Abstract
Hepatitis D virus (HDV) is a satellite of hepatitis B virus (HBV) for transmission and propagation and infects nearly 20 million people worldwide. The HDV genome is a compact circular single-stranded RNA genome with extensive intramolecular complementarity. Despite its different epidemiological and pathological patterns, the variability and geographical distribution of HDV are limited to three genotypes and two subtypes that have been characterized to date. Phylogenetic reconstructions based on the delta antigen gene and full-length genome sequence data show an extensive and probably ancient radiation of African lineages, suggesting that the genetic variability of HDV is much more complex than was previously thought, with evidence of additional clades. These results relate the geographic distribution of HDV more closely to the genetic variability of its helper HBV.
Hepatitis D virus (HDV) is a transmissible agent discovered 26 years ago (30) that requires helper functions from the hepatitis B virus (HBV) for virion assembly and propagation (37). Thus, HDV infection is necessarily associated with HBV infection because HDV ribonucleoprotein buds through the hepatitis B surface antigen (HBsAg) excretory pathway. The HDV genome is a circular single-stranded RNA genome of approximately 1,680 bases with extensive intramolecular complementarity (41). Part of the HDV genome might have historical homology to viroids or plant virus satellite RNA sequences (10, 15), and a rolling-circle model has been developed for viral RNA replication (reviewed in reference 39). However, in contrast to viroids, which do not code for any protein, the HDV antigenome contains an open reading frame that was probably acquired by HDV from a cellular ancestor transcript, leading to the expression of the delta protein (1, 20). Indeed, HDV mRNA is translated to sHD and LHD proteins, corresponding respectively to the “small-p24” and the “large-p27” hepatitis Delta proteins. The LHD amino acid sequence is identical to sHD with the addition of a carboxy-terminal extension of 19 to 20 amino acids following the editing of the sHD stop codon during the viral RNA replication cycle (23, 43). sHD is required for viral replication and might promote RNA polymerase II elongation of nascent HDV RNA (45), while LHD is essential for HDV particle assembly (5).
HDV-HBV coinfection and HDV superinfection of a patient chronically infected by HBV both lead to a liver disease more severe than that induced by HBV alone (2, 31). HDV is highly endemic in Mediterranean countries, the Middle East, Central Africa, and northern parts of South America. In contrast, in industrialized countries, its prevalence is low and its transmission is often associated with intravenous drug use. Intrafamilial transmission has been described in southern Italy, which has been considered an area of high endemicity (12, 28).
Although HDV was expected, like many RNA viruses, to exhibit considerable genetic variability, only three HDV genotypes have been characterized to date on the basis of a small number of complete genome sequences (3, 13, 41). Historically, the definition of “genotype” is based on the comparison of nucleotide similarity between pairs of sequences that have been discovered and characterized at the time. For HDV genomes that are known at present, the divergence in nucleotide sequence of the studied region is less than 14 to 15.7% among different isolates of the same genotype and ranges from 19 to 38% between sequences from different genotypes (3, 13, 34, 44). Genotype I includes the European, North American, African, and some Asian HDV isolates (6, 34, 41). Genotype II has been found in Japan, Taiwan, and Yakutia (Russia) (13, 14, 17, 44); some sequences from Taiwan and the Okinawa islands were tentatively assigned to a subtype of genotype II (i.e., genotype IIB) (33, 44). Genotype III has been found exclusively in South America (Peru, Colombia, and Venezuela) (3, 27).
So far, studies of HDV genome variability have been performed in non-African countries except for the description of two sequences (assigned to genotype I) from Ethiopia and Somalia (46). By studying samples that have been prospectively tested for HDV replication, we present an extensive analysis of African HDV sequences. Besides genotype I-like sequences, our phylogenetic analyses indicate that approximately 70% of the characterized African isolates (mostly from West and Central Africa) form highly divergent groups, suggesting an ancient African radiation and extending the known HDV genetic variability to at least seven clades, thus bringing the variability of this satellite closer to that of human HBV.
MATERIALS AND METHODS
Patients.
From a cohort of 227 individuals whose serum samples were prospectively collected for HDV genome replication analysis between 1999 and 2002, we selected 25 patients whose preliminary examination suggested that the HDV strains varied from previously described HDV genotypes. Twenty-two samples were obtained from patients born in Africa or who had traveled to Africa. Table 1 indicates the gender, age, place of birth, and biological and liver histological results of the infected patient. The male-to-female ratio was 0.8, and the mean age was 35 years (range, 15 to 53 years). Most patients had chronic active hepatitis or cirrhosis, and only one patient, aged 33 years, had acute HDV superinfection. One patient was treated with alpha interferon at the time of sampling.
TABLE 1.
Clinical data for the African HDV-infected patients
| Samplea | Sexb | Age (yr) | Country of origin | Anti-HDV IgM/IgGc | Level ofd:
|
Histologye | |
|---|---|---|---|---|---|---|---|
| ALT | AST | ||||||
| dFr-4 | F | 40 | Ghana | +/+ | N | N | ACH |
| dFr-45 | F | 45 | Cameroon | +/+ | 5N | 5N | ACH |
| dFr-47 | F | 40 | Guinea | +/+ | 3N | 2.5N | Cirrhosis |
| dFr-48 | F | 42 | Poland | +/+ | N | N | Cirrhosis |
| dFr-55 | F | 21 | Cameroon | +/− | 1.5N | 2N | ACH |
| dFr-56 | F | 21 | Cameroon | −/+ | N | N | NDf |
| dFr-59 | M | 33 | Egypt | +/+ | N | N | Cirrhosis |
| dFr-69 | M | 47 | Gambia | −/+ | N | N | Cirrhosis |
| dFr-70 | M | 35 | Egypt | +/+ | N | 1.5N | Cirrhosis |
| dFr-71 | F | 56 | Central African Republic | +/+ | 2N | 4N | Cirrhosis |
| dFr-73 | M | 34 | Ivory Coast | +/+ | 3N | 3N | ACH |
| dFr-74 | M | 49 | Democratic Republic of Congo | +/+ | 5.5N | 3.5N | Cirrhosis |
| dFr-644 | F | 32 | Congo-Brazzaville | +/+ | 2N | 4N | Cirrhosis |
| dFr-910 | F | 34 | Mali | +/+ | 2.6N | 2.6N | ACH |
| dFr-1594 | M | 33 | Angola | +/ND | 200N | 200N | Acute HDV superinfection |
| dFr-1843 | M | 21 | Gabon | +/+ | 1.2N | ND | Cirrhosis |
| dFr-1953 | M | 53 | Cameroon | +/+ | N | N | ACH |
| dFr-2020 | F | 28 | Ivory Coast | −/+ | 4N | 3N | ACH |
| dFr-2066 | M | 15 | Cameroon | −/+ | N | N | Cirrhosis |
| dFr-2204 | F | 23 | Ivory Coast | ND/+ | N | N | Cirrhosis |
| dFr-2301 | M | 40 | Ivory Coast | −/+ | 3N | 3N | ND |
| dFr-2317 | F | 25 | Guinea | +/+ | 2N | 3N | Cirrhosis |
Underlining indicates samples for which the complete HDV genome has been sequenced.
F, female; M, male.
IgM, IgG, immunoglobulins M and G.
ALT, alanine aminotransferase; AST, aspartic aminotransferase; N, ALT and AST normal upper limit values.
ACH, active chronic hepatitis.
ND, not determined.
HDV RNA extraction, reverse transcription, and amplification.
HDV RNA was extracted from 250 μl of serum, and region 6A-6S (237 nucleotides [nt]) was amplified as described in reference 7; primers 900s and 1280as encompass the R0 region (400 nt) covering the 3′ end of the HD gene (14). For all samples, two short reverse transcription-PCR-generated DNA fragments were directly sequenced.
Complete-genome molecular cloning.
Based on R0 sequence data, complete HDV cDNA was amplified using synthetic primers (Table 2). For four samples (dFr-45, dFr-47, dFr-48, and dFr-73), two overlapping cDNA fragments of 850 and 1,050 nt were used to obtain the entire HDV genome. Specific primers were also determined to amplify cDNA from samples dFr-644 and dFr-910. The PCR-amplified fragments were cloned into the TA Cloning pCRII vector (Invitrogen), and at least two clones were sequenced bidirectionally using the BigDye Terminator technology (ABI Prism 377; Perkin-Elmer Applied Biosystems).
TABLE 2.
Primers used for the PCR amplification of HDV African cDNAa
| Sample | Primer nameb | Sequence (position)c |
|---|---|---|
| dFr-45, dFr-47, dFr-48, dFr-73 | 305s | CTCCAGAGGACCCCTTCAGCGAAC (305-328) |
| 1161as | CCCGCGGGTTGGGGATGTGAACCC (1161-1138) | |
| 962s | GTACACTCGAGGAGTGGAAGGCG (962-984) | |
| 331as | TCTGTTCGCTGAAGGGGTCCT (331-311) | |
| dFr-644 | 307s | CCAGAGGACCCCTTCAGCGAAC (307-328) |
| 1289as | GAAGGAAGGCCCTCGAGAACAAGA (1289-1265) | |
| 889s | CATGCCGACCCGAAGAGGAAAG (889-911) | |
| 446as | AACACCCTCCTGCTAGCCCC (446-427) | |
| dFr-910 | 1206s | CCGGAGTTCCTCTTCCTCCTCC (1206-1227) |
| 929as | GTTCGCGTCCGAGTCCTTCTTTC (929-907) | |
| 1531s | GAGCTTTCTTCGATTCGGAC (1531-1550) | |
| 572as | GACTGGTCCCCTCATGTTCC (572-553) |
Primers used in cloning and complete African HDV genome sequence determination.
s, sense; as, antisense.
According to reference 41.
Secondary-structure prediction.
Secondary-structure prediction was performed using complete antigenome sequences of HDV genotypes I (Italy-A20), IIA (Japan-S), IIB (Taiwan-TW-2b), and III (Peru-1) and African isolates (dFr-45, dFr-47, dFr-48, dFr-73, dFr-644, and dFr-910). The mfold program version 3.1, predicting possible secondary structures for RNA sequences, is available at http://bioinfo.math.rpi.edu/∼mfold/rna. HD protein secondary-structure prediction was performed using the secondary-structure consensus from the network protein sequence interface available at the Pole Bio-Informatique Lyonnais (http://npsa-pbil.ibcp.fr).
Sequence alignments and phylogenetic analyses.
Alignment of HDV R0 sequences was generated with ClustalW 1.8 using a gap-opening penalty (GOP) of 15 and a gap extension penalty (GEP) of 6.66 with minimum manual corrections. Because different values of GOP and GEP gave different results, full-genome alignments were performed using the SOAP program (22). This program generates and compares alignments corresponding to 30 different sets of alignment parameters (GEP from 12 to 17 in steps of 1; GOP from 6 to 8 in steps of 0.5). Positions with different alignments can be excluded, or a proportion of GOP and GEP combinations yielding the same alignment can be studied. For example, with full-length genomes (26 taxa, approximately 1,800 characters), we compared the results of three phylogenetic analyses corresponding to three different consensus alignments: strict (734 characters excluded), 80% (690 characters excluded), and 50% (559 character excluded) consensus among the 30 different SOAP-generated alignments. We also used ProAlign (21), a program that provides a statistical approach to multiple-sequence alignment, such that a posterior probability is assigned to each aligned position. Positions with a posterior probability below a user-defined threshold can be excluded before phylogeny inference is made.
Maximum-parsimony (MP; using heuristic for the R0 region or branch-and-bound for the full genome and sHD searches) and neighbor-joining (NJ; maximum-likelihood [ML] distances) phylogenetic analyses were performed with PAUP*4.0b6 (38). ML analyses were carried out both with PAUP*4.0b6 (full-length and sHD data sets) and the metapopulation genetic algorithm (metaGA) (18) (all data sets) implemented in the program MetaPIGA (http://www.ulb.ac.be/sciences/ueg). MetaGA ML analyses were performed with the following settings: Hasegawa-Kishino-Yano (HKY) model, taking into account the proportion of invariable sites and rate heterogeneity (four categories), four populations of four individuals each, and probability consensus pruning. Bootstrap analyses (103 and 104 replicates for MP and NJ, respectively) and 10,000 metaGA samples (2,500 replicates with four populations, ML analyses) were used to assess the robustness and posterior probabilities of nodes, respectively. Due to computational limitations, ML inferences using PAUP*4.0b6 were performed only on taxon subsets (24 full-genome sequences and 33 sHD sequences). We used the HKY model and estimated the transition/transversion ratio, the proportion of invariable sites, and rate heterogeneity (gamma distribution with four categories) parameters from the data. Bootstrap analyses were limited to 100 replicates with the values of ML parameters constrained to those estimated from the original ML search.
Nucleotide sequence accession numbers.
The accession numbers of the sequences used were as follows: Afghanistan, AF008373; Albania-02, AF008309; Archangelos, AF008333; Cagliari, X85253; China, X77627; Colombia, L22061; Egypt, AF008375; Ethiopia, U81989; French-02, AF008372; Greek-19, AF008320; Italy- A20, X04451; Italy-35, AF008420; Japan-S, X60193; Lebanon, M84917; Miyako, AF309420; Nauru, M58629; Okinawa (Ok) 1-18, AB015442; Ok2-05, AB015443; Ok3-25, AB015444; Ok4-15, AB015445; Ok5-01, AB015446; Ok6-21, AB015447; Peru-1, L22063; Peru-2, L22064; Romania, AF008319; Russia, AF008374; Somalia, U81988; Taiwan, M92448; Taiwan-3, U19598; Taiwan-TW-2b, AF018077; TW2476, AF104264; Turkish-01, AF008347; US-1, D01075; US-23, AF008371; VnzD8349, AB037948; VnzD8375, AB037947; VnzD8624, AB037949; Woodchuck 5 (W5), AJ307077; Woodchuck 15 (W15), M21012; Yakut (Ya)-8, AJ309871; Ya-12, AJ309872; Ya-13, AJ309868; Yakut-26, AJ309879; Ya-29, AJ309869; Ya-30, AJ309873; Ya-51, AJ309876; Yakut-62, AJ30880; Ya-63, AJ309875; Ya-245, AJ309874; Ya-704, AJ309877; Ya-724, AJ309878; dFr-1594 (Angola), AJ583884; dFr-71 (Central African Republic), AJ583879; dFr-45 (Cameroon), AJ583868; dFr-48 (Cameroon), AJ583871; dFr-55 (Cameroon), AJ583872; dFr-56 (Cameroon), AJ583873; dFr-1953 (Cameroon); AJ583886; dFr-2066 (Cameroon), AJ583888; dFr-644 (Congo), AJ583882; dFr-74 (Democratic Republic of Congo), AJ583881; dFr-59 (Egypt), AJ583874; dFr-70 (Egypt), AJ583878; dFr-46 (France), AJ583869; dFr-1843 (Gabon), AJ583885; dFr-69 (Gambia), AJ583877; dFr-4 (Ghana), AJ583867; dFr-47 (Guinea), AJ583870; dFr-2317 (Guinea), AJ583891; dFr-73 (Ivory Coast), AJ583880; dFr-2020 (Ivory Coast), AJ583887; dFr-2204 (Ivory Coast), AJ583889; dFr-2301 (Ivory Coast), AJ583890; dFr-910 (Mali), AJ583883; dFr-60 (Romania), AJ583875; dFr-65 (Romania), AJ583876.
RESULTS
Phylogenetic analyses of HDV genomes.
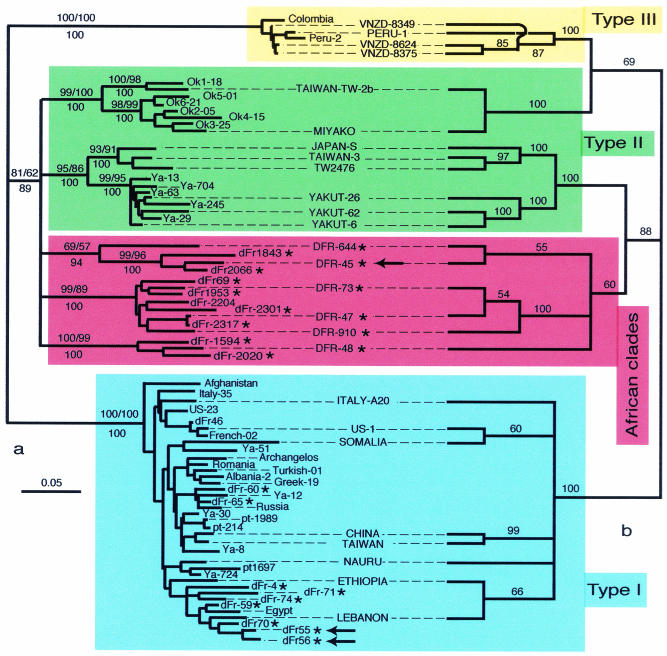
We used a 336-nt HDV cDNA fragment (here called R0), encompassing the 3′ end of the HD gene, to characterize a large number of clinical samples. From 227 patients monitored for HDV replication analyses between 1999 and 2001, we selected 25 samples (i) whose HDV cDNA could not be amplified by using previously described 6A and 6S primers although their HDV serology was positive (Table 1) or (ii) whose R0 DNA amplicon restriction pattern was atypical (data not shown). Of the 25 HDV-infected patients, 22 were later found to originate from African countries (Table 1). The 25 new R0 sequences were aligned with 16 sequences from Russia (13) and 36 R0 sequences from public databases. Analyses using MP, NJ, and ML methods yielded results compatible with the phylogenetic tree shown in Fig. 1a. Of the 22 newly characterized African sequences, 15 form at least three lineages (red box in Fig. 1) spanning a range of variability much larger than for type I or type III. The other seven African sequences are scattered within the type I clade.
FIG. 1.
Phylogenetic trees inferred from delta virus nucleotide sequences. (a) NJ phylogram obtained from the R0 data set; bootstrap values obtained for NJ and MP are indicated above the branches, whereas ML posterior probabilities inferred from 400 metaGA samplings are indicated below the branches. (b) ML cladogram obtained from the full genome data set (capital-lettered taxon); numbers at the branches indicate ML posterior probabilities inferred from 400 metaGA samplings. Of the 25 viral HDV sequences reported here, 15 form new clades (boxed in red), characterizing a variability range larger than that of type I or type III sequences. The remaining 10 new sequences cluster with type I. Asterisks indicate newly characterized HDV sequences. Arrows correspond to HDV genome sequences within a family, indicating that horizontal, rather than vertical, transmission from the mother (dFr-45) occurred. Scale is in percent expected substitution per position.
To test the validity of our results, we sequenced the full genome of isolates (Fig. 1b) spanning the range of the newly characterized clades (accession numbers AJ584844 to AJ584849). Alignments among these sequences showed approximately 25% divergence (Table 3), similar to that observed between type I and type II sequences, suggesting the existence of additional types. To avoid artifacts due to alignment ambiguities, we used the SOAP and ProAlign programs, the former to produce and compare 30 alignments (each corresponding to 1 of 30 different sets of alignment parameters) and the latter to provide statistical alignments so that each column has a posterior probability of being correctly aligned. The phylogenetic analyses were performed again after excluding positions (i) that are different among the 30 SOAP alignments or (ii) that have a posterior probability of <90% in the ProAlign alignments. These analyses yielded results (Fig. 1b) similar to those described above (Fig. 1a): the genetic variability of the HDV genus has more major monophyletic groups (i.e., clades) than previously thought.
TABLE 3.
Percent similarity between African full-length genomes and HDV complete sequence references
| Sample | % Similarity toa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Type I | Type II | TWD62/Miyako | Type III | dFr47 | dFr48 | dFr73 | dFr644 | dFr910 | |
| dFr45 | 75.6 (74.2-76.9) | 75.9 (74.9-76.6) | 76.6 (76.4-76.8) | 67.8 (67.5-68.4) | 76.8 | 74.3 | 76.7 | 77.8 | 75.5 |
| dFr47 | 75.8 (74.3-77.5) | 80.4 (79.2-81.6) | 77.9 (77.5-78.3) | 68.1 (67.9-68.5) | 78.2 | 90.8 | 76.9 | 89.9 | |
| dFr48 | 74.9 (73.2-75.7) | 77.6 (76.7-78.1) | 76.8 (75.5-78.0) | 68.3 (68.0-68.7) | 77.8 | 75.9 | 76.4 | ||
| dFr73 | 75.7 (74.5-77.0) | 79.8 (78.7-81.2) | 78.3 (77.7-78.9) | 68.6 (68.0-69.4) | 76.0 | 89.4 | |||
| dFr644 | 74.6 (73.2-75.7) | 77.3 (76.9-78.0) | 77.0 (76.4-77.7) | 66.9 (65.8-67.9) | 76.4 | ||||
| dFr910 | 75.9 (74.5-77.2) | 79.7 (78.7-81.0) | 78.3 (78.0-78.5) | 67.9 (67.0-68.7) | |||||
Mean values were obtained by aligning each African genome with genotype I (13 sequences), II (6 sequences), III (4 sequences), and the Taiwan TW-2b and Miyako sequences. Ranges are given in parentheses.
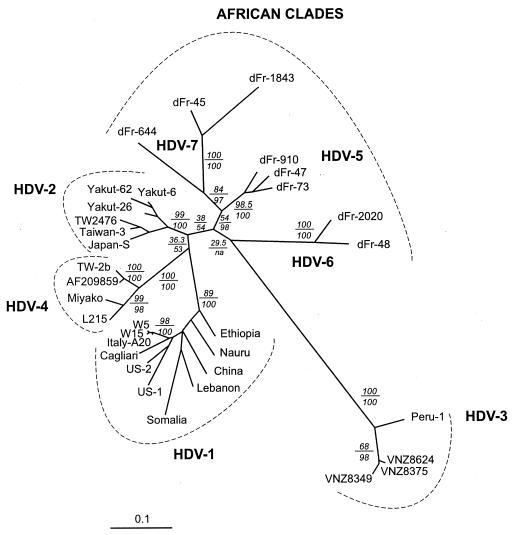
Finally, because there is functional evidence that the sHD protein trans-complements the corresponding HDV type more efficiently (4, 19), we phylogenetically analyzed the sHD coding sequences from 33 isolates including newly characterized strains from the island of Okinawa (24) and Taiwan (42) that had been tentatively identified as a genotype IIB and eight sHD gene sequences from African strains. The phylogenetic results obtained again confirmed that the African HDV clades account for a large proportion of HDV worldwide variability (Fig. 2). These analyses, including the African HDV sequences, also confirm that sequences from the “genotype IIB” complex form a distinct clade (proposed as HDV-4 in Fig. 2) which is not directly related to the genotype IIA sequences (cf. HDV-2 in Fig. 2). In all, at least seven major clades, including the well-defined genotype I (HDV-1 in Fig. 2) and genotype III (HDV-3 in Fig. 2), were identified; three of them (HDV-5, HDV-6, and HDV-7) correspond exclusively to African HDV sequences (Fig. 2).
FIG. 2.
ML analyses of 33 sHD cDNA sequences. Analyses were conducted using PAUP4b6 and MetaPIGA. Numbers above the horizontal lines indicate bootstrap values (100 replicates with PAUP*, parameters constrained to those obtained from the ML analysis of the original data set), and those below the lines indicate metaGA posterior probabilities (10,000 samples). Scale is in percent expected substitution per position. Suggested clade names are indicated.
HD protein and RNA secondary structure.
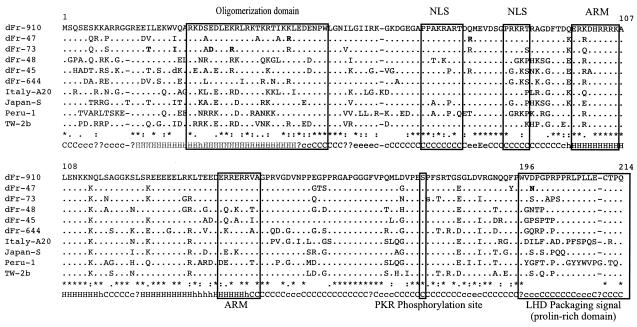
All the newly characterized complete HDV sequences exhibited the two expected overlapping open reading frames (sHD and LHD), and most of the conserved motifs were located in the central and carboxy-terminal regions of the sHD (Fig. 3). Since most patients studied had a clear antibody response to type I HD recombinant antigen (Table 2), we might expect immunogenic epitopes to correspond to these conserved regions. Analysis of the carboxy-terminal part of the LHD proteins reveals a highly variable proline-rich domain except for three conserved residues (Fig. 3).
FIG. 3.
Alignment of HDV amino acid sequences from African (dFr-910, dFr-47, dFr-73, dFr-48, dFr-45, and dFr-644,), Italian (A20, genotype I), Japanese (-S, genotype IIA), Peruvian (-1, genotype III), and Taiwanese (TW-2b, genotype IIB) isolates. Dots indicate identities to dFr-910, whereas asterisks indicate positions conserved across all aligned sequences. Bold type indicates polymorphisms. Motifs corresponding to functional properties are boxed (NLS, nuclear localization signal; ARM, arginine-rich motifs; PKR, protein kinase R). The carboxy-terminal part of LHD constitutes a highly variable proline-rich domain corresponding to the packaging signal except for three conserved residues, a tryptophan, a cysteine, and a glutamine, corresponding, respectively, to the UAG-to-UGG editing site, the farnesylation box CXXX, and the C-terminal residue. Letters on the bottom line indicate when all (capital letters) or 50 to 99% (lowercase letters) of the 10 aligned sequences yielded identical secondary-structure consensus predictions (H and h, helix; C and c, coiled; E and e, beta-sheet).
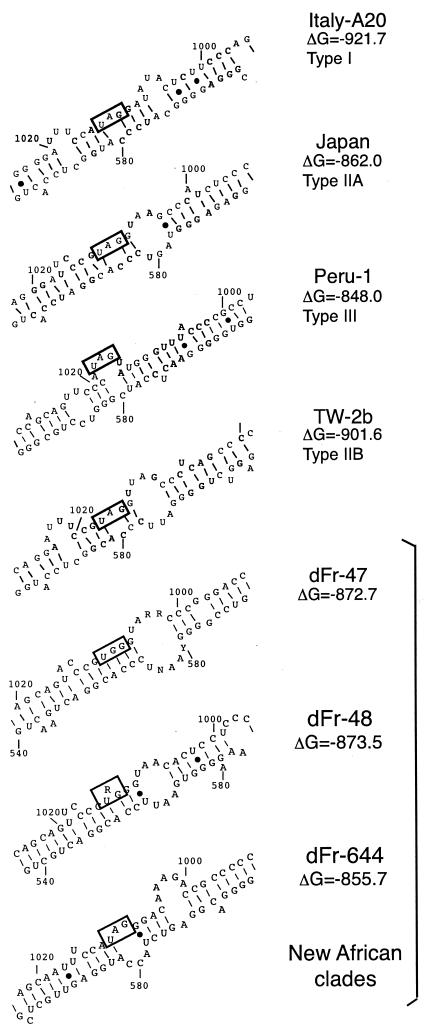
Predictions of HDV-RNA antigenomic secondary structure indicate that the characterized African isolates exhibit slightly different patterns in the vicinity of the RNA-editing site (at the amber/tryptophan codon) (Fig. 4). Each prototype sequence from each clade had a specific antigenomic RNA pseudo-double-strand secondary structure. Alternative branched secondary structures were also observed for some sequences (data not shown).
FIG. 4.
Pseudo-double-strand antigenomic HDV RNA structure, as inferred by the mfold program, in the vicinity of the editing site for representatives of HDV-1, HDV-2, HDV-3, and HDV-4 and three representatives of the new African clades (dFr-47, dFr-48, and dFr-644). Boxes indicate the stop codons for the small hepatitis delta protein. The base-paired structure required for edition (4 bp on each side of the central adenosine of the amber codon) is observed only for clade HDV-1, strain A20.
HDV phylogeography.
Due to historical geopolitical factors, and similar to hepatitis C virus (HCV) (26) and HBV, the delta viruses characterized in Paris (France) showed a wide African distribution (Table 1). For example, the six full-length viral RNA sequences were obtained from five patients from Western or Central sub-Saharan African countries (Cameroon, Guinea, Ivory Coast, Mali, and Republic of Congo) and from an adult Polish woman who had lived in Cameroon for 3 years. To determine whether the geographic distribution of the HDV isolates was correlated with their levels of sequence divergence, we compared the Kimura-2 parameter pairwise distances between full-genome sequences with the relative geographic distance matrix of the capitals of the countries where the patients had been infected. To compare two quantitative continuous variables, we decided to calculate the correlation coefficient to evaluate the degree of proportionality of data sets. When the ubiquitous type I sequences were removed, a significant statistical correlation was observed between the two matrices (Pearson correlation coefficient r = 0.791, P < 0.0001). Because r = 0.791, we can estimate that in this data set the Kimura-2 genetic distances and the corresponding relative geographic distances were directly proportional, with a minimal risk of error (0.0001).
DISCUSSION
The extensive genetic variability of HBV-related viruses suggests that hepadnaviruses have been infecting humans and other primates for a long time (11, 16, 25). HBV is endemic in sub-Saharan Africa, and chronically infected mothers transmit the virus to their children at birth, although intrafamilial spreading can occur later in childhood (9). The age of the HDV-HBV association needs to be clarified. Clearly, the classification of HDV into only three “genotypes” does not reflect the actual range of variability of the Deltavirus genus. Indeed, using the delta antigen gene and full genome sequence data, we identified a wide and probably ancient radiation of African lineages (as suggested by several clades branched inside the deepest part of the unrooted tree [Fig. 2]), making the genetic variability of HDV much wider than previously thought, although the South American sequences (HDV clade 3) remain the most divergent group. Furthermore, strains Taiwan-TW-2b, Miyako, L215, and AF209859 should be considered a specific clade, distinct from those including genotypes I, II, and III. We suggest that the sequence TW-2b (Fig. 1 and 2) be considered the clade 4 prototype due to the early date of the study by Wu et al. (44). Finally, the additional African lineages described here suggest that there are at least three new clades. In our geographic area (near Paris, France), HDV African clades 5, 6, and 7 represent 10.2, 2, and 2% of the HDV sequences characterized in the 2002, respectively (E. Gordien, unpublished data). This would mean that the Deltavirus genus includes at least seven clades, which, interestingly, is very similar to the human HBV genetic variability (which includes six distinct genotypes [A to F] [reviewed in reference 32] plus the recently characterized HBV genotype G [35]).
The non-type-I HDVs from patients infected in Cameroon (dFr-45 dFr-48, and dFr-1953 [Fig. 1]) are good representatives of the newly characterized African clades. Furthermore, HDV type 1 sequences have also been found in patients originating from Cameroon (dFr-55 and dFr-56 [Fig. 1]). A similar genetic diversity has been observed in other RNA viruses in this restricted area, e.g., the highly divergent human immunodeficiency virus type 1 group O, initially described in Cameroon (40). The general phylogeographic pattern revealed by our analyses suggests that HDV sequences constitute, besides some DNA virus genomes (36), efficient markers of human migrations. However, horizontal acquisitions of strains, rather than strict vertical familial transmission, can also occur. This is shown by the observation that the HDV type I sequences (dFr-55 and dFr-56) were obtained from two girls who are the daughters of a woman from whom sequence dFr-45 was characterized (arrows in Fig. 1). Since a wide range of HDV genetic variability is suspected, specific diagnostic tools (conserved versus clade-specific PCR primers) should be used.
Finally, the wide radiation reported here might explain the spectrum of disorders associated with HDV. For example, specific liver lesions, including morula cells, have been observed in African and Amazonian patients with severe hepatitis (3, 29). All African samples studied here came from screening of HDV replication markers in patients with liver disease who were immigrating to France. Most patients suffered from active chronic hepatitis or cirrhosis, and two of them underwent liver transplantation. Although the deltaviruses corresponding to the African radiation in this study are associated with severe liver-specific HDV histological lesions, it should be emphasized that these virus lineages might not necessarily be as pathogenic in the general population. Obviously, other factors such as the time and duration of infection, the genetic background of the patient (8), and the HBV helper strain may contribute to the pathogenicity.
Acknowledgments
We are particularly grateful for the help given by all physicians in collecting clinical data from patients. We thank Camille Sureau and Patrick Mardulyn for helpful comments on an earlier version of the manuscript.
N.R. was partly supported by the “Association Nationale de Recherches sur le Sida (ANRS)”, V.I. was supported by a “bonus qualité recherche” from “Université Paris 13”, and M.C.M. is supported by the National Fund for Scientific Research Belgium (FNRS) and the “Communauté Française de Belgique.” This work is part of the program of the “Laboratoire Associé au Centre National de Référence des Hépatites B et C pour le virus delta” (E.G. and P.D.) supported by the French Ministry of Health.
REFERENCES
- 1.Brazas, R., and D. Ganem. 1996. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science 274:90-94. [DOI] [PubMed] [Google Scholar]
- 2.Casey, J. L. 1996. Hepatitis delta virus. Genetics and pathogenesis. Clin. Lab. Med. 16:451-464. [PubMed] [Google Scholar]
- 3.Casey, J. L., T. B. Brown, F. S. Wignall, and J. L. Gerin. 1993. A genotype of hepatitis delta virus that occurs in northern South America. Proc. Natl. Acad. Sci. USA 90:9016-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey, J. L., and J. L. Gerin. 1998. Genotype-specific complementation of hepatitis delta virus RNA replication by hepatitis delta antigen. J. Virol. 72:2806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, F.-L., P.-J. Chen, S.-J. Tu, C.-J. Wang, and D.-S. Chen. 1991. The large form of hepatitis δ antigen is crucial for assembly of hepatitis δ virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, Y. C., M. F. Chang, I. Gust, and M. M. Lai. 1990. Sequence conservation and divergence of hepatitis delta virus RNA. Virology 178:384-392. [DOI] [PubMed] [Google Scholar]
- 7.Dény, P., G. Fattovich, F. Le Gal, G. Giustina, C. Lecot, G. Morsica, H. Poinsot, A. Alberti, and C. Brechot. 1994. Polymerase-chain-reaction-based semi-quantification of hepatitis D viraemia in patients treated with high doses of alpha 2b interferon. Res. Virol. 145:287-295. [DOI] [PubMed] [Google Scholar]
- 8.Dieye, A., V. Obami-Itou, M. F. Barry, G. Raphenon, A. Thiam, R. Ndiaye, M. Ndiaye, M. L. Diaw, and L. Diakhate. 1999. Association between class I HLA alleles and HBs antigen carrier status among blood donors in Senegal. Dakar Med. 44:166-170. [PubMed] [Google Scholar]
- 9.Dumpis, U., E. C. Holmes, M. Mendy, A. Hill, M. Thursz, A. Hall, H. Whittle, and P. Karayiannis. 2001. Transmission of hepatitis B virus infection in Gambian families revealed by phylogenetic analysis. J. Hepatol. 35:99-104. [DOI] [PubMed] [Google Scholar]
- 10.Elena, S. F., J. Dopazo, R. Flores, T. O. Diener, and A. Moya. 1991. Phylogeny of viroids, viroidlike satellite RNAs, and the viroidlike domain of hepatitis delta virus RNA. Proc. Natl. Acad. Sci. USA 88:5631-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fares, M. A., and E. C. Holmes. 2002. A revised evolutionary history of hepatitis B virus (HBV). J. Mol. Evol. 54:807-814. [DOI] [PubMed] [Google Scholar]
- 12.Gaeta, G. B., T. Stroffolini, M. Chiaramonte, T. Ascione, G. Stornaiuolo, S. Lobello, E. Sagnelli, M. R. Brunetto, and M. Rizzetto. 2000. Chronic hepatitis D: a vanishing disease? An Italian multicenter study. Hepatology 32:824-827. [DOI] [PubMed] [Google Scholar]
- 13.Imazeki, F., M. Omata, and M. Ohto. 1991. Complete nucleotide sequence of hepatitis delta virus RNA in Japan. Nuclleic Acids Res. 19:5439-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivaniushina, V., N. Radjef, M. Alexeeva, E. Gault, S. Semenov, M. Salhi, O. Kiselev, and P. Deny. 2001. Hepatitis delta virus genotypes I and II cocirculate in an endemic area of Yakutia, Russia. J. Gen. Virol. 82:2709-2718. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins, G. M., C. H. Woelk, A. Rambaut, and E. C. Holmes. 2000. Testing the extent of sequence similarity among viroids, satellite RNAs, and hepatitis delta virus. J. Mol. Evol. 50:98-102. [DOI] [PubMed] [Google Scholar]
- 16.Lanford, R. E., D. Chavez, K. M. Brasky, R. B. Burns III, and R. Rico-Hesse. 1998. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc. Natl. Acad. Sci. USA 95:5757-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, C. M., C. S. Changchien, J. C. Chung, and Y. F. Liaw. 1996. Characterization of a new genotype II hepatitis delta virus from Taiwan. J. Med. Virol. 49:145-154. [DOI] [PubMed] [Google Scholar]
- 18.Lemmon, A., and M. Milinkovitch. 2002. The metapopulation genetic algorithm: an efficient solution for the problem of large phylogeny estimation. Proc. Natl. Acad. Sci. USA 99:10516-10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, F. M., C. M. Lee, T. C. Wang, and M. Chao. 2003. Initiation of RNA replication of cloned Taiwan-3 isolate of hepatitis delta virus genotype II in cultured cells. Biochem. Biophys. Res. Commun. 306:966-972. [DOI] [PubMed] [Google Scholar]
- 20.Long, M., S. J. de Souza, and W. Gilbert. 1997. Delta-interacting protein A and the origin of hepatitis delta antigen. Science 276:824-825. [DOI] [PubMed] [Google Scholar]
- 21.Loytynoja, A., and M. C. Milinkovitch. 2003. A hidden Markov model for progressive multiple alignment. Bioinformatics 19:1505-1513. [DOI] [PubMed] [Google Scholar]
- 22.Loytynoja, A., and M. C. Milinkovitch. 2001. SOAP, cleaning multiple alignments from unstable blocks. Bioinformatics 17:573-574. [DOI] [PubMed] [Google Scholar]
- 23.Luo, G. X., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma, S.-P., H. Sakugawa, Y. Makino, M. Tadano, F. Kinjo, and A. Saito. 2003. The complete genomic sequence of hepatitis delta virus genotype IIb prevalent in Okinawa, Japan. J. Gen. Virol. 84:461-464. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald, D. M., E. C. Holmes, J. C. Lewis, and P. Simmonds. 2000. Detection of hepatitis B virus infection in wild-born chimpanzees (Pan troglodytes versus): phylogenetic relationships with human and other primate genotypes. J. Virol. 74:4253-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morice, Y., D. Roulot, V. Grando, J. Stirnemann, E. Gault, V. Jeantils, M. Bentata, B. Jarrousse, O. Lortholary, C. Pallier, and P. Deny. 2001. Phylogenetic analyses confirm the high prevalence of hepatitis C virus (HCV) type 4 in the Seine-Saint-Denis district (France) and indicate seven different HCV-4 subtypes linked to two different epidemiological patterns. J. Gen. Virol. 82:1001-1012. [DOI] [PubMed] [Google Scholar]
- 27.Nakano, T., C. N. Shapiro, S. C. Hadler, J. L. Casey, M. Mizokami, E. Orito, and B. H. Robertson. 2001. Characterization of hepatitis D virus genotype III among Yucpa Indians in Venezuela. J. Gen. Virol. 82:2183-2189. [DOI] [PubMed] [Google Scholar]
- 28.Niro, G. A., J. L. Casey, E. Gravinese, M. Garrubba, P. Conoscitore, E. Sagnelli, M. Durazzo, N. Caporaso, F. Perri, G. Leandro, D. Facciorusso, M. Rizzetto, and A. Andriulli. 1999. Intrafamilial transmission of hepatitis delta virus: molecular evidence. J. Hepatol. 30:564-569. [DOI] [PubMed] [Google Scholar]
- 29.Parana, R., F. Gerard, J. L. Lesbordes, C. Pichoud, L. Vitvitski, L. G. Lyra, and C. Trepo. 1995. Serial transmission of spongiocytic hepatitis to woodchucks (possible association with a specific delta strain). J. Hepatol. 22:468-473. [DOI] [PubMed] [Google Scholar]
- 30.Rizzetto, M., M. Canese, and S. Arico. 1977. Immunofluorescence detection of a new antigen antibody system (Delta/anti-Delta) associated to hepatitis B virus in liver and serum of HBs Ag carriers. Gut 18:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizzetto, M., and M. Durazzo. 1991. Hepatitis delta virus (HDV) infections. Epidemiological and clinical heterogeneity. J. Hepatol. 13:S116-S118. [DOI] [PubMed] [Google Scholar]
- 32.Robertson, B. H., and H. S. Margolis. 2002. Primate hepatitis B viruses—genetic diversity, geography and evolution. Rev. Med. Virol. 12:133-141. [DOI] [PubMed] [Google Scholar]
- 33.Sakugawa, H., H. Nakasone, T. Nakayoshi, Y. Kawakami, S. Miyazato, F. Kinjo, A. Saito, S. P. Ma, H. Hotta, and M. Kinoshita. 1999. Hepatitis delta virus genotype IIb predominates in an endemic area, Okinawa, Japan. J. Med. Virol. 58:366-372. [DOI] [PubMed] [Google Scholar]
- 34.Shakil, A. O., S. Hadziyannis, J. H. Hoofnagle, A. M. Di Bisceglie, J. L. Gerin, and J. L. Casey. 1997. Geographic distribution and genetic variability of hepatitis delta virus genotype I. Virology 234:160-167. [DOI] [PubMed] [Google Scholar]
- 35.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 81:67-74. [DOI] [PubMed] [Google Scholar]
- 36.Sugimoto, C., T. Kitamura, J. Guo, M. N. Al-Ahdal, S. N. Shchelkunov, B. Otova, P. Ondrejka, J. Y. Chollet, S. El-Safi, M. Ettayebi, G. Gresenguet, T. Kocagoz, S. Chaiyarasamee, K. Z. Thant, S. Thein, K. Moe, N. Kobayashi, F. Taguchi, and Y. Yogo. 1997. Typing of urinary JC virus DNA offers a novel means of tracing human migrations. Proc. Natl. Acad. Sci. USA 94:9191-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sureau, C., B. Guerra, and R. E. Lanford. 1993. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J. Virol. 67:366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swofford, D. 1998. PAUP*: Phylogenetic Analysis Using Parsimony (and other methods), version 4.0d64 (in progress). Sinauer Associates, Sunderland, Mass.
- 39.Taylor, J. M. 2003. Replication of human hepatitis delta virus: recent developments. Trends Microbiol. 11:185-190. [DOI] [PubMed] [Google Scholar]
- 40.Vanden Haesevelde, M., J. L. Decourt, R. J. De Leys, B. Vanderborght, G. van der Groen, H. van Heuverswijn, and E. Saman. 1994. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J. Virol. 68:1586-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, K.-S., Q.-L. Choo, A. J. Weiner, J.-H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta (∂) viral genome. Nature 323:508-514. (Erratum, 328:456, 1987.) [DOI] [PubMed] [Google Scholar]
- 42.Wang, T. C., and M. Chao. 2003. Molecular cloning and expression of the hepatitis delta virus genotype IIb genome. Biochem. Biophys. Res. Commun. 303:357-363. [DOI] [PubMed] [Google Scholar]
- 43.Weiner, A., Q.-L. Choo, K.-S. Wang, S. Govindarajan, A. Redeker, J. Gerin, and M. Houghton. 1988. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24∂ and p27∂. J. Virol. 62:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, J. C., T. Y. Chiang, and I. J. Sheen. 1998. Characterization and phylogenetic analysis of a novel hepatitis D virus strain discovered by restriction fragment length polymorphism analysis. J. Gen. Virol. 79:1105-1113. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi, Y., J. Filipovska, K. Yano, A. Furuya, N. Inukai, T. Narita, T. Wada, S. Sugimoto, M. M. Konarska, and H. Handa. 2001. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293:124-127. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y. Y., E. Tsega, and B. G. Hansson. 1996. Phylogenetic analysis of hepatitis D viruses indicating a new genotype I subgroup among African isolates. J. Clin. Microbiol. 34:3023-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]