested with restriction enzyme XbaI. The 10- to 20-kb genomic DNA fragments were collected from a sucrose gradient (10 to 40%), purified with phenol-chloroform, and cloned in preprepared lambda phage DASHII XbaI arms (Stratagene, Cedar Creek, Tex.). The positive plaques were screened by hybridization with a BH10 full-length genome probe and purified three times using a plaque-to-plaque purification method. The HIV-1 genomes were subcloned into the pBluescript 3.1 vector at the XbaI site. A primer walking method and an automatic ABI 377 sequencer (Applied Biosystems Inc., Foster City, Calif.) were used to sequence the entire HIV-1 genomes on both strands.
Construction of a green fluorescent protein (GFP) proviral clone.
To study the viral mutation rate in lymphocyte cell lines, we established a new reporter gene system by cloning enhanced green fluorescent protein (EGFP) genes in place of approximately 1,000 bp of the beginning of the coding region of gp120 within the HIV-1 molecular clone pNL4-3. Two PCR products were generated using Deep Vent polymerase (New England Biolabs, Beverly, Mass.). The 5′ fragment consisted of the region of HIV-1 NL4-3 from the unique SalI site to the beginning of the env gene open reading frame, and the 3′ fragment consisted of the EGFP gene amplified from pEGFP-N1 (Clontech, Palo Alto, Calif.), including the Kozak sequence upstream of the EGFP start codon. An NheI site was introduced at the 3′ end of the EGFP open reading frame and used for cloning into pNL4-3. A stop codon was introduced at the 5′ end of the EGFP gene in frame with the upstream partial vpu gene to prevent fusion with the unused parallel open reading frame present in the EGFP sequence. The two PCR products were joined by PCR sequence overlap extension to create a 1,165-bp fragment that was cloned directly into pNL4-3, replacing the original 1,465-bp pNL4-3 SalI-NheI fragment. The resulting construct, pNLEnvGFP2, expresses EGFP but lacks Env and Vpu expression. The vpr gene was inactivated by replacing the PflMI-EcoRI vpr deletion fragment from pNLpuroΔvpr (25), yielding the final construct pNLEnvGFP2Δvpr.
Establishment of Jurkat and THP-1 cell lines that a carry pNLEnvGFP2ΔVpr proviral genome.
Plasmids pNLEnvGFP2Δvpr and pVSV-G (vesicular stomatitis virus glycoprotein) were transfected together into 293T cells to produce pseudotyped infectious viruses. Viruses were harvested from the cell culture supernatant at 48 h and used to infect Jurkat or THP-1 cells in the presence of 10 μg of Polybrene/ml. Virus replication was limited to a single round of infection, owing to the lack of de novo Env production within the infected target cells. Infected Jurkat and THP-1 cells were identified by EGFP fluorescence analysis performed using a FACStarPlus apparatus (BD Biosciences, San Jose, Calif.), and individual cells were cloned (using the automatic cell deposition unit single-cell-sorting feature of the FACStarPlus) into 96-well round-bottom plates. The cells were maintained in RPMI 1640 supplemented with 2 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 10% heat inactivated fetal bovine serum, and 57 μM β-mercaptoethanol. After the colonies were grown in cultures for approximately 3 weeks, colonies positive for GFP expression were identified by fluorescence microscopy or flow cytometry and production of HIV-1 Gag antigen (p24) was confirmed. Four Jurkat and three THP-1 cell clones were selected for sequence analysis.
Long-range PCR amplification.
Genomic DNA from expanded cells was extracted with a Qiagen blood and tissue kit (Qiagen Inc.). Near-full-length HIV-1 genomes were amplified (using 0.2 μg of DNA from each cell line) with the primers UP1A (5′-AGTGGCGCCCGAACAGG-3′) and LOW1 (5′-CACAACAGACGGGCACACACTAC-3′) as previously described (9, 10). To avoid PCR error in any particular PCR amplification, ten PCRs were performed and combined for each sample. Nested primers UP2 (5′-GAGCTCTCTCGACGCAGGAC-3′) and LOW2 (5′-TGAGGCTTAAGCAGTGGGTTCC-3′) were used to obtain enough PCR products for direct sequencing in cases in which the PCR yield was low in the first round. Expand High Fidelity PCR system enzyme mix (Roche Molecular Biochemicals, Indianapolis, Ind.) was used to minimize misincorporation during PCR amplification. The combined PCR products were directly sequenced as described for the lambda phage clones.
Construction of a RT mutant.
A two-bp mutation (GG to CA) was introduced into the complementary primers G262A+ (5′-GTTAGTGGCAAAATTGAATTGG-3′) and G262A− (5′-CCAATTCAATTTTGCCACTAAC-3′). The mutation changed the Gly (GGG) codon to the Ala (GCA) codon. Primers PA (5′-CCAGAGGAGCTCTCTCGACGCA-3′) and PB (5′-CCATCCCCTAGCTTTCCCTGA-3′) were located outside of the BssHII and NdeI sites in the R7/3-gpt genome. To construct the RT mutant, two PCRs were performed with primers PA/G262A− and PB/G262A+ (with R7/3-gpt as the template) in a total volume of 100 μl containing 20 mM Tris HCl (pH 8.0), 10 mM KCl, 10 mM (NH4)2SO2, 0.1% Triton X-100, bovine serum albumin (0.1 mg/ml), 0.2 mM deoxynucleoside triphosphate, 10 ng of template DNA, 20 pmol of each primer, and 2.5 U of PFU polymerase (Stratagene, La Jolla, Calif.). The thermocycling conditions were as follows: one cycle of 94°C for 3 min and 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. For the second-round PCR, 1 μl of PCR product from each PCR and the PA and PB primers were used to connect the two PCR fragments under the same thermocycling conditions (with the exception of a 2-min extension at 72°C). We used high-fidelity enzyme (PFU) and 20 thermocycles during the PCR amplification to minimize misincorporation. The final PCR product was isolated from an agarose gel, digested with BssHII and NdeI, and ligated into the R7/3-gpt proviral clone. The insertion was confirmed by sequence analysis.
Inverted PCR amplification.
Genomic DNA (3 μg) was digested with PstI in a volume of 60 μl for 3 h at 37°C. The enzyme was inactivated at 65°C for 10 min. A total of 15 μl of the reaction mixture was subsequently self-ligated with T4 DNA ligase overnight at 16°C. The ligated DNA and two nested primer pairs (for first-round PCR, primer pair gagA [5′-ATCACCTAGAACTTTAAATGCATGGG-3′] and U3A [5′-AATCAGGGAAGTAGCCTTGTGTGTG-3′]; for second-round PCR, primer pair gagB [5′-GGAGCCACCCCACAAGATTTAA-3′] and U3B [5′-GTAGATCCACAGATCAAGG-3′]) were then used to amplify the junction sequence between the cellular DNA and the integrated proviral DNA. The thermocycling conditions for PCR were as follows: 1 cycle of 94°C for 3 min and 30 cycles of 94°C for 1 min, 64°C for 1 min, and 72°C for 4 min followed by 1 cycle of extension at 72°C for 10 min. The PCR products were visualized on a 0.7% agarose gel, purified with a QIAquick gel extraction kit (Qiagen Inc.), and sequenced either directly or from molecular clones.
Data analysis.
Statistical data analyses were performed using S-PLUS 6.1 for Windows, release 1 (Insightful Corp.) (for the calculation of contingency tables), and software written in Fortran 90 by one of the authors (T.B.K.) (for confidence intervals). To compute the expected values of the proportions of synonymous, nonsynonymous, and lethal mutants under the null hypothesis, we wrote a program (in Fortran 90) to examine all possible single-point mutations in the HIV genome. In computing the expected values, transition mutations were weighted more heavily (by 4:1) than transversions. All genes containing the mutation were examined for each point mutation, since there are several areas of gene overlap. A lethal change in at least one gene was scored lethal, a nonsynonymous change in at least one gene was scored nonsynonymous, and all others were scored synonymous.
RESULTS
Mutation rates of the HIV-1 genome in HeLa cells.
To study the HIV-1 mutation rate on the basis of the results of complete viral genome analysis in a single-round infection system, we cotransfected both R7/3-gpt and pMuLV-env DNA into 293T cells and infected HeLa cells with the pseudotyped viruses. After selection and expansion of Gpt-resistant cell colonies, genomic DNA was extracted, digested with XbaI, purified on a sucrose gradient, and ligated into preprepared lambda phage arms. Two clones and four other clones from a previous study were included for sequence analysis (8). The full-length genomes from lambda phage clones were subcloned into pBluescript 3.1 vector and sequenced for both DNA strands. The six full-length HIV-1 genome sequences were compared to the R7/3-gpt sequence. To ensure a precise comparison between the transfected proviral clone and newly integrated proviral DNA, the R7/3-gpt clone was resequenced.
The mutations of the HIV-1 genomes in HeLa cells are summarized in Table 1. One mutation was identified in both the Gpt1 and the M2.1 clones, while two mutations were identified in the Gpt2 clone. Among these mutations, three were transition mutations (G to A) and one was a transversion mutation (T to A). All site mutations resulted in either amino acid changes or stop codons (nonsynonymous mutations). Three other clones (M2.3, M2.B, and H12A) did not have any mutations. No deletions or insertions were found in any clones.
TABLE 1.
The mutation rate of the HIV-1 genome in HeLa cellsa
| Clone | Genome lengths (in bp) (RNA/DNA) | Nucleotide mutation | Posi- tion | Gene | Amino acid change | No. of substi- tutionsb |
|---|---|---|---|---|---|---|
| Gpt1 | 8,638/9,174 | G to A | 8947 | nef | M to I | 1 |
| Gpt2 | 8,638/9,174 | T to A | 2894 | pol | Y to stop | |
| G to A | 5611 | vif | G to R | |||
| vpr | W to stop | 2 | ||||
| M2.1 | 8,480/9,021 | G to A | 2194 | gag | V to I | |
| pol | G to D | 1 | ||||
| M2.3 | 8,480/9,021 | None | 0 | |||
| M2.B | 8,480/9,021 | None | 0 | |||
| H12A | 8,480/9,021 | None | 0 |
The mutation rate was calculated per base per replication cycle. The gpt gene (459 bp) was subtracted from the genome.
Total number of mutations, 4; mutation rate, 3.0 × 10−5.
To calculate the mutation rate in the single-round infection system, three stages in the viral-replication cycle should be taken into consideration. First, after the proviral DNA was transfected into 293T cells the viral genome (R to R; 9,097 bp) was transcribed by the host cell RNA polymerase II and packaged into the virions. Second, viral RNA was reverse transcribed by the viral RT into double-stranded proviral DNA with complete long terminal repeat (LTR) sequences at each end (2 × 9,633 bp). Third, the proviral DNA was then integrated into chromosomes and replicated together with the host genome during cell division. Since the fidelity of DNA polymerase in the host cell is approximately a millionfold higher than that of the RT (7, 23), misincorporation in the proviral DNA in the host cells can be neglected. Because the selective gpt gene was under strong selection pressure, the gpt gene sequence (459 bp) was excluded and 8,638 and 9,174 bp were used for mutation rate estimation for RNA and cDNA genomes, respectively (Table 1). In all later calculations, selective genes (gpt or EGFP) were deducted from the genomes sequenced. Mutations that occurred during plus-strand DNA synthesis had a 50% chance of detection in this system, and the overall mutations were detected using two-and-a-half replication steps. Therefore, a total of 22,399 bp (8,638 and 1.5 × 9,174 bp) were synthesized during one viral-replication cycle. The RNA genome length of the other clones (M2.1, M2.3, M2.B, and H12A) was shorter, because it contains a deletion in the integrase gene. Its viral genome is 8,480 bp long (R to R), and the proviral DNA length is 9,021 bp. The total number of base pairs synthesized is 22,012 bp per viral-replication cycle. Therefore, the average mutation rate per base per replication cycle is 3.0 × 10−5 [3/(22,399 × 2) and 1/(22,012 × 4)]. The genome mutation rate per HIV-1 infection cycles is 0.67 per genome (four mutations in six genomes).
To investigate whether each clone represented one independent infection (especially for the clones that have no mutations [M2.3, M2.B, and H12A]), the cellular sequences adjacent to the viral LTR were compared. The sequences from all six clones had unique cellular flanking sequences, and they were all different from the sequence of the original plasmid DNA used for transfection. This confirmed that all the clones were from independent integration events.
Mutation rates of the HIV-1 genome in lymphocytes.
The lambda phage library method is a reliable way to study the mutation rate of the HIV genome in a single-round infection system, since only high-fidelity DNA polymerase is used to obtain the entire proviral genome. However, this method is time consuming and labor intensive. Recently, we used the long-range PCR technique to obtain near-full-length HIV-1 genomes for the study of viral variation and evolution (9-12). Only 191 bp of the HIV-1 genome is missing in the amplified PCR product. We tested whether the same method could be used to quickly evaluate the nature and rate of mutations of the HIV-1 genome in lymphocyte cell lines, since most previous studies of RT mutation rates were performed on adherent cells (HeLa) that are not the HIV-1 target cells in vivo (28-30, 33).
A selection system for single-round-infected lymphocyte cells was established by replacing the beginning of the env gene in the HIV-1 genome with the EGFP gene. Plasmids pNLEnvGFP2Δvpr and pVSV-G were cotransfected into 293T cells. At 2 days later, pseudotyped viruses were harvested and used to infect either Jurkat (T cell) or THP-1 (monocyte) cells. Since plasmid pNLEnvGFP2Δvpr did not contain the env gene, no de novo Env protein would be synthesized after infection and only one round of infection was completed, as seen with our Gpt selection system. Individual cells expressing GFP were sorted with a FACStarPlus cell sorter and seeded into single wells of a 96-well plate. Following expansion of the clones, GFP expression and the presence of p24 protein were confirmed with fluorescence microscopy and a commercial enzyme-linked immunosorbent assay kit, respectively.
Four Jurkat and three THP-1 cell clones were expanded, and the genomic DNA was extracted. Near-full-length HIV-1 genomes were amplified by nested PCR amplifications. To limit the misincorporation generated during PCR amplification, the following steps were adopted. First, a high-fidelity PCR system was used for PCR amplification (Expand High Fidelity PCR system enzyme mix). This enzyme mix has high efficiency for long-range PCR and higher fidelity than standard Taq polymerase. Second, high copy numbers (at least 32,000 copies) of proviral DNA were used for each PCR. Therefore, random misincorporation in any one template at the initial PCR cycles was diluted out and was not detected by directly sequencing of the PCR products. Third, 10 or more independent PCR amplification reactions were carried out for each sample and were combined before sequencing was performed. Finally, all mutations were confirmed by additional independent PCR amplifications (covering only the mutation regions) and sequence analysis.
Near-full-length HIV-1 genomes from four Jurkat and three THP-1 cell clones were sequenced, and the resulting sequences were compared to the original pNLEnvGFP2Δvpr sequence. Three proviral genomes from THP-1 cell clones were sequenced, and mutations were found in all of them. THP-1.15 had two single-base mutations (Table 2). THP-1.4 had a single-base mutation and two deletions. The first deletion (118 bp) was in the vif gene, and the second deletion (23 bp) was in the tat/rev first exon-overlap region. Direct repeats (ACAG and CA) were found for two deletion mutations in THP-1.4 and THP-1.8 clones (Fig. 1), suggesting that both deletions were the result of template misalignment between direct repeats in the genome, as previously reported (37). Among four HIV-1 genomes from Jurkat cell clones, two (Jurkat.1 and Jurkat.3) were found to have two and three base mutations, respectively, while two others (Jurkat.2 and Jurakat.4) did not carry any mutations. No deletions or insertions were found in the proviral genomes from the Jurkat cell clones (Table 2).
TABLE 2.
Mutation rates of the HIV-1 genome in lymphocyte cellsa
| Clone | Genome length (bp) | Nucleotide mutation | Position(s) | Gene(s) | Amino acid change | No. of substitutionsb | No. of deletionsc | Total no. of mutationsd |
|---|---|---|---|---|---|---|---|---|
| THP-1.8 | 7,537 | dele | 5524-5823 | vif, vpr, tat | Δ100f | 1 | 1 | |
| THP-1.15 | 7,837 | A to T | 942 | gag | L to F | |||
| G to T | 5381 | vif | C to F | 2 | 2 | |||
| THP-1.4 | 7,817 | C to A | 1766 | gag | A to E | |||
| del | 5492-5609 | vif | Δ40g | |||||
| del | 5871-5893 | tat, rev | Δ8g | 1 | 2 | 3 | ||
| Jurkat.1 | 7,837 | C to T | 3000 | pol | Q to stop | |||
| G to A | 5203 | vif | V to I | 2 | 2 | |||
| Jurkat.3 | 7,837 | T to C | 654 | LTR | NAh | |||
| A to T | 3293 | pol | E to D | |||||
| T to A | 5282 | vif | L to stop | 3 | 3 | |||
| Jurkat.2 | 7,837 | None | 0 | |||||
| Jurkat.4 | 7,837 | None | 0 |
The mutation rate was calculated per base per replication cycle. The EGFP gene (720 bp) was subtracted from the genome.
Total number of mutations, 8; mutation rate, 5.9 × 10−5.
Total number of mutations, 3; mutation rate, 2.2 × 10−5.
Total number of mutations, 11; mutation rate, 8.1 × 10−5.
Each deletion (del) was treated as one mutation for the calculation of mutation rates.
In-frame deletion.
Non-in-frame deletion.
NA, not applicable.
FIG. 1.
Sequence alignments of deletion mutations. The top lines represent wild-type sequences; the bottom lines represent sequences with deletion mutations. Dashes stand for deleted bases. Direct repeats at the deletion sites are highlighted with boldface letters. The underlined bases indicate the mismatches in the repeats.
A total of eight site mutations and three deletions were identified in seven proviral genomes. A total of 7,837 bp were sequenced for most of the proviral genomes, with the exception of the THP-1.4 and THP-1.8 genomes (which are 7,817 and 7,537 bp long, respectively). The EGFP gene (720 bp) was excluded from the genomes. As described above, each viral life cycle consists of two-and-a-half genome replication steps. For these HIV-1 genomes, a total of 136,348 bp were synthesized. The site mutation rate and the deletion mutation rate in lymphocyte cells were 5.9 × 10−5 and 2.2 × 10−5 per base per replication cycle, respectively. The total mutation rate (site mutation and deletion) was 8.1 × 10−5. Although this mutation rate is nearly threefold higher than that observed in HeLa cells (3.0 × 10−5), the difference was not statistically significant (P = 0.073). The genome mutation rate in lymphocyte cell lines was 1.6 mutations per genome per infection cycle (11 mutations in seven genomes).
Since PCR-amplified HIV genomes did not contain cellular flanking sequences, it was impossible to determine whether they represented independent clones, especially for the clones that were identical to each other. To rule out this possibility, inverted PCRs were performed to amplify the cellular sequences flanking the 5′ LTR of the integrated HIV-1 proviral genomes and to investigate whether each clone was from an independent infection (5). The sequence analysis of the PCR products indicated that all cellular sequences from the studied clones were unique. Thus, each clone indeed represented an independent infection event.
The mutation rate of the HIV-1 genome with a mutation in the RT gene.
The mutation (G262A) in HIV-1 RT can dramatically increase the dissociation rate constant for templates and primers and decrease its fidelity (1). To investigate whether the mutation in RT would result in an increase of mutations in the single-round infection system, we used a recombinant PCR technique to introduce a 2-bp mutation (GG to CA) at position 262 of RT in the R7/3-gpt genome. The mutation changed Gly to Ala in wild-type RT. The PCR product containing the mutated RT was subsequently cloned into the R7/3-gpt proviral clone at two unique restriction enzyme sites (BssHII and NdeI). Sequence analysis of the mutated clone (R7/3-gptG262A) confirmed the mutations, and no additional mutation was introduced. R7/3-gptG262A and pVSV-G were cotransfected into 293T cells, and pseudotyped viruses were used to infect HeLa cells.
Six clones were expanded, and genomic DNA was extracted. Since the long-range PCR technique proved to be a reliable and much faster means for the study of mutations in a single-round infection system, near-full-length proviral genomes were amplified and sequenced to evaluate the nature and rate of mutations in all six clones (Table 3). Mutations were found in four genomes (G262A.1, G262A.3, G262A.4, and G262A.5); two other genomes (G262A.2 and G262A.6) contained no mutation. G262A.4 and G262A.5 had identical 72-bp deletions. There were two 72-bp long direct repeats in the region, and the mutation deleted one complete repeat. All deletion mutations occurred at the direct repeat sites (Fig. 1).
TABLE 3.
Mutation rates of the HIV-1 genome with an RT mutant (G262A) in HeLa cellsa
| Clone | Genome length (bp) | Nucleotide mutation | Position(s) | Gene(s) | Amino acid change | No. of substitutionsb | No. of insertions or deletionsc | Total no. of mutationsd |
|---|---|---|---|---|---|---|---|---|
| G262A.1 | 7,470 | dele | 6046-6914 | vpu, env, noncoding region | NAf | 1 | 1 | |
| G262A.2 | 8,339 | None | 0 | |||||
| G262A.3 | 8,340 | G to T | 1264 | gag | V to L | 1 | ||
| ins (A)g | 759 | LTR | NA | 1 | 2 | |||
| G262A.4 | 8,267 | A to T | 8939 | nef | R to stop | 1 | ||
| del | 6500-6571 | Noncoding region | NA | 1 | 2 | |||
| G262A.5 | 8,267 | del | 6500-6571 | Noncoding region | NA | 1 | 1 | |
| G262A.6 | 8,339 | None | 0 |
The mutation rate was calculated per base per replication cycle. The gpt gene (459 bp) was subtracted from the genome.
Total number of mutations, 2; mutation rate, 1.6 × 10−5.
Total number of mutations, 4; mutation rate, 3.3 × 10−5.
Total number of mutations, 6; mutation rate, 4.9 × 10−5.
Each deletion (del) was treated as one mutation for the calculation of mutation rates.
NA, not applicable.
ins, insertion.
A total of 49,022 bp were sequenced and analyzed from six HIV-1 genomes. Given the nature of two-and-a-half genome replication steps in each viral life cycle, the site mutation rate per base per replication cycle was 1.6 × 10−5 (2/122,555). The deletion and insertion mutation rate per base per replication cycle (each deletion and insertion was treated as one mutation) was 3.3 × 10−5. The total mutation rate was 4.9 × 10−5 (Table 3). The genome mutation rate was one mutation per genome per infection cycle. Although the overall mutation rate in the HIV-1 genome with the G262A mutation was similar to that in the genome with wild-type RT (P > 0.1), the deletion and insertion mutation rate was significantly increased (P = 0.015).
DISCUSSION
Genetic analysis of HIV genomes has shown the extraordinary heterogeneity among viral isolates (22, 34). HIV genomes show not only high levels of genetic variability between the different viral strains but also significant genetic variation within infected individuals. Groups of closely related viruses within an individual are termed quasispecies (15, 43). It can be envisioned that such a high level of genetic divergence will have a strong impact on viral biology, drug resistance, immune escape, and vaccine development. To fully understand what the driving force is for this high level of genetic variation among HIV-1 genomes, HIV-1 RT has been widely studied for its fidelity. Because different assays were used, however, a broad range of mutation rates (3.4 × 10−5 to 6 × 10−4) has been reported. To limit the variable factors that affect the estimation of RT fidelity in in vitro assays, Mansky and Temin determined that the HIV-1 mutation rate was 3.4 × 10−5 per base per replication cycle in a single-round infection system (32). In all previous studies, however, the mutation rate could only be extrapolated on the basis of small viral or nonviral templates and no information could be obtained on how these mutations affect viral biological phenotypes. To directly estimate the mutation rate of the HIV-1 genome and understand the potential impact of mutations on viral biological phenotypes, we studied the mutation rate of the HIV-1 genome by analyzing complete or near-full-length genomes obtained using either a lambda phage library method or a long-range PCR method.
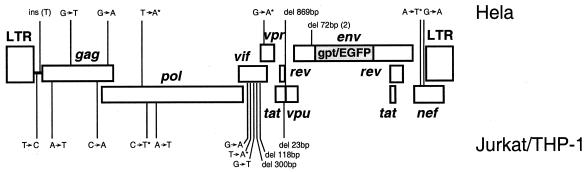
We sequenced 19 complete and near-full-length HIV genomes and found at least one mutation in 12 clones. Seven other clones contained no mutation. The number of the genomes with or without mutations were Poisson distributed. The highest number of mutations identified in any HIV-1 genome was three. A total of 157,993 bases were sequenced, and 21 mutations (14 site mutations, one insertion, and six deletions) were identified (Table 4). The locations of all the mutations are summarized in Fig. 2. The overall mutation rate was estimated at 5.4 × 10−5 per base per replication cycle (21/391,749 bp), which was similar to the rate (3.4 × 10−5) estimated by using the lacZ gene as a template in a single-round infection system (28, 29, 32, 40). The genome mutation rate was estimated at 1.1 mutations per genome per infection cycle (21 mutations among 19 genomes), which was similar to that previously estimated by using small nonviral sequences as target templates (32).
TABLE 4.
Summary of mutations in HIV-1 genomes
| Mutation | Nucleotide change | No. of mutations |
|---|---|---|
| Substitution | G to A | 4 |
| A to T | 3 | |
| T to A | 2 | |
| G to T | 2 | |
| C to A | 1 | |
| C to T | 1 | |
| T to C | 1 | |
| Deletion | Δ23 bp | 1 |
| Δ72 bp | 2 | |
| Δ118 bp | 1 | |
| Δ300 bp | 1 | |
| Δ869 bp | 1 | |
| Insertion | A | 1 |
| Total | 21 |
FIG. 2.
Distribution of mutations in the HIV-1 genome. Asterisks indicate mutations that introduce in-frame stop codons. The deletions and insertions are indicated by del and ins, respectively.
The mutation rate (8.1 × 10−5) in lymphocytes in this study was similar to that (3.0 × 10−5) in adherent HeLa cells (P = 0.073). However, the threefold-higher mutation rate in the lymphocytes suggests different mutation rates among different cells. Jurkat and THP-1 cells are immortalized cells and not natural host cells for HIV-1 infection. The actual mutation rates of the HIV-1 genome should be evaluated in peripheral blood mononuclear cells. Recent studies showed that cells that expressed CEM15/APOBEC3G significantly increased G-to-A hypermutations in HIV-1 genomes (13, 26, 47). This observation strongly supports the notion that mutation rates may be different in different cell types.
We used the long-PCR technique to amplify 13 near-full-length HIV-1 proviral genomes from single-round-infected lymphocyte or HeLa cells. Seventeen mutations (10 site mutations, six deletions, and one insertion) were identified, and all were confirmed by additional independent PCR amplification and sequence analysis. With Expand High Fidelity Taq polymerase, a combination of multiple PCRs, the input of high copy numbers of templates, and the confirmation of the mutations by independent PCRs, the long-range PCR method proved to be a reliable and quick method for the study of the mutation rates of the HIV-1 genome.
Other viral proteins (NC and Vpr) have been reported to influence viral RT fidelity and mutation rates (27, 29, 36). By comparing Vpr+ and Vpr− retroviral vectors, Mansky found that the mutation rate in a Vpr− viral clone was fourfold higher than that in a Vpr+ viral clone (12 × 10−5 versus 3.4 × 10−5) (29). In our system, all proviral clones contained a defective vpr gene but the mutation rate (5.4 × 10−5) was similar to that for a Vpr+ viral vector (P = 0.081). To evaluate whether the mutation rate is higher with Vpr protein when a complete HIV-1 genome is used as the template, a functional vpr gene should be restored in our vectors to reevaluate the mutation rates. The presence of the cellular protein CEM15/APOBEC3G was found to severely diminish retrovirus infectivity through deamination of dC→dU during the minus-strand cDNA synthesis (13, 26, 47). The deamination led to highly biased G-to-A hypermutation and resulted in a high rate of defective genomes and decreased infectivity. The HIV-1 vif gene might be able to counteract the CEM15/APOBEC3G-mediated inhibition of retroviral infections. However, it was not clear whether the G-to-A hypermutation was prevented by the presence of the vif gene. Since CEM15/APOBEC3G-negative cells (293T and HeLa) were used for most of the experiments in this study and all the proviral clones contained the functional vif gene, mutations caused by CEM15/APOBEC3G should be negligible in our analysis.
Near-full-length retroviral genomes and either fingerprint or denaturing gradient gel methods (24, 35) were used to estimate the viral mutation rate for Rouse sarcoma virus and murine leukemia virus. Since no sequence data were obtained in these studies, however, mutations could not be precisely located and the biological impact of mutations could not be evaluated. By sequencing complete viral genomes after single-round infection, we identified 14 single-base mutations. A total of 13 site mutations occurred in the viral protein-coding regions, and all led to amino acid changes. Among 13 nonsynonymous mutations (8 nonsynonymous mutations and five stop codons), the mutation frequencies at each position of the codons were very similar. We found six, five, and four mutations (two extra mutations from the gene overlap regions) at the first, second, and third positions of amino acid codons, respectively. Therefore, the high nonsynonymous mutation rate cannot be explained by a preference for mutations at the first and second positions in amino acid codons. Since (in general) about two-thirds of random mutations lead to amino acid changes, about 8 mutations (among 13 site mutations) were expected to be nonsynonymous mutations. This expectation is significantly different from what was observed in this study (P = 0.0001). The reason why no synonymous mutation was observed is not clear. It is possible that some synonymous mutations will be identified when more genomes are analyzed.
More than one-third of nonsynonymous mutations (5/13) resulted in dysfunctional genes due to premature stop codons (Table 1, Table 2, and Table 3). All six deletions (23, 72, 118, 300, and 869 bp) were either big or not in frame, and they also led to defective genes. These deleterious mutations yielded a lethal mutation rate as high as 52% (11/21). Many other nonsynonymous mutations were changes between very different amino acids or at highly conserved positions. These mutations might affect the protein conformation and/or biological functions. Some of them might result in nonfunctional genes and further increase the ratio of defective genomes.
Amino acid residues 257 to 266 are highly conserved among retroviral pol genes and form part of an α-helix in the thumb subdomain that interacts with DNA (18). To investigate whether G262A mutation would increase the mutation rate in the complete genome analysis, we replaced Gly with Ala in the R7/3-gpt clone. We amplified and sequenced six proviral genomes from HeLa cell clones. A total of six mutations (three deletions, one insertion, and two site mutations) were identified. In the analysis of six proviral genomes in the R7/3-gpt genome in HeLa cells, in contrast, no deletion or insertion mutation was detected with wild-type RT. For deletions or insertion to occur, the primer and template need to dissociate and realign to the template. Therefore, the presence of higher-level deletion and insertion mutations in R7/3-gptG262A clones could be due to the higher dissociation rate between the template and the primer, as reported previously (1). G262A mutations could increase the dissociation rate constant for the template and primer by over 200-fold and could increase the mutation rate by fourfold (1). In our study, however, the site mutation rate with the R7/3-gptG262A clone (1.6 × 10−5) was only about twofold lower than that observed with the wild-type R7/3-gpt clone (Table 1), and the differences in mutation rates between R7/3-gptG262A and R7/3-gpt genomes were not statistically significant (P > 0.1). More sequences are needed from both R7/3-gpt and R7/3-gptG262A clones to clarify the discrepancies between these two studies.
The G262A mutation in RT is also resistant to izovudine (1). The results of this and other studies indicated that HIV-1 genome with drug-resistant mutations may change the mutation pattern and possibly the mutation rate. Many drug resistance-related mutations have been found to modify RT fidelity (4, 20, 31, 45). Since complete genome analysis in a single-round infection system can directly estimate the mutation rate and potential biological influence of the mutations in the HIV-1 genome, viruses that carry mutations resistant to RT inhibitors can be studied in this system to understand how drug-resistant mutations affect the nature and rates of mutations and what their potential biological influence is on viral biological phenotypes.
Acknowledgments
We thank Beatrice Hahn for support and Barton Haynes and Fredric Bibollet-Roche for critical reading of the manuscript.
F.G. was supported by an NIH/NIGMS grant, GM065057.
REFERENCES
- 1.Beard, W. A., S. J. Stahl, H. R. Kim, K. Bebenek, A. Kumar, M. P. Strub, S. P. Becerra, T. A. Kunkel, and S. H. Wilson. 1994. Structure/function studies of human immunodeficiency virus type 1 reverse transcriptase. Alanine scanning mutagenesis of an alpha-helix in the thumb subdomain. J. Biol. Chem. 269:28091-28097. [PubMed] [Google Scholar]
- 2.Bebenek, K., J. Abbotts, J. D. Roberts, S. H. Wilson, and T. A. Kunkel. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 264:16948-16956. [PubMed] [Google Scholar]
- 3.Boyer, J. C., K. Bebenek, and T. A. Kunkel. 1992. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc. Natl. Acad. Sci. USA 89:6919-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer, P. L., and S. H. Hughes. 2000. Effects of amino acid substitutions at position 115 on the fidelity of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 74:6494-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carteau, S., C. Hoffmann, and F. Bushman. 1998. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol. 72:4005-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake, J. W. 1993. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. USA 90:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echols, H., and M. F. Goodman. 1991. Fidelity mechanisms in DNA replication. Annu. Rev. Biochem. 60:477-511. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher, T. M., III, M. A. Soares, S. McPhearson, H. Hui, M. Wiskerchen, M. A. Muesing, G. M. Shaw, A. D. Leavitt, J. D. Boeke, and B. H. Hahn. 1997. Complementation of integrase function in HIV-1 virions. EMBO J. 16:5123-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barre-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, F., D. L. Robertson, S. G. Morrison, H. Hui, S. Craig, J. Decker, P. N. Fultz, M. Girard, G. M. Shaw, B. H. Hahn, and P. M. Sharp. 1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, F., S. A. Trask, H. Hui, O. Mamaeva, Y. Chen, T. S. Theodore, B. T. Foley, B. T. Korber, G. M. Shaw, and B. H. Hahn. 2001. Molecular characterization of a highly divergent HIV type 1 isolate obtained early in the AIDS epidemic from the Democratic Republic of Congo. AIDS Res. Hum. Retrovir. 17:1217-1222. [DOI] [PubMed] [Google Scholar]
- 12.Gao, F., N. Vidal, Y. Li, S. A. Trask, Y. Chen, L. G. Kostrikis, D. D. Ho, J. Kim, M. D. Oh, K. Choe, M. Salminen, D. L. Robertson, G. M. Shaw, B. H. Hahn, and M. Peeters. 2001. Evidence of two distinct subsubtypes within the HIV-1 subtype A radiation. AIDS Res. Hum. Retrovir. 17:675-688. [DOI] [PubMed] [Google Scholar]
- 13.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 14.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 15.Holland, J. J., J. C. De La Torre, and D. A. Steinhauer. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 176:1-20. [DOI] [PubMed] [Google Scholar]
- 16.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 87:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubner, A., M. Kruhoffer, F. Grosse, and G. Krauss. 1992. Fidelity of human immunodeficiency virus type I reverse transcriptase in copying natural RNA. J. Mol. Biol. 223:595-600. [DOI] [PubMed] [Google Scholar]
- 18.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, et al. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji, J., and L. A. Loeb. 1994. Fidelity of HIV-1 reverse transcriptase copying a hypervariable region of the HIV-1 env gene. Virology 199:323-330. [DOI] [PubMed] [Google Scholar]
- 20.Kim, B., J. C. Ayran, S. G. Sagar, E. T. Adman, S. M. Fuller, N. H. Tran, and J. Horrigan. 1999. New human immunodeficiency virus, type 1 reverse transcriptase (HIV-1 RT) mutants with increased fidelity of DNA synthesis. Accuracy, template binding, and processivity. J. Biol. Chem. 274:27666-27673. [DOI] [PubMed] [Google Scholar]
- 21.Kim, T., R. A. Mudry, Jr., C. A. Rexrode II, and V. K. Pathak. 1996. Retroviral mutation rates and A-to-G hypermutations during different stages of retroviral replication. J. Virol. 70:7594-7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuiken, C. L., B. Foley, B. H. Hahn, P. A. Marx, F. McCutchan, J. W. Mellors, J. I. Mullins, J. Sodroski, S. Wolinksy, and B. Korber. 2000. Human retroviruses and AIDS 2000: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 23.Kumar, S., and S. Subramanian. 2002. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. USA 99:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leider, J. M., P. Palese, and F. I. Smith. 1988. Determination of the mutation rate of a retrovirus. J. Virol. 62:3084-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy, D. N., L. S. Fernandes, W. V. Williams, and D. B. Weiner. 1993. Induction of cell differentiation by human immunodeficiency virus 1 vpr. Cell 72:541-550. [DOI] [PubMed] [Google Scholar]
- 26.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 27.Mansky, L. M. 1997. Accessory replication proteins and the accuracy of reverse transcription: implications for retroviral genetic diversity. Trends Genet. 13:134-136. [DOI] [PubMed] [Google Scholar]
- 28.Mansky, L. M. 1996. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS Res. Hum. Retrovir. 12:307-314. [DOI] [PubMed] [Google Scholar]
- 29.Mansky, L. M. 1996. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology 222:391-400. [DOI] [PubMed] [Google Scholar]
- 30.Mansky, L. M., A. E. Krueger, and H. M. Temin. 1995. The bovine leukemia virus encapsidation signal is discontinuous and extends into the 5′ end of the gag gene. J. Virol. 69:3282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansky, L. M., E. Le Rouzic, S. Benichou, and L. C. Gajary. 2003. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J. Virol. 77:2071-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansky, L. M., and H. M. Temin. 1994. Lower mutation rate of bovine leukemia virus relative to that of spleen necrosis virus. J. Virol. 68:494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14:S31-S44. [PubMed] [Google Scholar]
- 35.Monk, R. J., F. G. Malik, D. Stokesberry, and L. H. Evans. 1992. Direct determination of the point mutation rate of a murine retrovirus. J. Virol. 66:3683-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Negroni, M., and H. Buc. 2000. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl. Acad. Sci. USA 97:6385-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc. Natl. Acad. Sci. USA 87:6024-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl. Acad. Sci. USA 87:6019-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 40.Preston, B. D., and J. P. Dougherty. 1996. Mechanisms of retroviral mutation. Trends Microbiol. 4:16-21. [DOI] [PubMed] [Google Scholar]
- 41.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171-1173. [DOI] [PubMed] [Google Scholar]
- 43.Saag, M. S., B. H. Hahn, J. Gibbons, Y. Li, E. S. Parks, W. P. Parks, and G. M. Shaw. 1988. Extensive variation of human immunodeficiency virus type-1 in vivo. Nature 334:440-444. [DOI] [PubMed] [Google Scholar]
- 44.Sharp, P. M. 2002. Origin of human virus diversity. Cell 108:305-312. [DOI] [PubMed] [Google Scholar]
- 45.Wainberg, M. A., W. C. Drosopoulos, H. Salomon, M. Hsu, G. Borkow, M. Parniak, Z. Gu, Q. Song, J. Manne, S. Islam, G. Castriota, and V. R. Prasad. 1996. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 271:1282-1285. [DOI] [PubMed] [Google Scholar]
- 46.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]