Abstract
During the export of flavivirus particles through the secretory pathway, a viral envelope glycoprotein, prM, is cleaved by the proprotein convertase furin; this cleavage is required for the subsequent rearrangement of receptor-binding E glycoprotein and for virus infectivity. Similar to many furin substrates, prM in vector-borne flaviviruses contains basic residues at positions P1, P2, and P4 proximal to the cleavage site; in addition, a number of charged residues are found at position P3 and between positions P5 and P13 that are conserved for each flavivirus antigenic complex. The influence of additional charged residues on pr-M cleavage and virus replication was investigated by replacing the 13-amino-acid, cleavage-proximal region of a dengue virus (strain 16681) with those of tick-borne encephalitis virus (TBEV), yellow fever virus (YFV), and Japanese encephalitis virus (JEV) and by comparing the resultant chimeric viruses generated from RNA-transfected mosquito cells. Among the three chimeric viruses, cleavage of prM was enhanced to a larger extent in JEVpr/16681 than in YFVpr/16681 but was slightly reduced in TBEVpr/16681. Unexpectedly, JEVpr/16681 exhibited decreased focus size, reduced peak titer, and depressed replication in C6/36, PS, and Vero cell lines. The reduction of JEVpr/16681 multiplication correlated with delayed export of infectious virions out of infected cells but not with changes in specific infectivity. Binding of JEVpr/16681 to immobilized heparin and the heparin-inhibitable infection of cells were not altered. Thus, diverse pr-M junction-proximal sequences of flaviviruses differentially influence pr-M cleavage when tested in a dengue virus prM background. More importantly, greatly enhanced prM cleavability adversely affects dengue virus export while exerting a minimal effect on infectivity. Because extensive changes of charged residues at the pr-M junction, as in JEVpr/16681, were not observed among a large number of dengue virus isolates, these results provide a possible mechanism by which the sequence conservation of the pr-M junction of dengue virus is maintained in nature.
The genus Flavivirus within the family Flaviviridae comprises about 73 enveloped RNA viruses that are transmitted by either mosquitoes or ticks or without a known vector (11). For these viruses, a single-stranded RNA genome of about 11 kb encodes a polyprotein, which is cleaved by cellular and viral enzymes into three structural proteins (C, prM/M, and E) and seven nonstructural proteins (54). Virions consist of two envelope proteins, E and prM/M, and an internal C protein, which binds genomic RNA. Differences in antigenicities of E allow the subdivision of flaviviruses into eight antigenic complexes and a number of unclassified viruses, which include the prototype yellow fever virus (YFV) (12). More recent assignments based on nucleotide sequence variations of the nonstructural gene NS5 generally agree with antigenic classifications (49).
The assembly of flaviviruses in the endoplasmic reticulum is followed by modification of the two envelope proteins, E and prM, and virion export through the secretory pathway (54). In addition to N-glycosylation and subsequent modifications, prM (approximately 19 to 23 kDa) is cleaved into a soluble pr peptide and a virion-associated M protein (approximately 8 to 8.5 kDa) by trans-Golgi resident furin (72), resulting in two different forms of virion: the intracellular form, E- and prM-containing virions, and the extracellular from, E- and M-containing virions. The spatial arrangement of E and prM/M is known for both forms and also for a recombinant subviral particle. On the surfaces of extracellular virions of dengue virus, head-to-tail dimers of E lie parallel to the lipid bilayer and 30 sets of three parallel dimers are arranged in a herringbone pattern over a layer of nonexposed M protein (48). A different arrangement (T=1) of E dimers is found on the icosahedral surfaces of subviral particles of another flavivirus, tick-borne encephalitis virus (TBEV) (22). In contrast, three prM-E heterodimers are organized into each of the 60 icosahedrally arranged spikes that project the pr portion of prM out of the surfaces of intracellular dengue virions (85). The pr portion of prM also covers the fusion peptide at the tip of domain II of E in intracellular virions (85). Functionally, E binds cell surface receptors, is involved in envelope fusion to the cellular membrane, and serves as the major target for neutralization by antibodies. The direct role of prM/M during the early phase of infection is not known (11, 54, 65). In the case of dengue virus, some anti-prM monoclonal antibodies cause a low level of neutralization in vitro, but this activity may be due to their cross-reactivities with E (20, 40). On the other hand, it is well established that dengue virus prM and M actively induce a protective immune response, and passively administered anti-prM antibodies protect mice against a lethal challenge (9, 20, 40, 80). The protective effect of a nonneutralizing anti-prM monoclonal antibody was also observed for a Langat virus challenge (39).
Current evidence implicates prM as a chaperone for E during intracellular virion assembly and maturation (54). After cleavage of the flaviviral polyprotein at the C-prM, prM-E, and E-NS1 junctions by host signalase, prM and E noncovalently associate in the endoplasmic reticulum; prM-E heterodimers are subsequently incorporated into immature virions (82). The heterodimeric interaction between prM and E is important for the proper folding and transport of E (47), whereas folding of prM does not require the presence of E (55). Sites located within the predicted α-helical regions of the stem and the membrane-spanning region of E from TBEV are required for stabilization of the prM-E heterodimer (2). During the transport of immature virions through the secretory pathway, prM prevents E from undergoing premature conformational changes and oligomeric rearrangements induced by the acidic pH of the trans-Golgi network (1, 28, 33, 76).
Intracellular virions of flaviviruses remain in the immature form until shortly before release and are then converted to the mature form by cleavage of prM by the cellular proprotein convertase furin, which localizes in the trans-Golgi network (72). In TBEV, cleavage of prM dissociates the prM-E heterodimers and allows E to undergo further structural arrangements, resulting in the acquisition of the ability to induce cell fusion, agglutinate red blood cells, and efficiently infect susceptible cells (72). The cleavage of prM is incomplete in certain preparations of flavivirus, including Langat virus (27, 39), Japanese encephalitis virus (JEV) (43), and Kunjin virus (42). Most notably, cleavage of the dengue virus pr-M junction was consistently found to be ineffective in mosquito cells (3, 34, 61, 64, 65) and Vero cells (32, 61, 64, 81). When prM cleavage is blocked by the treatment of infected cells with acidotropic reagents, the resultant extracellular flavivirus virions with higher proportions of prM are less infectious and do not induce cell fusion at acidic pHs (28, 33, 64). Depending on the specific flaviviruses and cells employed, the infectivity of either intracellular virions or extracellular virions that are released from amine-treated cells diminishes by varying degrees. From several reports, the reduction of specific infectivity of the immature form ranged from 6- to 8-fold for dengue virus (64) to 10-fold for Murray Valley encephalitis virus (28), 50-fold for TBEV (33), and 62-fold for West Nile virus (82). When variations inherent to virus plaque titration are taken into account, acidotropic amines appear to exert a comparatively small effect on the infectivity of dengue virus. Whether this small effect of an acidotropic agent on dengue virus infectivity is related to its incomplete prM cleavage remains unclear.
The cleavage of cellular proproteins by the proprotein convertase furin and other members of the mammalian subtilisin and Kex2p-like serine endoprotease family generally requires basic amino acids with the consensus sequence Arg-Xaa-(Lys/Arg)-Arg (where Xaa is any amino acid) proximal to the cleavage site (reviewed in references 56, 62, 74, 79, and 86). Similar to many furin substrates, the prM of flaviviruses with known insect vectors contains three basic amino acids at positions P1, P2, and P4 of the cleavage site (Table 1). Between positions P5 and P13, additional basic residues are found at locations that are quite conserved in each antigenic complex, ranging from one in dengue virus to four in the JEV antigenic complex. In the latter, some of these conserved basic residues form the minimal furin motif, Arg-Xaa-Xaa-Arg, that is known to be cleaved by furin in a few precursors (10, 31, 45, 57, 59). Moreover, two acidic residues which are highly conserved at positions P3 and P7 in dengue virus are lacking in other flaviviruses (Table 1). It is not known whether these sequence variations affect the prM cleavage efficiency and, consequently, the infectivity of extracellular virions. It also remains unclear how and to what extent such variations in prM cleavage efficiency could affect other in vivo properties of flaviviruses, including specific receptor-mediated cellular entry and tissue tropism. For investigation of the first possibility, the 13-amino-acid region proximal to the pr-M cleavage junction of dengue virus, which contains the lowest positive charge and net charge contents among the three vector-borne flavivirus antigenic complexes with a known prM sequence and YFV, was replaced with those of TBEV, YFV, and JEV, with a full-length cDNA clone of a dengue serotype 2 virus used as the recipient (71). Chimeric viruses were generated by transfecting mosquito cells with capped in vitro transcripts of altered full-length cDNA clones in parallel with that of an unaltered plasmid. A comparison of their in vitro characteristics revealed the extent to which diverse pr-M cleavage junctions of vector-borne flaviviruses affected prM cleavage and other viral properties when tested in a dengue virus background.
TABLE 1.
Comparison of pr-M junction of insect-borne flaviviruses and pr-M junction chimeras
| Virusd | Amino acid sequencea | P1 to P13 chargeb
|
Accession no.e | |||
|---|---|---|---|---|---|---|
| + | − | Net | Setc | |||
| Dengue virus type 1 | YGTCS QTGEH RRDKR↓SVALA PHVGL | 4 | 2 | +2 | 1 | P33478 |
| Dengue virus type 2 | YGTCT TMGEH RREKR SVALV PHVGM | U87411 | ||||
| Dengue virus type 3 | YGTCN QAGEH RRDKR SVALA PHVGM | NC 001475 | ||||
| Dengue virus type 4 | YGTCT QSGERRREKR SVALT PHSGM | M14931 | ||||
| TBEV | YGRCG KQEGS -RTRR SVLIP SHAQG | 5 | 1 | +4 | 1 | U27495 |
| Langat | YGRCG RREGS -RSRR SVLIP SHAQR | P29837 | ||||
| Sofjin | YGRCG KQEGS -RTRR SVLIP SHAQG | X03870 | ||||
| KFDV | YGRCG KPAGG -RNRR SVSIP VHAHS | X74111 | ||||
| Louping ill | YGRCG KQEGS -RTRR SVLIP THAQG | M59376 | ||||
| Powassan | YGRCG RQAGS -RGKR SVVIP THAQK | L06436 | ||||
| Yellow fever | YGKCD SAGRS RRSRR AIDLP THENH | 6 | 1 | +5 | 1 | AF094612 |
| JEV | YGRCT RTRHS KRSRR SVSVQ THGES | 7 | 0 | +7 | 2 | M55506 |
| Kunjin | YGRCT KTRHS RRSRR SLTVQ THGES | D00246 | ||||
| MVEV | YGRCT RARHS KRSRR SITVQ THGES | AF161266 | ||||
| SLEV | YGRCT RMGHS RRSRR SISVQ HHGDS | M16614 | ||||
| West Nile | YGRCT KTRHS RRSRR SLTVQ THGES | NC 001563 | ||||
| 16681Nde(+) | YGTCT TMGEH RREKR SVALV PHVGM | 4 | 2 | +2 | 1 | NA |
| TBEVpr/16681 | YGRCG KQEGS -RTRR SVALV PHVGM | 5 | 1 | +4 | 1 | NA |
| YFVpr/16681 | YGKCD SAGRS RRSRR SVALV PHVGM | 6 | 1 | +5 | 1 | NA |
| JEVpr/16681 | YGRCT RTRHS KRSRR SVALV PHVGM | 7 | 0 | +7 | 2 | NA |
| Cleavage positionf | 15 11 6 1 1′ 6′ 10′ | |||||
Amino acids in bold represent additional charged residues between positions P1 and P13. The arrow indicates the pr-M cleavage site.
For antigenic complexes with known sequences from multiple members, charge contents represent those of the consensus sequence.
Set of nonoverlapping Arg-Xaa-(Lys/Arg)-Arg and Arg-Xaa-Xaa-Arg sequences.
KFDV, Kyasanur Forest disease virus; MVEV, Murray Valley encephalitis virus; SLEV, St. Louis encephalitis virus.
NA, not applicable.
Numbers refer to the positions of the amino acids relative to the pr-M cleavage site in the proximal direction (without apostrophe) and distal direction (with apostrophe).
MATERIALS AND METHODS
Virus and cell lines.
Dengue serotype 2 virus strain 16681, isolated from a patient with dengue hemorrhagic fever in Thailand in 1964, was provided by Bruce Innis and Ananda Nisalak, Department of Virology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand. The virus was grown at 29°C in the C6/36 mosquito cell line in Leibovitz's L-15 medium (GIBCO BRL, Carlsbad, Calif.) supplemented with 1.5% fetal bovine serum (FBS), 0.26 g% tryptose phosphate broth (GIBCO BRL), and glutamine-penicillin-streptomycin solution and was stored in 20% FBS at −70°C. The PS clone D cell line (83) was maintained in L-15 medium containing 10% FBS, 0.26 g% tryptose phosphate broth, and glutamine-penicillin-streptomycin solution at 37°C. Vero cells were maintained in Dulbecco's modified Eagle medium supplemented with 5% FBS and glutamine-penicillin-streptomycin solution in 5% CO2 in humidified air at 37°C.
Antibodies.
A pool of high-titer sera taken during convalescence from patients with dengue hemorrhagic fever at Siriraj Hospital, Mahidol University, Bangkok, Thailand, was used as an anti-dengue virus polyclonal antibody. A rabbit anti-dengue virus serotype 2 antiserum was a gift of Toshihiko Fukunaga, University of the Ryukyus, Nishihara, Okinawa, Japan. Monoclonal antibodies specific for flavivirus E protein (4G2 and 1D10), dengue virus serotype 2 E protein (3H5), prM protein (2H2 and 4C1), and NS1 protein (1B2 and 1A4) (35, 69; W. Kasinrerk and P. Malasit, unpublished results) were used in the form of ascites or culture supernatant.
Virus titration and plaque size measurement.
Virus plaque titration was performed in 24-well tissue culture plates with the PS cell line (83) as described previously (4). A focus immunoassay for virus enumeration was performed with PS cells or C6/36 cells in 96-well cell culture plates as described previously (70), with the following modifications. The virus was serially diluted with L-15 medium containing 3% FBS, and 50 μl of each dilution was added to each well of 3-day-old confluent cell monolayers for 2 h at 37°C (PS cells) or 29°C (C6/36 cells), with intermittent manual shaking. An overlay of L-15 medium containing 1.5% FBS, 0.26 g% tryptose phosphate broth, penicillin-streptomycin-glutamine solution, and 1.5% carboxymethylcellulose (Sigma Chemicals, St. Louis, Mo.) was added, and plates were incubated further at 37°C for 3 days (PS cells) or 29°C for 4 days (C6/36 cells). Foci of infected PS cells were visualized after fixing with 3.7% formaldehyde in phosphate-buffered saline (PBS) and permeabilizing with 2% Triton X-100 in PBS by reacting them successively either with monoclonal antibody 4G2, rabbit anti-mouse immunoglobulin antibody, sheep anti-rabbit immunoglobulin antibody, peroxidase-rabbit antiperoxidase complex, and H2O2-diaminobenzidine or with 4G2, alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (IgG) heavy-plus-light-chain antibody, and BCIP-NBT mixture (5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium) (Promega, Madison, Wis.). The visualization of infected C6/36 foci was performed similarly, but with either 4G2, alkaline phosphatase-conjugated goat anti-mouse IgG heavy-plus-light-chain antibody, and BCIP-NBT mixture or with 4G2 and fluorescein isothiocyanate-conjugated rabbit anti-mouse immunoglobulin antibody (68). Virus concentrations were expressed in focus-forming units (FFU) per milliliter.
For measurement of plaque size, infected PS cell monolayers in 24-well tissue culture plates were incubated under a 1.5% carboxymethylcellulose overlay for 8 days, washed three times with PBS, fixed and stained with glacial acetic acid-naphthol blue-black solution, and dried in air. Plaque diameters were determined by using a vernier caliper with a 0.05-mm accuracy (Mitutoyo, Tokyo, Japan). Focus size measurements were performed with infected PS cells under a 1.5% carboxymethylcellulose overlay after 3 days of incubation. Infected foci were fixed, permeabilized, and visualized by successively reacting them with 4G2, rabbit anti-mouse immunoglobulin antibody, sheep anti-rabbit immunoglobulin antibody, peroxidase-rabbit antiperoxidase complex, and H2O2-diaminobenzidine as for the focus immunoassay, but the virus inoculum was adjusted to result in 15 to 20 foci or less per well for the parent virus, TBEVpr/16681, and YFVpr/16681 chimeras and 50 foci or less per well for the JEVpr/16681 chimera. Isolated foci were then photographed by using the 10× objective lens of an inverted microscope, and the number of infected cells per focus was determined.
Generation of pr-M cleavage junction chimeras.
A full-length cDNA clone of dengue virus type 2, strain 16681, based on pBluescript II KS (Stratagene, La Jolla, Calif.) was described previously (71). It was assembled by ligation of a 5′ half-genome containing viral cDNA nucleotides (nt) 1 to 4497 with a 3′ half-genome containing the nt 4497 to 10723 sequence (numbering was according to references 7 and 44) and propagation in Escherichia coli strain DH5α F′ at 22 to 25°C in Luria broth containing 25 μg of ampicillin/ml. Two restriction sites, for NdeI and BamHI, were introduced into a cDNA subclone containing nt 1 to 1547 of the strain 16681 cDNA at nt 666 and 709, respectively, by using a PCR-based site-directed mutagenesis scheme (QuikChange; Stratagene) (Table 2). The introduction of the NdeI recognition sequence did not alter the amino acid sequence, whereas the BamHI modification caused an arginine-to-glycine substitution at amino acid position 205. The oligonucleotides used are listed in Table 2. Recombinant plasmids with the desired restriction sites were screened by restriction enzyme digestion, confirmed by nucleotide sequence analysis, and used for chimera construction. Oligonucleotide pairs encoding amino acid positions P1 to P14 of the pr-M cleavage junction of dengue virus strain 16681 (GenBank accession no. U87411), YFV (GenBank no. AF094612), TBEV (GenBank no. U27495), and JEV (GenBank no. M55506) were synthesized with a 5′ NdeI end and a 3′ BamHI end and then were ligated into the NdeI/BamHI-digested plasmid subclone; ligation regenerated the NdeI site, but the BamHI site was abolished and amino acid 205 reverted to arginine. Restriction sites were included in oligonucleotides to aid in the screening of recombinant clones (Table 2). Following nucleotide sequence verification of the inserted oligonucleotides, the chimeric sequences were transferred to the 5′ half-genome by first cutting a 1.3-kb PstI fragment containing cDNA nt 212 to 1535 out of the chimeric cDNA subclones and then ligating it into a 6-kb PstI-digested 5′ half-genome plasmid. Following a check for the correct orientation of the inserted PstI fragment, the chimeric 5′ half-genome was digested with KpnI (nt 4497) and ligated with a 6.2-kb KpnI fragment containing nt 4497 to 10723 of a 3′ half-genome to generate the full-length chimeric cDNA plasmid. Transformation of the full-length plasmid into E. coli strain DH5α F′, selection, preparation of plasmid DNA, and in vitro transcription with SP6 RNA polymerase were done as described previously (71). In vitro transcripts were digested with RNase-free DNase (Promega) and purified with an RNeasy mini kit (Qiagen, Valencia, Calif.), and the total RNA concentration was measured by using a spectrophotometer. For quantitation of the full-length transcripts, DNase-digested, affinity column-purified in vitro transcripts were mixed with loading buffer containing 62.5% (vol/vol) formamide and 1.14 M formaldehyde, heated at 65°C for 10 min, electrophoresed in 0.7% agarose gels containing 2 M formaldehyde, and stained with ethidium bromide. Stained gels were photographed under UV light, and the proportion of full-length transcript to total RNA was determined by scanning the photograph at a high resolution (GS-700 imaging densitometer; Bio-Rad, Hercules, Calif.) and analyzing it with Molecular Analyst software (Bio-Rad). The concentration of full-length in vitro transcripts was then calculated from the total RNA concentration and the proportion of full-length transcripts.
TABLE 2.
Oligonucleotides for site-directed mutagenesis and chimera construction
| Oligonucleotide | Type of oligonucleotide | Nucleotide sequencea (5′→3′) |
|---|---|---|
| NdeI addition | Coding | GGGTAACATATGGGACGTGTACCACCATGGC |
| Noncoding | CCCATATGTTACCCACGTGGACGTAGAGTTGC | |
| BamHI addition | Coding | AAAAGGATCCGTGGCACTCGTTCCACATG |
| Noncoding | CCACGGATCCTTTTTCTCTTCTATGTTCTC | |
| Dengue pr-M | Coding | TATGGGACGTGTACCACGATGGGAGAACATAGAAGAGAAAAAA |
| Noncoding | GATCTTTTTTCTCTTCTATGTTCTCCCATCGTGGTACACGTCCCA | |
| TBEV pr-M | Coding | TACGGACGCTGTGGGAAACAGGAAGGCTCACGGACGCGTA |
| Noncoding | GATCTACGCGTCCGTGAGCCTTCCTGTTTCCCACAGCGTCCG | |
| YFV pr-M | Coding | TATGGTAAGTGTGACTCCGCGGGCAGGTCTAGGAGGTCAAGAA |
| Noncoding | GATCTTCTTGACCTCCTAGACCTGCCCGCGGAGTCACACTTACCA | |
| JEV pr-M | Coding | TATGGACGGTGCACGCGGACCAGGCATTCCAAGAGATCTAGGA |
| Noncoding | GATCTCCTAGATCTCTTGGAATGCCTGGTCCGCGTGCACCGTCCA |
Bold bases indicate a point mutation introduced to abolish the NcoI site. Underlined sequences in the TBEV pr-M, YFV pr-M, and JEV pr-M oligonucleotides indicate engineered restriction enzyme sites for MluI, SacII and BglII, respectively.
Transfection of the C6/36 cell line was performed by mixing 1 μg of capped, full-length in vitro transcripts with 5 μl of Lipofectin (GIBCO BRL) in 1 ml of L-15 medium before adding them onto twice-washed confluent cell monolayers in 35-mm-diameter dishes. After a 4-h incubation at room temperature, the RNA-Lipofectin mixture was removed and L-15 maintenance medium containing 1.5% FBS was added. Transfected monolayers were incubated at 29°C for up to 2 weeks. The resultant plasmid-derived 16681 virus containing an additional NdeI site and the three pr-M junction chimeric viruses were amplified for 1 to 2 passages in C6/36 cells and were then titrated. When the entire structural genes (for C, prM, and E) of the plasmid-derived parent virus and chimeric viruses were reversed transcribed, amplified, and sequenced, the presence of appropriate pr-M junctions was confirmed and no spurious mutation of the three genes was detected. The viruses were designated 16681Nde(+), YFVpr/16681, TBEVpr/16681, and JEVpr/16681.
Virus purification and immunoblot analysis.
Virions present in the culture fluid of infected C6/36 cells were precipitated with 7 g% polyethylene glycol 8000 and 400 mM NaCl. Viral pellets were collected by centrifugation at 15,000 × g at 4°C for 20 min and were suspended in 10 mM Tris (pH 7.2)-2 mM EDTA-150 mM NaCl. The concentrated virus suspension was laid onto a 10 to 50 g% linear sucrose gradient in the same buffer and centrifuged at 210,000 × g at 4°C for 22 h in a Kontron TST 41.14 or Beckman SW40Ti rotor. Fifteen fractions of 750 μl each were collected by the upward displacement method, and the presence of infectious virions was determined by a focus immunoassay on PS cells. Virions present in the fraction with the highest titer were disrupted by the addition of sodium dodecyl sulfate (SDS) to 1 g%, without boiling. Viral proteins were separated by electrophoresis in a 0.1% SDS-15% polyacrylamide gel (50) in the absence of a reducing agent and were blotted onto a nitrocellulose or polyvinyl difluoride membrane by use of a semidry blotting apparatus. Detection was performed with pooled, convalescent-phase human sera, rabbit anti-dengue virus antiserum, or murine monoclonal antibodies against E, prM, and NS1 proteins and with appropriate enzyme-conjugated anti-IgG antibodies and substrates. A prestained protein ladder (Benchmark; GIBCO BRL) was employed as a molecular weight marker.
Metabolic labeling and quantitation of prM/M content.
A monolayer of C6/36 cells was infected with dengue virus at a multiplicity of infection of 1 FFU/cell. Twenty-four hours after infection, the cells were starved for 1 h at 37°C in methionine- and cysteine-free Dulbecco's modified Eagle medium containing 1.5% FBS and subsequently supplemented with an l-[35S]methionine and l-[35S]cysteine in vitro cell labeling mixture (Redivue Pro-mix; Amersham, Piscataway, N.J.) to a concentration of 50 μCi/ml. After 16 h of labeling, the culture medium was harvested for use in copanning of virion-associated prM and M with an anti-E monoclonal antibody. The culture medium was first incubated with normal mouse serum, which was immobilized on a rabbit anti-mouse immunoglobulin antibody-coated 96-well plate, at 4°C for 3 h. The precleared culture fluid was then transferred to another 96-well plate, which was coated successively with rabbit anti-mouse immunoglobulin antibody and 3H5, to selectively capture dengue virus. After nonspecific proteins were removed by extensive washing with PBS, captured viruses were lysed with 1× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (without the reducing agent), boiled for 5 min, and analyzed on a 16.5% Tris-Tricine SDS-PAGE gel (67). The gel was fixed with isopropanol-acetic acid-water (25:10:65), immersed in Amplify fluorographic reagent (Amersham), dried, and exposed to X-ray film (XAR-5; Kodak, Rochester, N.Y.). Alternatively, viral proteins were visualized and quantitated by exposing the dried gel to a storage phosphor screen and analyzing it with a phosphorimager (Typhoon 9210; Amersham). Labeled bands were identified as E, prM, and M proteins based on their electrophoretic mobilities and sizes. The background signal was subtracted from E, prM, and M bands in all cases.
Comparison of infectivities.
The specific infectivities of viral samples were determined by comparing infectious virus titers with the total number of virions. The infectious titer was determined by using a focus immunoassay in PS or C6/36 cells as described above. A quantitative reverse transcriptase (RT) PCR method (51) was used to determine the number of virions by measuring the viral RNA content. Briefly, genomic RNA present in a 140-μl viral sample was extracted with a viral RNA mini kit (QIAGEN), reverse transcribed, and amplified in the presence of a dengue virus type 2-specific TaqMan fluorogenic probe in an ABI 7700 thermocycler (Applied Biosystems). An in-house dengue virus type 2 RNA standard was prepared by in vitro transcription of a cDNA subclone containing nt 7737 to 7874 directly linked with nt 9659 to 10723 of strain 16681 by use of T7 RNA polymerase, digestion with RNase-free DNase, and purification with an RNeasy mini kit (Qiagen). Visual inspection of an ethidium bromide-stained RNA standard after electrophoresis in a 1% formaldehyde-agarose gel revealed that the great majority of RNA molecules run as a single band with a very faint smear, indicating that most of them are intact. From the known sequence of the RNA standard and the RNA concentration, as determined by UV absorption, the molecular concentration of the RNA standard was determined. The RNA standard was then diluted to 1013 molecules/ml and stored at −80°C in small aliquots for single use. From 104 to 1012 molecules of RNA standard/ml were employed in the RT-PCR, along with unknown RNA samples, with the following parameters: reverse transcription, 50°C for 30 min; denaturation, 94°C for 5 min; and 45 cycles of 94°C for 15 s, 55°C for 1 min, and 72°C for 30 s. At the end of the reaction, a relationship between the input standard RNA concentrations and the cycle numbers at which the fluorescence signal reached a threshold level was plotted and then used to determine the concentrations of genomic RNA in unknown samples. The specific infectivity of a viral sample (in FFU per copy) was calculated by dividing the virus titer by the virion concentration. Since viruses were grown in either C6/36 or PS cells and then titrated in at least one of the two cell lines, specific infectivities were determined separately for four multiplication-titration categories: (i) viruses grown in C6/36 cells and titrated in C6/36 cells (C6-C6), (ii) viruses grown in C6/36 cells and titrated in PS cells (C6-PS), (iii) viruses grown in PS cells and titrated in PS cells (PS-PS), and (iv) viruses grown in PS cells and titrated in C6/36 cells (PS-C6). For an assessment of changes in the specific infectivities of pr-M junction chimeras from that of the 16681Nde(+) parent, a ratio of specific infectivity between each chimera and 16681Nde(+) was derived for each of the multiplication-titration categories. Comparisons were made only among sets of the three chimeric viruses and 16681Nde(+) that were grown concurrently and quantitated in the same plates during virus titration and genome measurement in order to minimize variations.
Measurement of cell-associated and extracellular virus production.
Two-day-old PS cell monolayers in 35-mm-diameter plastic dishes (106 cells/dish) were infected with dengue virus at a multiplicity of infection of 1 FFU/cell for 2 h. Unbound viruses were washed off extensively with L-15 medium; 1 ml of L-15 medium supplemented with 3% FBS was then added to infected monolayers, which were incubated at 37°C in humidified air for 0, 4, 12, 14, 16, 20, 24, 36, and 48 h. At the end of each incubation period, the culture fluid was removed, centrifuged to sediment detached cells, supplemented with FBS to a concentration of 20%, and stored in aliquots at −70°C. Concurrently, infected monolayers were washed extensively to remove remaining extracellular viruses and loosely bound viruses. Cell-associated viruses were then liberated by adding 1 ml of L-15 medium supplemented with 20% FBS, followed by three cycles of freezing at −70°C and thawing at 4°C. After the cellular debris was removed by spinning, the cell lysate was either used directly for virus titration or stored in small aliquots at −70°C until use. Virus titers in the culture fluid and cell lysate were assessed by using a focus immunoassay with PS cells. Titration experiments with strain 16681 revealed that one, two, or three cycles of freezing and thawing of infected PS monolayers yielded similar virus titers, and further storage of cell lysates in small aliquots at −70°C did not reduce infectious virus titers.
Comparison of virus-heparin binding.
For a comparison of the affinities of virus binding to immobilized heparin, 108 FFU were concentrated by centrifugation (23,000 × g or, in the case of JEVpr/16681, 210,000 × g) at 4°C for 90 min, suspended gently in 1 ml of 0.5% bovine serum albumin (BSA) in 5 mM sodium phosphate (pH 7.5)-100 mM NaCl, loaded via a peristaltic pump into a 1-ml porcine heparin-Sepharose column (HiTrap heparin; Amersham), and eluted with a 0.1 to 1 M linear gradient of NaCl in 0.5% BSA-5 mM phosphate buffer, pH 7.5. One-milliliter fractions were collected, and the NaCl concentration was determined by using a cryoscopic osmometer (Osmomat 030; Gonotec GmbH, Berlin, Germany). Viruses in the column fractions were detected by dot immunoassays or enzyme-linked immunoassays. For dot immunoassays, 150 μl of sample was applied onto a nitrocellulose membrane by using a 96-well suction manifold. The dotted membrane was blocked with 5% skim milk in PBS for 1 h and reacted successively with 3H5 or 4G2 monoclonal antibodies and horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin antibody, H2O2, and diaminobenzidine-CoCl2 in PBS. The color signal from the membrane was recorded and stored digitally without data compression by using a digital still camera (MVC-CD1000; Sony, Tokyo, Japan). The signal densities from dotted areas were then compared in a gray-scale format by using Molecular Analyst software (Bio-Rad) after subtracting the background density of undotted areas. For ease of comparison, a column fraction displaying the highest dot density in a membrane was assigned an arbitrary value of 50 and the density of other fractions was adjusted proportionally.
For enzyme-linked immunoassays, a 96-well plate (F96 MaxiSorp; Nunc, Naperville, Ill.) was coated with 1D10, an anti-dengue virus E antibody, in PBS at 4°C overnight and blocked with 0.5% BSA in PBS before use. A 150-μl portion of the column fraction was applied to each well, incubated at 37°C for 1 h, and washed with PBS containing 0.05% Tween 20. Captured viruses were detected by reacting them successively with a dengue convalescent-phase serum, horseradish peroxidase-conjugated anti-human immunoglobulin antibody, H2O2, and o-phenylenediamine HCl in sodium citrate buffer (pH 5.0), and the color intensity was recorded with a microplate reader (EL311s; Bio-Tek, Winooski, Vt.). As a negative control and a positive control, the culture medium of C6/36 cells and a serially diluted dengue virus prepared in C6/36 cells, respectively, were employed in each plate.
Inhibition of virus infection by heparin.
A fixed amount of dengue virus was mixed with porcine intestinal mucosal heparin (188 U/mg; Sigma) at 0, 6.25, 12.5, 25, 50, 100, 200, and 400 μg/ml and incubated on ice for 5 min. The virus-heparin mixture was then diluted 200-fold with ice-cold L-15 medium and transferred to PS monolayers in 96-well plates (a 12-well row was used for each heparin concentration) for an additional 5 min on ice to allow for virus-cell binding. After unbound viruses were washed away extensively with L-15 medium, a 1.5% carboxymethylcellulose overlay was added and the plates were incubated at 37°C for 3 days. Foci of infected cells were then visualized by a focus immunoassay and counted. For each virus, the optimal amount of input virus was predetermined such that, in the absence of heparin, no more than 60, 70, and 85 foci were present in wells infected with 16681Nde(+) and YFVpr/16681, TBEVpr/16681, and JEVpr/16681, respectively. The concentrations of heparin which caused 50 and 80% reduction of infected foci compared with the row receiving no heparin were then determined by the method of Reed and Muench (25).
RESULTS
Influence of cleavage-proximal sequences on dengue virus prM cleavage.
Comparison of the 13-amino-acid sequence proximal to the pr-M cleavage site of many vector-borne flaviviruses revealed sets of charged amino acids that are differently conserved. Basic residues at positions P1, P2, and P4, or the consensus furin motif, were conserved in all viruses examined, whereas additional charged residues at positions P3, P5, P7, P8, P10, and P13 were found only within one or a few antigenic complexes. The sequence diversity manifests a gradation of positive charge, net charge, and furin cleavage motif, ranging from the smallest in the dengue virus antigenic complex to the largest in the JEV antigenic complex (Table 1). The influence of these diverse sequences on prM cleavage was tested by replacing the dengue virus sequence with those of TBEV, YFV, and JEV and by comparing the prM and M content of extracellular viruses. When extracellular virions from infected C6/36 cultures were disrupted with SDS and subjected to SDS-PAGE and immunoblot analysis using rabbit anti-dengue virus type 2 antiserum, comparatively less prM was observed for YFVpr/16681 and JEVpr/16681 than for TBEVpr/16681 and the 16681Nde(+) parent (Fig. 1A). In this immunoblot analysis, an additional band of <9 kDa was also observed. This protein was a secreted protein from C6/36 cells, but was not dengue virus M protein, as several monoclonal antibodies raised against this protein reacted with uninfected C6/36 cells in an indirect immunofluorescence assay and a fluorescence-activated cell sorter analysis (data not shown). As previously documented, dengue virus C and M were not detectable in an immunoblot analysis of virions prepared by SDS lysis (20). When viruses were purified by sucrose density gradient centrifugation, immunoblot analysis of the fractions with the highest virus titers yielded the same results, but with minimal contamination with the secreted C6/36 protein (Fig. 1B). Subsequent tests of other sucrose gradient fractions by immunoblot analysis and enzyme-linked immunoassay revealed that subviral particles were present in negligible quantities in these virus preparations (data not shown). The observed reduction of prM in the gradient-purified, infectious fractions of YFVpr/16681 and JEVpr/16681 indicated that a reduced prM content is an inherent property of these virions. The minimal level of subviral particles, together with a very similar reduction in prM reactivity in YFVpr/16681 and JEVpr/16681 when tested by using the infected culture media (Fig. 1A) and the purified fractions (Fig. 1B), strongly suggested that the changes in prM content detected in the infected culture media were mainly contributed by the virions rather than by the variation in the level of subviral particles, which may contain different amounts of prM from virions. Further comparisons revealed that the reduction of prM in YFVpr/16681 and JEVpr/16681 was more evident in viruses harvested from an early stage of infection, which was associated with a low level of mosquito cell fusion, than from a late stage of infection.
FIG. 1.
Comparison of prM and M by immunoblot analysis and phosphorimager analysis. (A) Extracellular virions in infected C6/36 culture fluid were pelleted (50,000 × g for 90 min), disrupted with SDS, separated in a nondenaturing SDS-polyacrylamide gel, and transferred to a polyvinyl difluoride membrane. Viral proteins were visualized with rabbit anti-dengue 2 antiserum and alkaline phosphatase-conjugated anti-rabbit immunoglobulin antibody. Molecular sizes (in kilodaltons) are indicated on the left. NS1 dimer, E, and prM bands were identified based on size and reactivity with specific monoclonal antibodies. P, strain 16681Nde(+). In contrast to previous findings (23, 52, 84), NS1 was routinely detected in infected C6/36 culture medium in our laboratories, probably reflecting the heterogeneity of the C6/36 cell line employed. This observation agreed with findings that dengue virus NS1 and JEV NS1 can be secreted from Spodoptera frugiperda (Sf9) cells (24, 52). (B) Virions were first purified by sucrose density gradient centrifugation. The fractions with the highest virus titers were chosen for SDS-PAGE and immunoblot analysis. (C) Infected C6/36 cells were labeled with [35S]methionine and [35S]cysteine for 16 h. Extracellular virions were captured on 3H5-coated plates and disrupted with SDS. A total of 3,000 cpm of labeled proteins was electrophoresed in an SDS-polyacrylamide gel and visualized by fluorography.
To confirm that reduced prM in YFVpr/16681 and JEVpr/16681 resulted from enhanced cleavage rather than an alteration of prM antigenicity or diminished prM incorporation into virions, coimmunoprecipitations were performed. Twenty-four hours after C6/36 cell infection, chimeric viruses and the parent virus were radiolabeled and then subjected to panning with 3H5, which recognizes a linear epitope on domain III of the dengue virus serotype 2 E protein (36). Following lysis of the captured virions with SDS, an analysis of disrupted viral proteins with SDS-PAGE and fluorography (Fig. 1C) or phosphorimager detection (data not shown) revealed that the reduction of prM in YFVpr/16681 and JEVpr/16681 was accompanied by a reciprocal increase of M protein. The results indicated that cleavage of prM is indeed enhanced in YFVpr/16681 and JEVpr/16681. For this SDS-PAGE analysis, the Tris-Tricine buffer system was selected to optimize the quality of the M protein band, but the apparent mobilities of E and prM were lower than in Laemmli's Tris-glycine system. Quantitation of the radioactivity from viral protein bands was then attempted with phosphorimager analysis under conditions that depicted closely the parallel relationship between the total input radioactivity and the signal, as registered by a phosphorimager, with E and M proteins (data not shown). The average prM and M virion contents were then calculated from the prM/(prM plus M) and M/(prM plus M) ratios, with the assumptions of uniform labeling of prM and M and comparable contributions from labeled methionine and cysteine. When the total number of prM plus M was taken to be 180 molecules/virion, as was demonstrated for E (48), we found that prM was reduced from about 40 to 60 molecules/virion in the 16681Nde(+) parent to the lowest level, 9 molecules/virion, in JEVpr/16681 (Table 3). Cleavage of prM in YFVpr/16681 was also enhanced, but not as much as JEVpr/16681, whereas cleavage of TBEVpr/16681 prM was slightly reduced. In two separate phosphorimager analyses, the E/(prM plus M) ratio varied between 0.74 and 1.10 for the four viruses, consistent with the equimolar relationship between E and prM plus M. These results indicated that prM cleavage in dengue virus is affected by cleavage-proximal sequences other than the conserved basic P1, P2, and P4 residues and that replacement with a 13-residue, cleavage-proximal sequence derived from JEV, and to a lesser extent, YFV, results in enhanced prM cleavage.
TABLE 3.
Quantitation of prM and M proteins in anti-E antibody-captured, radiolabeled virions
| Expt or virus | Signala
|
Ratio of protein to prM plus M
|
Avg content (molecules/virion)
|
|||||
|---|---|---|---|---|---|---|---|---|
| E | prM | M | E | prM | M | prM | M | |
| 1b | ||||||||
| 16681Nde(+) | 162,891.5209 | 48,397.6100 | 164,219.2740 | 0.7661 | 0.2276 | 0.7724 | 41.0 | 139.0 |
| TBEVpr/16681 | 48,565.0791 | 16,890.5533 | 48,409.1940 | 0.7437 | 0.2587 | 0.7413 | 46.6 | 133.4 |
| YFVpr/16681 | 117,955.0130 | 13,305.1527 | 115,688.1220 | 0.9144 | 0.1031 | 0.8969 | 18.6 | 161.4 |
| JEVpr/16681 | 147,356.1212 | 8,114.1520 | 153,033.3740 | 0.9144 | 0.0504 | 0.9496 | 9.1 | 170.9 |
| 2b | ||||||||
| 16681Nde(+) | 71,379.9700 | 28,677.6775 | 56,867.4420 | 0.8344 | 0.3352 | 0.6648 | 60.3 | 119.7 |
| TBEVpr/16681 | 46,002.1997 | 19,127.6053 | 22,866.5080 | 1.0954 | 0.4555 | 0.5445 | 82.0 | 98.0 |
| YFVpr/16681 | 42,981.8736 | 5,613.1967 | 33,388.4380 | 1.1021 | 0.1439 | 0.8561 | 25.9 | 154.1 |
| JEVpr/16681 | 64,252.4861 | 3,758.6333 | 67,755.3180 | 0.8985 | 0.0526 | 0.9474 | 9.5 | 170.5 |
Signal from an equal area of E, prM, and M bands was subtracted from the background and corrected with the following methionine-plus-cysteine contents: E, 33; 16681Nde(+) prM, 16; TBEVpr/16681 prM, 15; YFVpr/16681 prM, 15; JEVpr/16681 prM, 15; and M, 5.
A total of 5,000 and 3,000 cpm of labeled viral proteins was employed in the first and second experiments, respectively.
Alteration of plaque size and titer and replication kinetics.
Quantitation of chimeric viruses generated by transfection and subsequent expansion in mosquito cells by plaque assays unexpectedly revealed a reduction in plaque size that was most pronounced in JEVpr/16681 (Fig. 2 and Table 4). Two independent sets of chimeric viruses were used to infect PS cells, and the infected foci were visualized by immunological staining. The mean number of infected cells within an isolated focus (focus size) showed as much as 88 or 91% reduction with JEVpr/16681 compared to those with the 16681Nde(+) parent (Table 4). A marked reduction of plaque size and focus size in JEVpr/16681 correlated with a lower virus titer, as determined with either PS cells or C6/36 cells. For TBEVpr/16681 and YFVpr/16681, some reductions in focus size were observed, but they were not accompanied by a consistent decrease in virus titer. It is notable that foci of JEVpr/16681 and YFVpr/16681 appeared to be more compact than those of TBEVpr/16681 and the parent virus (Fig. 2). JEVpr/16681 was passaged up to four times in C6/36 cells and the reduction of focus size and titer was consistently observed (data not shown).
FIG. 2.
Representative dengue virus-infected PS foci. PS cells were infected with 16681Nde(+) and the pr-M junction chimeras for 3 days under an L-15 medium-carboxymethylcellulose overlay and were visualized by four-step immunological staining. Representative foci were photographed under a 10× objective lens.
TABLE 4.
Plaque size and titer of 16681Nde(+) and pr-M junction chimeras
| Set or virus | Plaque size (mm, [n])a | Focus size (no. of cells/ focus [% reduction])b | Titer (FFU/ml) in cell line
|
|
|---|---|---|---|---|
| PS | C6/36 | |||
| 1 | ||||
| 16681Nde(+) | 2.64 ± 0.53 (32) | 190.8 ± 18.0 | 2.04 × 108 | 1.58 × 108 |
| TBEVpr/16681 | 1.11 ± 0.50 (54) | 70.5 ± 6.8 (63.0) | 1.60 × 108 | 7.05 × 107 |
| YFVpr/16681 | 1.56 ± 0.35 (52) | 99.2 ± 7.2 (48.0) | 7.74 × 107 | 6.73 × 107 |
| JEVpr/16681 | <0.50 | 22.2 ± 1.2 (88.4) | 2.10 × 106 | 2.52 × 106 |
| 0.71 ± 0.09c (26) | ||||
| 2 | ||||
| 16681Nde(+) | NDd | 178.8 ± 22.4 | 1.52 × 108 | 1.80 × 108 |
| TBEVpr/16681 | ND | 80.1 ± 28.2 (55.2) | 1.40 × 107 | 5.09 × 107 |
| YFVpr/16681 | ND | 138.8 ± 16.0 (22.4) | 1.54 × 108 | 1.01 × 108 |
| JEVpr/16681 | ND | 15.4 ± 1.4 (91.4) | 2.12 × 106 | 1.38 × 106 |
Data are means ± standard deviations.
Data are means and standard errors of the means calculated from four and three separate experiments for set 1 and set 2, respectively. The reduction of focus size was calculated from the equation 100 × [1 − mean focus size of chimera/mean focus size of 16681Nde(+)].
Plates were incubated for 2 additional days more than the usual 8 days.
ND, not done.
The kinetics of multistep virus multiplication in vitro were assessed by infecting C6/36, PS, and Vero cells at multiplicities of infection of 0.001, 0.01, and 1, respectively. The different multiplicities were selected from titration experiments for a gradual accumulation of viruses during a period of several days. During a 2-week period of replication in the three cell lines, there was no consistent difference in virus production among 16681Nde(+), TBEVpr/16681, and YFVpr/16681 (Fig. 3). In contrast, JEVpr/16681 displayed a slower rise and reduced levels during the entire period with the two mammalian cell lines and during days 3 to 9 of infection of mosquito cells. Thus, replacement of the dengue virus pr-M junction with those of TBEV and YFV results only in some decrease in plaque size, whereas replacement with the JEV pr-M junction causes a consistent reduction of plaque size and titer and replication kinetics.
FIG. 3.
Kinetics of virus multiplication in C6/36, PS, and Vero cell lines. C6/36, PS, and Vero cells were infected with 16681Nde(+) and the pr-M junction chimeras at multiplicities of infection of 0.001, 0.01, and 1 FFU/cell, respectively. The culture fluid was collected at the indicated days for 2 weeks and was titrated by a focus immunoassay. The means and standard errors of the means for virus titers were determined from three separate experiments. (A) C6/36 cells. (B) PS cells. (C) Vero cells.
Effect of enhanced prM cleavage on infectivity.
For assessment of the influence of diverse pr-M cleavage-proximal junctions on dengue virus infectivity, parent and chimeric viruses were grown concurrently as sets in both C6/36 and PS cell lines. Infectious virus titers, as determined by a focus immunoassay using either PS or C6/36 cells, were then compared with the numbers of virions obtained by measuring viral RNA by a quantitative RT-PCR method (51). The influence of cellular sources employed for virus growth and infectivity titration was initially explored by measuring the specific infectivity of 16681Nde(+) in four categories (C6-C6, C6-PS, PS-PS, and PS-C6) according to which cells were used for virus growth and subsequent titration. The variation of specific infectivity was limited to within a 1-log10 range in three of four categories, and the mean specific infectivities for all categories were comparable (Fig. 4A), indicating that infectivity is minimally affected by the cell lines employed for virus multiplication and titration. Alteration of the infectivity of the pr-M junction chimeras was then determined by comparing their specific infectivity values only against that of the 16681Nde(+) preparation that was grown and titrated concurrently. Average specific infectivity ratios were then determined for each chimeric virus from the comparisons with 16681Nde(+) in several sets of virus preparations and were displayed in separate multiplication-titration categories, as shown in Fig. 4B. These comparisons revealed that the mean specific infectivity of the pr-M junction chimeras differed <10-fold from that of 16681Nde(+). Although there was a tendency for increased specific infectivity in three categories for JEVpr/16681 and YFVpr/16681, the mean specific infectivity ratios were <3.2 for all six comparisons. When the variation inherent to virus titration is taken into account, there appear to be minimal changes in the specific infectivity of the pr-M junction chimeras compared with the 16681Nde(+) parent. The reduction in JEVpr/16681 multiplication cannot be explained by its altered ability to initiate a productive infection.
FIG. 4.
Comparison of specific infectivity. (A) Specific infectivity of 16681Nde(+) compared in four categories (C6-C6, C6-PS, PS-PS, and PS-C6) according to the cell sources used for virus multiplication and subsequent titration. Individual values are shown together with the means and standard deviations. (B) Change of specific infectivity of pr-M junction chimeras compared with 16681Nde(+). For each pr-M junction chimera, the specific infectivity ratio was determined by dividing its specific infectivity value with that of the 16681Nde(+) preparation that was grown and titrated concurrently. The number of comparisons in each set is indicated in parentheses. The mean infectivity ratios and standard deviations were assessed separately for the four virus multiplication-titration categories. *, statistically significant difference; P < 0.01 (analysis of variance).
Comparison of cell-associated and extracellular virus production.
Since prM is involved in virion formation and its cleavage coincides with viral release, it is possible that the assembly of JEVpr/16681 within infected cells is diminished or that its release is defective. For assessment of these possibilities, the levels of infectious viruses within the infected cells and those that were released into the culture medium were compared with a single-step multiplication experiment. When PS cells were infected with 16681Nde(+) at a multiplicity of infection of 1 FFU/cell for 2 h and both cell-associated and extracellular virus titers were determined at various intervals, an increase in the cell-associated virus titer was first observed at 16 h and reached a plateau at 36 h (Fig. 5A). Changes in the extracellular virus titer followed the same trend, but at slightly lower levels from 16 to 36 h after infection. Following infection with TBEVpr/16681 and YFVpr/16681, virus levels in the intracellular and extracellular compartments were comparable to those of 16681Nde(+) (Fig. 5B and C). In contrast, JEVpr/16681 exhibited distinct kinetics. Twelve, 14, and 16 h after infection, the intracellular level of JEVpr/16681 was slightly lower than that of 16681Nde(+), whereas at 24, 36, and 48 h, cell-associated JEVpr/16681 titers were similar to the others (Fig. 5D). At all time points, the extracellular JEVpr/16681 titer was markedly lower than the cell-associated virus titer. The disparity was largest 20 and 24 h after infection, when there were 669- and 797-fold, respectively, fewer extracellular JEVpr/16681 viruses than the cell-associated counterparts, compared with only 7.7- and 2.4-fold for 16681Nde(+). Based on the previous finding that there was no major difference in the specific infectivities of extracellular JEVpr/16681 and 16681Nde(+), the results indicated that delayed export of cell-associated virus into the extracellular compartment contributes to the reduced level of extracellular JEVpr/16681. It should be noted that the enhanced cleavage of prM induced by the altered pr-M junction could render the cell-associated form of JEVpr/16681 more infectious than the other chimeric viruses and the 16681Nde(+) parent, causing a selective overestimation of the intracellular virions by the focus immunoassay and thereby diminishing the quantitative aspect of the comparison of changes in cell-associated virus titers. However, this should not affect the magnitude and temporal relationship of extracellular viruses.
FIG. 5.
Comparison of cell-associated and extracellular viruses in a single-step multiplication study. PS cells were infected at a multiplicity of infection of 1 FFU/cell for 2 hours and washed extensively. Extracellular and cell-associated viruses were collected concurrently at various time points after infection. Virus titers were determined with PS cells by using a focus immunoassay. The means and standard errors of the means for virus titers were determined from four separate experiments. (A) 16681Nde(+). (B) TBEVpr/16681. (C) YFVpr/16681. (D) JEVpr/16681.
Minimal change in virus-GAG interaction.
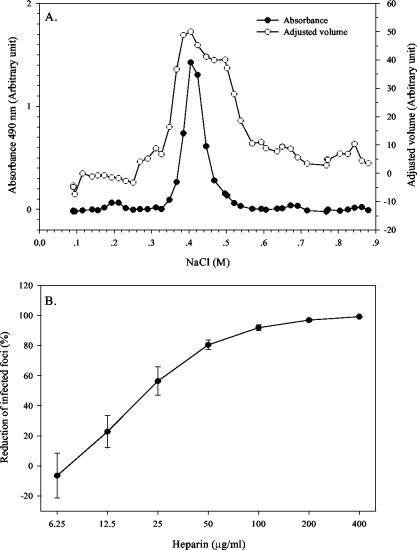
Besides serving as a target for furin within infected cells, the pr-M junction may play an additional role when present in uncleaved prM on the extracellular virion. As observed with cell culture-adapted Sindbis virus, the cluster of positive charges at the furin cleavage site of the PE2 envelope glycoprotein can mediate virus binding to cell surface heparan sulfate (46). Dengue virus binds heparan sulfate glycosaminoglycan (GAG) at the cell surface, and the E protein is implicated in the binding (16). If the pr-M junction provides a supplementary heparan sulfate binding site, its contribution may be detectable by comparing the GAG binding activities of dengue viruses with varied prM contents. For an exploration of the changes in virus-GAG binding affinity, 16681Nde(+) and the chimeras were applied onto a heparin-Sepharose column and eluted with a linear NaCl gradient. Determination of E protein levels by both ELISA and dot immunoassay revealed a single peak of eluted viruses, although the shoulders tended to be broader with the latter assay (Fig. 6A). Elution with a heparin gradient did not result in a recognizable peak. Comparisons between 16681Nde(+) and pr-M junction chimeras showed that peak virus elution occurred at comparable NaCl concentrations for 16681Nde(+), at 403.9 ± 0.4 mM (n = 3), and the three chimeras, at 407.2 ± 9.2 mM (n = 4) for TBEVpr/16681, 422.1 ± 13.9 mM (n = 3) for YFVpr/16681, and 428.3 ± 12.8 mM (n = 3) for JEVpr/16681. The binding affinities of pr-M junction chimeras for immobilized heparin appeared to be similar to that of 16681Nde(+). The failure to detect a reduction in GAG binding affinity for chimeric viruses with reduced prM content suggests that prM is not involved in virus-GAG binding. Alternatively, any reduction in GAG binding secondary to diminished prM content of YFVpr/16681 and JEVpr/16681 may be counteracted by binding enhancement from extra furin target sequences in the uncleaved prM.
FIG. 6.
(A) Elution profile of 16681Nde(+) from heparin-Sepharose column. Strain 16681Nde(+) was concentrated by centrifugation, resuspended in buffered solution containing 0.1 M NaCl and 0.5% BSA, and applied to a heparin-Sepharose column. Bound viruses were eluted with a 0.1 to 1 M NaCl gradient and detected by enzyme-linked immunosorbent assay (expressed as the arbitrary absorbance at 490 nm) and dot immunoassay (expressed as an arbitrary adjusted volume unit). (B) Dose-dependent reduction of PS cell infection by heparin. PS cells were infected with 16681Nde(+) that was pretreated with increasing concentrations of heparin. The reduction of infected foci was calculated from the equation (1-Hn/Ho) × 100, where Hn is the number of infected foci at n micrograms of heparin per milliliter and Ho is the number of infected foci in the absence of heparin. The mean reductions and standard errors of the means were determined from three separate experiments with Ho values of 256, 609, and 687 foci/12 wells, respectively.
Changes in binding affinities between viruses and cell surface GAG were studied by comparing the concentrations of heparin required for 50 and 80% inhibition (IC50 and IC80, respectively) of PS cell infection. Preincubation of fixed quantities of 16681Nde(+) and chimeras with increasing concentrations of heparin resulted in a dose-dependent reduction of 16681Nde(+)-infected PS foci between 12.5 and 50 μg of heparin/ml (Fig. 6B). As shown in Table 5, the mean IC50 and mean IC80 of heparin for the pr-M junction chimeras were comparable to those for 16681Nde(+), indicating that for this set of pr-M junction chimeras, the GAG-dependent pathway of PS cell infection is not affected by alteration of the prM content of dengue virions.
TABLE 5.
Concentration of heparin required for 50 and 80% inhibition of dengue virus infection of PS cells
| Virus | Heparin IC50 (μg/ml)a | Heparin IC80 (μg/ml)a |
|---|---|---|
| 16681Nde(+) | 26.34 ± 5.13 | 49.11 ± 7.76 |
| TBEVpr/16681 | 24.69 ± 4.74 | 44.48 ± 8.17 |
| YFVpr/16681 | 22.02 ± 0.38 | 41.52 ± 0.68 |
| JEVpr/16681 | 22.89 ± 4.11 | 39.36 ± 7.65 |
Data are means ± standard errors of the means determined from three separate experiments.
DISCUSSION
Cleavage of a viral glycoprotein precursor by proprotein convertases in the secretory pathway is an obligatory step in the multiplication of many enveloped RNA viruses (26). For flaviviruses, prM cleavage occurs after virus assembly, which takes place in the rough endoplasmic reticulum (54), in contrast to cleavage of individual glycoproteins prior to assembly at the cell membrane, as occurs with human immunodeficiency virus type 1 (HIV-1) gp160 (17, 29, 30, 58), HIV-2 gp140 (29), the HA0 precursor of some influenza A virus strains (37, 38, 77), and others. Moreover, cleavage serves to terminate the chaperone function of prM for E, allowing E to participate in cell binding and fusion, rather than activating the potential functional capability of prM. A recent study showed that the failure to cleave prM in a TBEV mutant with an engineered loss of P2 arginine results in the release of totally noninfectious virions which can be revived only temporarily by trypsin digestion (19). In light of this finding, previous results which revealed varying degrees of reduction of virus specific infectivity following treatment of cells infected with several flaviviruses with acidotropic agents or bafilomycin (28, 33, 64) are likely to reflect different levels of residual virus with cleaved prM. Such varying results may be secondary to the use of different doses and treatment schedules of acidotropic agents, the diversity of cells employed for virus multiplication and cleavage inhibition, and the characteristics, including the intracellular level and sensitivity to inhibitory high pH, of cellular proprotein convertases that are involved. Yet another possible cause is the difference in furin target sequence in prM among members of the genus Flavivirus and the resultant variation in prM cleavability.
An assessment of the influence of the cleavage-proximal sequence from three flaviviruses on the cleavage of dengue virus prM in this study showed that the 13-amino-acid sequence derived from YFV and JEV, but not TBEV, results in enhancement of prM cleavage over the level observed with the dengue virus parent. These three viruses share with dengue virus the conserved basic residues at positions P1, P2, and P4 which are known to be required for efficient cleavage by the proprotein convertase of many target proteins (56, 62, 74, 79, 86). Although lysine is found at position P2 of the dengue virus sequence, compared with arginine in the others, the P2 lysine is common among cellular and viral targets of proprotein convertases (56, 62). Moreover, a dengue virus mutant with an arginine substitution at position P2 displayed a similar peak titer, focus size, and multiplication kinetics to those of the parent virus after transfection of mosquito cells (13). The alteration of prM cleavage that was detected with these pr-M junction chimeras is therefore likely to reflect variation of non-P1, -P2, and -P4 sequences.
While all three pr-M junction chimeras contain more basic residues in the 13-amino-acid cleavage-proximal sequence than dengue virus and also lack the P3 and P7 acidic residues which are conserved in dengue virus, it is striking that only TBEVpr/16681 lacks the P5 basic residue. The 3-bp deletion, which causes an absence of P5 arginine, appears to be common to all members of the tick-borne encephalitis (TBE) antigenic complex with known prM sequences. This deletion likely reflects the adaptation of these viruses for efficient multiplication in ticks but may yield a less than optimal target for cleavage in mosquito cells. It is well documented, especially from studies of the influenza virus HA0 precursor, that factors other than the presence of the P1, P2, and P4 basic residues can affect the cleavage efficiency of the furin target sequence. The furin motif of the influenza virus HA0 precursor is located in an exposed loop that is accessible to proteases (15, 75). The presence of a nearby carbohydrate moiety reduces cleavage efficiency, whereas a polybasic amino acid insertion within the loop or replacement of an uncharged amino acid with a basic residue serves to nullify the adverse influence of the carbohydrate, possibly by reversing the poor accessibility of the target sequence to protease (41, 75). For HIV-1 gp160, a loop configuration for the furin cleavage motif was also proposed (60, 63). If the dengue virus pr-M junction is located in a similar loop, the P5 deletion in TBEVpr/16681 may shorten or otherwise alter the local conformation of this loop in dengue virus prM in such a way that basic residues at more distal positions cannot compensate for the lack of P5 arginine for efficient cleavage. It should be noted that the prM cleavage defect in a TBEV mutant with an engineered loss of P2 arginine (19) also coincides with further shortening of the cleavage-proximal sequence.
A distinguishing feature between JEVpr/16681 and other pr-M junction chimeric viruses is the presence in JEVpr/16681 of a minimal furin cleavage motif, Arg-Xaa-Xaa-Arg, in addition to the consensus motif at the nominal cleavage site. JEV and other members of the JE antigenic complex always contain an additional minimal motif, although its position may vary, e.g., at positions P10 to P13 in JEV, Murray Valley encephalitis virus, and St. Louis encephalitis virus and at positions P5 to P8 in Kunjin virus and West Nile virus. The minimal motif is also found at positions P4 to P7 of YFV but partially overlaps the consensus motif; this minimal motif is, with the exception of Langat and Powassan viruses, absent from TBEV and other members of the TBE antigenic complex. Duplication of the furin cleavage motif was observed previously for the HA0 precursor of certain virulent H5N1 and H5N3 strains of avian influenza A virus (38, 66, 78) and for HIV-1 gp160 (14), but with different organizations. Two furin cleavage motifs are present as a tandem repeat in the HA0 precursor of some H5N1 and H5N3 avian influenza virus strains, whereas there are four amino acid residues separating the two motifs in HIV-1 gp160. Tandem duplication of the furin motif as observed in the influenza virus HA0 precursor resulted in about fivefold enhancement of in vitro cleavage by furin when compared with a single motif sequence; cleavage occurred primarily at the physiologic site (5). Similarly, a recombinant HIV-1 envelope glycoprotein precursor (rgp140) which lacked the transmembrane and intracytoplasmic domains of gp41 but contained an engineered tandem repeat of the furin motif was cleaved better than the wild-type protein when expressed in cells both in the absence and presence of coexpressed furin (6). For the wild-type HIV-1 gp160 with two separated furin motifs, the effect of sequence duplication on cleavage efficiency is not known. Nevertheless, cleavage of HIV-1 gp160 by furin occurred preferentially at the cleavage-proximal physiological site both in vivo (21, 58) and in vitro (8, 18), as was the case for the influenza virus HA0 precursor. This cleavage site preference was further demonstrated in vitro with two other proprotein convertases for the influenza virus HA0 precursor (5) and HIV-1 gp160 (8, 18). The pronounced cleavage of prM in JEVpr/16681 is consistent with this role of duplicated furin motifs in enhancing the cleavability of the physiologic cleavage site, as documented for the influenza virus HA0 precursor and possibly HIV-1 gp160. While the actual cleavage site in JEVpr/16681 prM has not been defined experimentally in this study, the possibility that the minimal furin motif affects prM cleavability by simply providing an additional target for furin is unlikely since the Arg-Xaa-Xaa-Arg sequence is a relatively poor target for furin compare to the consensus Arg-Xaa-(Lys/Arg)-Arg sequence (31, 45, 57). The other possibility, that an additional feature of JEVpr/16681 and YFVpr/16681 prM, such as the absence of P3 and P7 acidic residues, also contributes to better prM cleavage, cannot yet be ruled out and is under investigation.
Since prM cleavage is not a prerequisite for the export of flavivirus particles, it was not expected that the unusually high level of prM cleavage observed with JEVpr/16681 would be associated with a delay of dengue virion export. To the best of our knowledge, this is the first example of a virion export defect manifested as a result of a modification of the furin cleavage sequence in a viral envelope protein. Previous studies examining the effect of alteration of the furin motif in a viral glycoprotein precursor on protein export and subcellular localization were performed with the HIV-1 rgp140 precursor, which forms trimers upon expression in transfected cells but does not assemble into a viral particle (6, 73). These studies revealed that certain changes of the furin motif in HIV-1 rgp140, the presence of coexpressed furin, or both differentially affect the extent of cleavage of HIV-1 rgp140 and the extracellular level of cleaved products that are secreted from transfected cells (6, 73). Specifically, an alteration of the wild-type furin cleavage motif in HIV-1 rgp140 to the tandem repeat form, which is analogous to the change in the pr-M junction in JEVpr/16681, caused the extracellular level of the cleaved products to diminish substantially, despite a significant enhancement of cleavage (6). Alternatively, the presence of coexpressed furin, which enhanced cleavage of both the wild-type furin motif and the tandem repeat motif, reduced the extracellular level of cleaved products from the former and did not reverse the poor secretion of the latter (6). Coexpression of furin also altered the localization of HIV-1 rgp140 with a wild-type furin motif to the same subcellular compartment as furin (73). These changes required an interaction between furin and its target sequence in the precursor protein but were not dependent on the membrane association of furin (6). Because the extracellular expression of the cleaved products of more optimized furin target sequences was not affected (6), the reduction of product secretion in the case of HIV-1 rgp140 containing the tandem repeat motif (or rgp140 containing the wild-type motif expressed in the presence of coexpressed furin) was thought to reflect complexing of furin with the nonoptimal target sequence and the retention of such a complex in the trans-Golgi network (6, 73). Our observations of a low extracellular virus titer and a delay in virus export from cells infected with JEVpr/16681 strongly suggested that, at the virion level, certain changes of the furin target sequence on the virus envelope may similarly alter the interaction between furin and virion, leading to the analogous consequence of export retardation. Alternatively, it is possible that enhanced cleavage of prM could cause the premature expression of fusion-competent virions, which, in the presence of a sufficiently low pH in the Golgi apparatus and secretory vesicles, may result in fusion of the virus to the intracellular membrane and the apparent export retardation. For JEVpr/16681, the other possibility for a prM conformational change due to the chimeric pr-M junction sequence and the resultant enhancement of virion transport to the lysosome cannot be tested directly due to the paucity of monoclonal antibody reagents. However, the lack of a major change of virus specific infectivity and the comparable levels of cell-associated virus titers with those of the parent virus and other chimeric viruses during the late stage of single-step virus multiplication render this possibility unlikely. While previous studies established that the export of flavivirus virions is independent of prM cleavage (19, 28, 33, 64), our results extend these findings by revealing the negative impact of enhanced pr-M cleavage on virus export in the case of dengue virus. Enhanced cleavage at the N terminus of prM by signalase was also shown to be counterproductive for the replication of YFV, but by affecting virus production (53).
During the evolution of dengue virus, the export defect associated with enhanced prM cleavage may have contributed to the restriction of sequence variation at the pr-M junction. Our examination of the pr-M junction sequence of 100 dengue virus isolates deposited in the GenBank database confirmed the highly conserved nature of the pr-M junction. For dengue virus serotypes 1, 2 and 3, variation of the charged amino acids within the 13-residue cleavage-proximal region was detected in only 4 out of 85 isolates (Table 6). In three isolates, an increase in the net positive charge occurred with the loss of the P3 acidic residue (two isolates) and the acquisition of a P5 arginine (one isolate). The other isolate lost the P5 basic residue, but this was compensated for by the P12 arginine. None acquired duplicated furin target motifs. Dengue virus serotype 4 is distinct with its P6 arginine residue; however, no additional charge variation was detected. Conceivably, further viral changes that result in enhanced prM cleavability can be selected against by virus export delay as demonstrated in this study. It thus appears to be advantageous for dengue virus to retain some prM on the envelope of extracellular virions. The advantage must be sufficiently substantial so as to withstand the selective pressure imposed by specific antibody responses. The host protective effect of anti-prM antibodies has been documented for dengue virus (9, 20, 40, 80). prM is not known to be involved in receptor binding, and the lack of a major change in specific infectivity that was observed with JEVpr/16681 reinforces this notion. A recent structural analysis indicated that the pr portion of dengue virus prM projects out of the surface of immature virions and covers the fusion peptide at the distal end of the E glycoprotein (85). Based on this model and a previous result showing the fusion defect in prM-containing TBE virions (27), the remaining prM proteins in dengue virions may contribute to the regulation of fusion in the acidic endosomal compartment of newly infected cells by directly preventing the E molecule of the same prM-E heterodimer to engage in fusion. Alternatively, the remaining prM proteins may collectively hinder or retard the general outward expansion of the virion envelope, which is thought to be required for the rearrangement of E dimers on the viral envelope in the acidic endosome (48). Currently, it is not clear whether all extracellular dengue virions retain some prM molecules or if two subpopulations of intermingling M-only virions and prM-only virions exist. For a full understanding of the functional role of prM in the replication of dengue virus and other flaviviruses, it will be crucial to determine the distribution of prM in the pr-M junction chimeras at both the single virion level and the virus population level and to correlate such variations with fusion capability and infectivity.
TABLE 6.
Variation of charged residues at the pr-M junction of dengue viruses
| Serotype | Strain | Amino acid sequencea | P1 to P13 charge
|
Accession no. | |||
|---|---|---|---|---|---|---|---|
| + | − | Net | Setb | ||||
| 1 | Mochizuki | YGTCS QTGEH RRGKR↓SVALA PHVGL | 4 | 1 | +3 | 1 | BAB72261 |
| 2 | 16681 | YGTCT TMGEH RRQKR SVALV PHVGM | 4 | 1 | +3 | 1 | AAA73185 |
| 2 | D2-04 | YGTRT TTGEH GREKR SVALV PHVGM | 4 | 2 | +2 | 1 | P30026 |
| 3 | 68784 | YGTCN QAGER RRDKR SVALT PHSGM | 5 | 2 | +3 | 1 | AAK74146 |
| Cleavage positionc | 15 11 6 1 1′ 6′ 10′ | ||||||
Amino acids in bold represent variations of charged residues between positions P1 and P13 of the pr-M junction. The arrow indicates the pr-M cleavage site.
Set of nonoverlapping Arg-Xaa-(Lys/Arg)-Arg and Arg-Xaa-Xaa- Arg sequences.
Numbers refer to the positions of the amino acids relative to the pr-M cleavage site in the proximal direction (without apostrophe) and distal direction (with apostrophe).
Acknowledgments
We are grateful to Bruce Innis and Ananda Nisalak, Department of Virology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand; Toshihiko Fukunaga, University of the Ryukyus, Nishihara, Okinawa, Japan; and Robert Anderson, University of Dalhousie, Halifax, Canada, for gifts of strain 16681 and other reagents and to Surinda Kawichai, Research Institute for Health Sciences, Chiang Mai University, for help with statistical analysis. We thank Ruenkaew Praput and Seangdeun Moonsom for excellent technical assistance and Nuntarudee Juabsamai, Perkin-Elmer (Thailand), and Wipawanee Kasemworaphoom, Amersham Biosciences (Thailand), for instrument support.
This investigation received financial support from the Thailand Tropical Diseases Research (T-2) Program (99-2-DEN-03-008) and from the Medical Biotechnology Unit Network from the National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Bangkok, Thailand. P.M. is a Senior Research Scholar supported by the Thailand Research Fund.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R., S. Wang, C. Osiowy, and A. C. Issekutz. 1997. Activation of endothelial cells via antibody-enhanced dengue virus infection of peripheral blood monocytes. J. Virol. 71:4226-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avirutnan, P., P. Malasit, B. Seliger, S. Bhakdi, and M. Husmann. 1998. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J. Immunol. 161:6338-6346. [PubMed] [Google Scholar]
- 5.Basak, A., M. Zhong, J. S. Munzer, M. Chretien, and N. G. Seidah. 2001. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem. J. 353:537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley, J. M., R. W. Sanders, A. Master, C. S. Cayanan, C. L. Wiley, L. Schiffner, B. Travis, S. Kuhmann, D. R. Burton, S.-L. Hu, W. C. Olson, and J. P. Moore. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76:2606-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blok, J., S. M. McWilliam, H. C. Butler, A. J. Gibbs, G. Weiller, B. L. Herring, A. C. Hemsley, J. G. Aaskov, S. Yoksan, and N. Bhamarapravati. 1992. Comparison of a dengue-2 virus and its candidate vaccine derivative: sequence relationships with the flaviviruses and other viruses. Virology 187:573-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brakch, N., M. Dettin, C. Scarinci, N. G. Seidah, and C. Di Bello. 1995. Structural investigation and kinetic characterization of potential cleavage sites of HIV gp160 by human furin and PC1. Biochem. Biophys. Res. Commun. 213:356-361. [DOI] [PubMed] [Google Scholar]
- 9.Bray, M., and C. J. Lai. 1991. Dengue virus premembrane and membrane proteins elicit a protective immune response. Virology 185:505-508. [DOI] [PubMed] [Google Scholar]
- 10.Brennan, S. O., and K. Nakayama. 1994. Cleavage of proalbumin peptides by furin reveals unexpected restrictions at the P2 and P′1 sites. FEBS Lett. 347:80-84. [DOI] [PubMed] [Google Scholar]
- 11.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strauss (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Calisher, C. H., N. Karabatsos, J. M. Dalrymple, R. E. Shope, J. S. Porterfield, E. G. Westaway, and W. E. Brandt. 1989. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 70:37-43. [DOI] [PubMed] [Google Scholar]
- 13.Chaichoun, K. 2002. M.S. thesis. Chiang Mai University, Chiang Mai, Thailand.
- 14.Chakrabarti, L., M. Guyader, M. Alizon, M. D. Daniel, R. C. Desrosiers, P. Tiollais, and P. Sonigo. 1987. Sequence of simian immunodeficiency virus from macaque and its relationship to other human and simian retroviruses. Nature 328:543-547. [DOI] [PubMed] [Google Scholar]
- 15.Chen, J., K. H. Lee, D. A. Steinhauer, S. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 16.Chen, Y., T. Maguire, R. E. Hileman, J. R. Fromm, J. D. Esko, R. J. Linhardt, and R. M. Marks. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 3:866-871. [DOI] [PubMed] [Google Scholar]
- 17.Decroly, E., M. Vandenbranden, J.-M. Ruysschaert, J. Cogniaux, G. S. Jacob, S. C. Howard, G. Marshall, A. Kompelli, A. Basak, F. Jean, C. Lazure, S. Bedannet, M. Chretien, R. Day, and N. G. Seidah. 1994. The convertases furin and PC1 can both cleave the human immunodeficiency virus (HIV-1) envelope glycoprotein gp160 into gp120 (HIV-1 SU) and gp41 (HIV-1 TM). J. Biol. Chem. 269:12240-12247. [PubMed] [Google Scholar]
- 18.Decroly, E., S. Wouters, C. Di Bello, C. Lazure, J.-M. Ruysschaert, and N. G. Seidah. 1996. Identification of the paired basic convertases implicated in HIV gp160 processing based on in vitro assays and expression in CD4+ cell lines. J. Biol. Chem. 271:30442-30450. [DOI] [PubMed] [Google Scholar]
- 19.Elshuber, S., S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84:183-191. [DOI] [PubMed] [Google Scholar]
- 20.Falconar, A. K. I. 1999. Identification of an epitope on the dengue virus membrane (M) protein defined by cross-protective monoclonal antibodies: design of an improved epitope sequence based on common determinants present in both envelope (E and M) proteins. Arch. Virol. 144:2313-2330. [DOI] [PubMed] [Google Scholar]
- 21.Fenouillet, E., and J.-C. Gluckman. 1992. Immunological analysis of human immunodeficiency virus type 1 envelope glycoprotein proteolytic cleavage. Virology 187:825-828. [DOI] [PubMed] [Google Scholar]
- 22.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 23.Flamand, M., F. Megret, M. Mathieu, J. Lepault, F. A. Rey, and V. Deubel. 1999. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J. Virol. 73:6104-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flamand, M., V. Deubel, and M. Girard. 1992. Expression and secretion of Japanese encephalitis virus nonstructural protein NS1 by insect cells using a recombinant baculovirus. Virology 191:826-836. [DOI] [PubMed] [Google Scholar]
- 25.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Principles of virology, p. 402-436. American Society for Microbiology, Washington, D.C.
- 26.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Principles of viology, p. 418-421. American Society for Microbiology, Washington, D.C.
- 27.Guirakhoo, F., F. X. Heinz, C. W. Mandl, H. Holzmann, and C. Kunz. 1991. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J. Gen. Virol. 72:1323-1329. [DOI] [PubMed] [Google Scholar]
- 28.Guirakhoo, F., R. A. Bolin, and J. T. Roehrig. 1992. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology 191:921-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallenberger, S., M. Moulard, M. Sordel, H.-D. Klenk, and W. Garten. 1997. The role of eukaryotic subtilisin-like endoproteases for the activation of human immunodeficiency virus glycoproteins in natural host cells. J. Virol. 71:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallenberger, S., V. Bosch, H. Angliker, E. Shaw, H.-D. Klenk, and W. Garten. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358-361. [DOI] [PubMed] [Google Scholar]
- 31.Hatsuzawa, K., M. Nagahama, S. Takahashi, K. Takada, K. Murakami, and K. Nakayama. 1992. Purification and characterization of furin, a Kex2-like processing endoprotease, produced in Chinese hamster ovary cells. J. Biol. Chem. 267:16094-16099. [PubMed] [Google Scholar]
- 32.He, R.-T., B. L. Innis, A. Nisalak, W. Usawattanakul, S. Wang, S. Kalayanarooj, and R. Anderson. 1995. Antibodies that block virus attachment to Vero cells are a major component of the human neutralizing antibody response against dengue virus type 2. J. Med. Virol. 45:451-461. [DOI] [PubMed] [Google Scholar]
- 33.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 34.Henchal, E. A., J. M. McCown, D. S. Burke, M. C. Seguin, and W. E. Brandt. 1985. Epitope analysis of antigenic determinants on the surface of dengue-2 virions using monoclonal antibodies. Am. J. Trop. Med. Hyg. 34:162-169. [DOI] [PubMed] [Google Scholar]
- 35.Henchal, E. A., M. K. Gentry, J. M. McCown, and W. E. Brandt. 1982. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 31:830-836. [DOI] [PubMed] [Google Scholar]
- 36.Hiramatsu, K., M. Tadano, R. Men, and C.-J. Lai. 1996. Mutational analysis of a neutralization epitope on the dengue type 2 virus (DEN2) envelope protein: monoclonal antibody resistant DEN2/DEN4 chimeras exhibit reduced mouse neurovirulence. Virology 224:437-445. [DOI] [PubMed] [Google Scholar]
- 37.Horimoto, T., K. Nakayama, S. P. Smeekens, and Y. Kawaoka. 1994. Proprotein-processing endoproteases PC6 and furin both activate hemagglutinin of virulent avian influenza viruses. J. Virol. 68:6074-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horimoto, T., and Y. Kawaoka. 2001. Pandemic threat posed by avian influenza A viruses. Clin. Microbiol. Rev. 14:129-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iacono-Connors, L. C., J. F. Smith, T. G. Ksiazek, C. L. Kelley, and C. S. Schmaljohn. 1996. Characterization of Langat virus antigenic determinants defined by monoclonal antibodies to E, NS1 and preM and identification of a protective, non-neutralizing prM-specific monoclonal antibody. Virus Res. 43:125-136. [DOI] [PubMed] [Google Scholar]
- 40.Kaufman, B. M., P. L. Summers, D. R. Dubois, W. Houston Cohen, M. K. Gentry, R. L. Timchak, D. S. Burke, and K. H. Eckels. 1989. Monoclonal antibodies for dengue virus prM glycoprotein protect mice against lethal dengue infection. Am. J. Trop. Med. Hyg. 41:576-580. [DOI] [PubMed] [Google Scholar]
- 41.Kawaoka, Y., and R. G. Webster. 1989. Interplay between carbohydrate in the stalk and the length of the connecting peptide determines the cleavability of influenza virus hemagglutinin. J. Virol. 63:3296-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 72:5967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura-Kuroda, J., and K. Yasui. 1983. Topographical analysis of antigenic determinants on envelope glycoprotein V3 (E) of Japanese encephalitis virus, using monoclonal antibodies. J. Virol. 45:124-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinney, R. M., S. Butrapet, G.-J. Chang, K. R. Tsuchiya, J. T. Roehrig, N. Bhamarapravati, and D. J. Gubler. 1997. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230:300-308. [DOI] [PubMed] [Google Scholar]
- 45.Klimpel, K. R., S. S. Molloy, G. Thomas, and S. H. Leppla. 1992. Anthrax toxin protective antigen is activated by a cell surface protease with the sequence specificity and catalytic properties of furin. Proc. Natl. Acad. Sci. USA 89:10277-10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klimstra, W. B., H. W. Heidner, and R. E. Johnston. 1999. The furin protease cleavage recognition sequence of Sindbis virus PE2 can mediate virion attachment to cell surface heparan sulfate. J. Virol. 73:6299-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konishi, E., and P. W. Mason. 1993. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J. Virol. 67:1672-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuno, G., G.-J. J. Chang, K. R. Tsuchiya, N. Karabatsos, and C. B. Cropp. 1998. Phylogeny of the genus Flavivirus. J. Virol. 72:73-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 51.Laue, T., P. Emmerich, and H. Schmitz. 1999. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the TaqMan automated amplification system. J. Clin. Microbiol. 37:2543-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leblois, H., and P. R. Young. 1995. Maturation of the dengue-2 virus NS1 protein in insect cells: effects of downstream NS2A sequences on baculovirus-expressed gene constructs. J. Gen. Virol. 76:979-984. [DOI] [PubMed] [Google Scholar]
- 53.Lee, E., C. E. Stocks, S. M. Amberg, C. M. Rice, and M. Lobigs. 2000. Mutagenesis of the signal sequence of yellow fever virus prM protein: enhancement of signalase cleavage in vitro is lethal for virus production. J. Virol. 74:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 55.Lorenz, I. C., S. L. Allison, F. X. Heinz, and A. Helenius. 2002. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 76:5480-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molloy, S. S., E. D. Anderson, F. Jean, and G. Thomas. 1999. Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 9:28-35. [DOI] [PubMed] [Google Scholar]
- 57.Molloy, S. S., P. A. Bresnahan, S. H. Leppla, K. R. Klimpel, and G. Thomas. 1992. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 267:16396-16402. [PubMed] [Google Scholar]
- 58.Morikawa, Y., E. Barsov, and I. Jones. 1993. Legitimate and illegitimate cleavage of human immunodeficiency virus glycoproteins by furin. J. Virol. 67:3601-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morsy, J., W. Garten, and R. Rott. 1994. Activation of an influenza virus A/Turkey/Oregon/71 HA insertion variant by the subtilisin-like endoprotease furin. Virology 202:988-991. [DOI] [PubMed] [Google Scholar]
- 60.Moulard, M., L. Chaloin, S. Canarelli, K. Mabrouk, H. Darbon, and L. Challoin. 1998. Retroviral envelope glycoprotein processing: structural investigation of the cleavage site. Biochemistry 37:4510-4517. [DOI] [PubMed] [Google Scholar]
- 61.Murray, J. M., J. G. Aaskov, and P. J. Wright. 1993. Processing of the dengue virus type 2 proteins prM and C-prM. J. Gen. Virol. 74:175-182. [DOI] [PubMed] [Google Scholar]
- 62.Nakayama, K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oliva, R., M. Leone, L. Falcigno, G. D'Auria, M. Dettin, C. Scarinci, C. Di Bello, and L. Paolillo. 2002. Structural investigation of the HIV-1 envelope glycoprotein gp160 cleavage site. Chemistry 8:1467-1473. [DOI] [PubMed] [Google Scholar]
- 64.Randolph, V. B., G. Winkler, and V. Stollar. 1990. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology 174:450-458. [DOI] [PubMed] [Google Scholar]
- 65.Roehrig, J. T., R. A. Bolin, and R. G. Kelly. 1998. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246:317-328. [DOI] [PubMed] [Google Scholar]
- 66.Rohm, C., T. Horimoto, Y. Kawaoka, J. Suss, and R. G. Webster. 1995. Do hemagglutinin genes of highly pathogenic avian influenza viruses constitute unique phylogenetic lineages? Virology 209:664-670. [DOI] [PubMed] [Google Scholar]
- 67.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 68.Schoepp, R. J., and B. J. Beaty. 1984. Titration of dengue viruses by immunofluorescence in microtiter plates. J. Clin. Microbiol. 20:1017-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sittisiri, K. 1994. M.S. thesis. Mahidol University, Bangkok, Thailand.
- 70.Sittisombut, N., N. Maneekarn, A. Kanjanahaluethai, W. Kasinrerk, K. Viputtikul, and J. Supawadee. 1995. Lack of augmenting effect of interferon-γ on dengue virus multiplication in human peripheral blood monocytes. J. Med. Virol. 45:43-49. [DOI] [PubMed] [Google Scholar]
- 71.Sriburi, R., P. Keelapang, T. Duangchinda, S. Pruksakorn, N. Maneekarn, P. Malasit, and N. Sittisombut. 2001. Construction of infectious dengue 2 cDNA clones using high copy number plasmid. J. Virol. Methods 92:71-82. [DOI] [PubMed] [Google Scholar]
- 72.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Staropoli, I., C. Chanel, M. Girard, and R. Altmeyer. 2000. Processing, stability, and receptor binding properties of oligomeric envelope glycoprotein from a primary HIV-1 isolate. J. Biol. Chem. 275:35137-35145. [DOI] [PubMed] [Google Scholar]
- 74.Steiner, D. F. 1998. The proprotein convertases. Curr. Opin. Chem. Biol. 2:31-39. [DOI] [PubMed] [Google Scholar]
- 75.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1-20. [DOI] [PubMed] [Google Scholar]
- 76.Stiasny, K., S. L. Allison, A. Marchler-Bauer, C. Kunz, and F. X. Heinz. 1996. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 70:8142-8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stieneke-Grober, A., M. Vey, H. Angliker, E. Shaw, G. Thomas, C. Roberts, H. D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11:2407-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 79.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3:753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vazquez, S., M. G. Guzman, G. Guillen, G. Chinea, A. B. Perez, M. Pupoa, R. Rodriguez, O. Reyes, H. E. Garay, I. Delgado, G. Garcia, and M. Alvarez. 2002. Immune response to synthetic peptides of dengue prM protein. Vaccine 20:1823-1830. [DOI] [PubMed] [Google Scholar]
- 81.Wang, S., R. He, and R. Anderson. 1999. PrM- and cell-binding domains of the dengue virus E protein. J. Virol. 73:2547-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wengler, G., and G. Wengler. 1989. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 63:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Westaway, E. G. 1966. Assessment and application of a cell line from pig kidney for plaque assay and neutralization tests with twelve group B arboviruses. Am. J. Epidemiol. 84:439-456. [DOI] [PubMed] [Google Scholar]
- 84.Winkler, G., V. B. Randolph, G. R. Cleaves, T. E. Ryan, and V. Stollar. 1988. Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology 162:187-196. [DOI] [PubMed] [Google Scholar]
- 85.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou, A., G. Webb, X. Zhu, and D. F. Steiner. 1999. Proteolytic processing in the secretory pathway. J. Biol. Chem. 274:20745-20748. [DOI] [PubMed] [Google Scholar]