Abstract
Nucleos is a web server for the identification of nucleotide-binding sites in protein structures. Nucleos compares the structure of a query protein against a set of known template 3D binding sites representing nucleotide modules, namely the nucleobase, carbohydrate and phosphate. Structural features, clustering and conservation are used to filter and score the predictions. The predicted nucleotide modules are then joined to build whole nucleotide-binding sites, which are ranked by their score. The server takes as input either the PDB code of the query protein structure or a user-submitted structure in PDB format. The output of Nucleos is composed of ranked lists of predicted nucleotide-binding sites divided by nucleotide type (e.g. ATP-like). For each ranked prediction, Nucleos provides detailed information about the score, the template structure and the structural match for each nucleotide module composing the nucleotide-binding site. The predictions on the query structure and the template-binding sites can be viewed directly on the web through a graphical applet. In 98% of the cases, the modules composing correct predictions belong to proteins with no homology relationship between each other, meaning that the identification of brand-new nucleotide-binding sites is possible using information from non-homologous proteins. Nucleos is available at http://nucleos.bio.uniroma2.it/nucleos/.
INTRODUCTION
The majority of cellular key processes involves a transfer of energy and genetic information. These processes have in common the same biological currency, represented by nucleotides. Different types of nucleotides exist, but all share the same chemical groups, or modules: the nucleobase, the carbohydrate and the phosphate group. Given the ubiquitous nature of nucleotides, it is not surprising that they were among the earliest cofactors bound by proteins during evolution (1). The interaction between nucleotides and proteins has been extensively studied so that many features that proteins must possess to interact with a nucleotide have been discovered (2–4), such as the P-loop and the Walker A motifs. Some structural features have been also derived such as the acceptor–donor–acceptor environment necessary for the binding of the nucleobase group (5) and several phosphate-binding structural motifs (6,7). However, the binding site of a nucleotide cannot be simply reduced to these features, as some studies highlighted the large amount of possible conformations, even not energetically favorable, that can be presented by nucleotides when bound by proteins (8). Therefore the identification of binding sites for nucleotides in protein structures is not an easy task. Different web servers are available for the identification of nucleotide-interacting residues in protein sequences, mostly based on machine learning approaches, like ATPint, GTPbinder, NADbinder and NsitePred (9–12). From the structural point of view, no web server has been dedicated to the identification of nucleotide-binding sites in protein structures. Some methods have been developed for the identification of carbohydrate- and nucleobase-binding sites (13,14), but no related web services have been produced. We developed in the past years, a method and a web server for the identification of phosphate-binding sites in protein structures (15,16), called Phosfinder. Given this scenario, we decided to build a web server for the identification of nucleotide-binding sites based on the concept of nucleotide modularity, described by Gherardini et al. (17) and used to predict nucleotide-binding sites in protein structures (18). This concept is based on the observation that nucleotides, and their binding sites, are composed of modules shared by evolutionary unrelated proteins and combinable in different ways to form binding sites even for different types of nucleotides. This web server, called Nucleos, searches for structural similarities between the query protein structures and a dataset of template binding sites for nucleotide modules: the nucleobase, the carbohydrate and the phosphate. Each similarity identifies a putative binding site for a nucleotide module, evaluated according to its position in space with respect of the protein surface and taking into account the conservation of the involved residues. Complete nucleotide-binding sites are built combining predicted nucleotide modules following distance thresholds observed in crystallized structures of bound nucleotides. Nucleos allows the biologist user to scan protein structures of interest for binding sites for different types of nucleotides directly on the web, at the address http://nucleos.bio.uniroma2.it/nucleos/.
MATERIALS AND METHODS
The Nucleos web server is based on a previously developed methodology (18) for the identification of nucleotide-binding sites in protein structures based on the concept of nucleotide modularity. Binding sites for nucleotide modules (the nucleobase, the carbohydrate and the phosphate) are predicted independently; subsequently, they are joined together to build complete nucleotide-binding sites.
Identification of potential binding sites for nucleotide modules
The Superpose3D (19) structural comparison algorithm is used to find structural similarities between the query protein structure and a dataset of template-binding sites for nucleobase, carbohydrate and phosphate modules (4657, 3073 and 10 185, respectively). The template-binding sites are composed of at least three residues of a binding pocket interacting with at least one atom of the ligand. Structural similarities are evaluated by the Root Mean Square Deviation (RMSD) of the matching residue atoms and by the BLOSUM62 substitution value of the residues involved in the similarity. Whenever a structural similarity is found, the nucleotide module bound by the template-binding site is transposed onto the query protein structure following the structural match with the residues of the query protein. Any predicted module-binding site placed inside the protein or at less then a specified distance from the solvent accessible surface of the protein is discarded. These distances are derived after analyzing the minimum distances observed by nucleotide modules from the protein surface in nucleotide–protein complexes; therefore, a threshold for each nucleotide module is derived. The remaining predictions of the same type are clustered together with a hierarchical clustering procedure.
Scoring of predicted binding sites
A clustering score is assigned to each prediction as the amount of predictions in its cluster. A conservation score is assigned to each prediction as the sum of the conservation value of the query protein structure residues involved in its structural similarity. This conservation value is calculated from the PFAM multiple alignments of the domains contained in the query protein structures, and represents the percentage of similar residues (BLOSUM62 substitution value ≥1) in the alignment column. This percentage is then normalized using percentiles of the distribution of values for each PFAM domain; this provides comparable conservation scores between different protein domains and different proteins. The final score of a prediction is the sum of the clustering and conservation score.
Identification of complete nucleotide-binding sites
Finally, predicted nucleotide modules are joined to form complete nucleotide-binding sites for a particular nucleotide type: e.g., an ADP-binding site will be reconstructed using predicted binding sites for a nucleobase, a carbohydrate and two phosphates, in this order. Modules are joined following a set of empirically derived distance thresholds between nucleotide modules obtained by analyzing crystallized structure of bound nucleotides. If a full reconstruction is not possible, the method tries to build the largest sub-architecture possible, e.g. the ADP is a sub-architecture of the ATP. The minimum allowed sub-architecture is composed of two consecutive nucleotide modules (nucleobase–carbohydrate and carbohydrate–phosphate). Nucleos is based on scripts written in Python, C and C++ that are linked to the web interface using CGI.
Usage of Nucleos
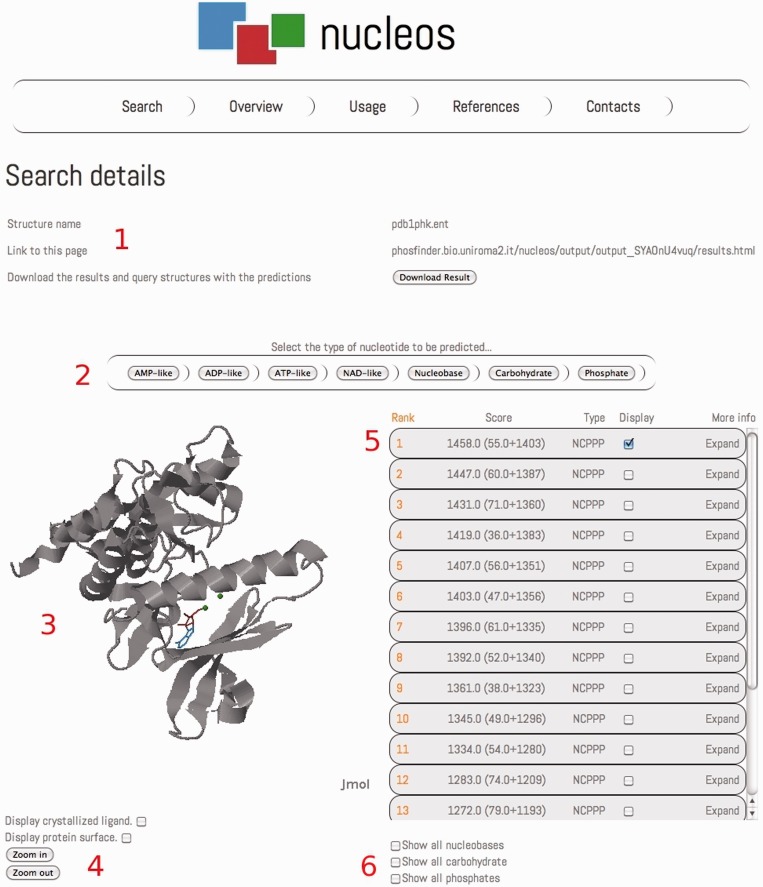
The Nucleos search page accepts as input query protein structures both as PDB codes and as PDB structures uploaded by the user. In both cases, the chain of the protein structure to analyze can be specified; in the case of an uploaded structure, the user can specify a reference PDB structure to use to calculate the residue conservation of the query structure. In this page, the user can also provide an email address to which links to the results will be sent, although providing an email address is not compulsory. A loading page will report the real-time status of each query. A typical Nucleos analysis takes few minutes for each protein (2 minutes for a 300-residues protein structure). The output of Nucleos reports the details of the query, the link to the results page that the user can bookmark and a downloadable folder containing results in the form of both a table and a PDB structure containing the predictions (Figure 1). Predictions are grouped by the their type (e.g. AMP-like), and the output reports information about the structural similarity and the residues involved in the query structure and in the template-binding site, and the sequence identity between the query protein and the protein containing the template-binding site (Figure 2). A checkbox is provided for each module in the prediction to highlight the residues involved in the binding of the module directly on the Jmol graphical applet (an open-source Java-based viewer for 3D chemical structures available at http://www.jmol.org/). The template-binding site can be viewed and highlighted in a popup Jmol applet in its original structure. All the predictions are displayed together with the query protein structure in a Jmol applet in the results page (Figure 1). A menu is provided to switch the results in the table and in the Jmol applet depending on the type of the predictions. Several buttons are provided to show/hide the protein surface, any crystallized ligand and predicted bound nucleotide modules. A page describing the usage of Nucleos is provided to the user together with explanatory images. The user can also explore an example output page in the Usage page and can know more about the core methodology behind the Nucleos server in the Overview page. The Web site also includes help pages that guide the user with pre-computed examples (a complete and interactive output page is given as example). The Nucleos web server is freely available at http://nucleos.bio.uniroma2.it/nucleos/ and does not require any registration.
Figure 1.
An example of the Nucleos output page. The details of the query are displayed on the top of the page (1), reporting the name of the structure, the address of the results page and a download link to get a folder containing results in form of table and of PDB structures containing the predictions. A menu is provided to quickly navigate the results, switching among the different nucleotide predictions (2). A Jmol applet displays the query protein structure with the prediction (3), when switching to results for a different nucleotide type, the first prediction in the ranking is displayed. Buttons are provided to interact with the Jmol applet (4): zoom in/out, show/hide protein surface and crystallized ligand. Results for ATP-like predictions are ranked in the results table on the right (5). Buttons are provided to quickly show/hide all the predicted nucleobase, carbohydrate and phosphate binding sites (6).
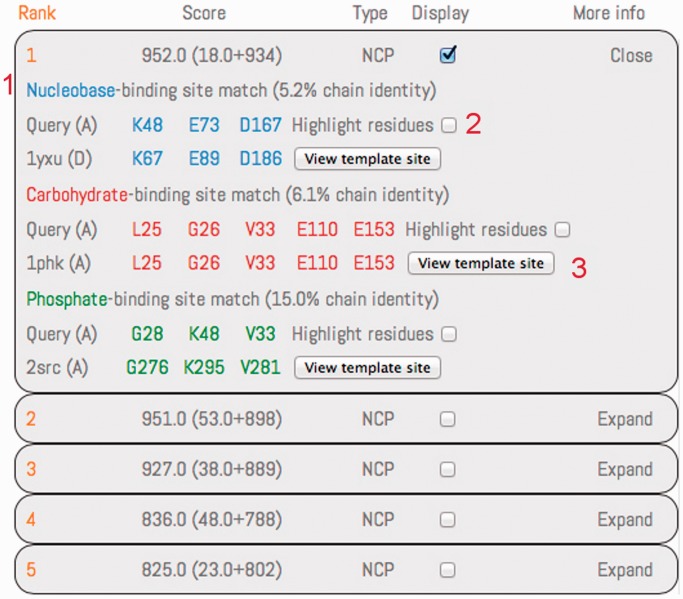
Figure 2.
The results section for the first ranked prediction has been expanded using the button on the top right. Predictions are ranked by their score, with the scoring components reported within brackets (clustering score plus conservation score). The type of the prediction is reported using the ‘N’, ‘C’ and ‘P’ letters for the nucleobase, carbohydrate and phosphate, respectively. Results are displayed for all the modules composing the prediction, following a color code (blue for nucleobase, red for the carbohydrate and green for the phosphate). The structural similarity between residues of the query protein and a template binding site is detailed with the involved residues, paired following the structural match. The sequence identity, between the query protein structure and the protein containing the template-binding match involved in the similarity, is reported for each module. A checkbox is provided to highlight/bleach the residue in the Jmol applet. The template-binding site, highlighted in its original PDB structure, can be displayed in a popup Jmol applet.
RESULTS
The method (18) has been tested on a set of 924 high-quality non-redundant (30% of sequence identity as threshold) proteins binding one of the following nucleotide types: AMP, ADP, ATP, GDP, GTP, ANP, GNP, FAD, FMN, NAD and NAP.
Evaluation of predicted binding sites for nucleotide modules
We evaluated the performance of the method in predicting binding sites for nucleotide modules independently with a 10-fold cross validation test. For each of the 10 tests, the training and the test set respected a 9:1 ratio; an optimal scoring threshold is chosen on the training set and applied to the test set. The results were evaluated with precision, recall and F-score that is the harmonic mean of precision and recall. The method gained an average F-score of 0.48, 0.47 and 0.64 on nucleobases-, carbohydrates- and phosphates-binding sites, respectively. Importantly, we found that the performance of the method can vary significantly among different nucleotide modules and nucleotide types. For example, the method obtained an average F-score of 0.87, 0.62 and 0.92 on GNP-binding sites and 0.43, 0.6 and 0.71 on FAD-binding sites for the nucleobase, the carbohydrate and the phosphate. We also evaluated the method by ranking the predictions by their score; therefore, we measured the percentage of protein structures in which the method placed a correct prediction in the top one, three, five and ten predictions. Considering only the first-ranked prediction, the method was able to identify the binding site in 48%, 48% and 68% of the proteins for the nucleobase, carbohydrate and phosphate, respectively, while considering the top five predictions the performance changes to 71%, 65% and 86%. We observed again a different performance depending on the type of the nucleotide considered. Moreover, we observed that the method predicts carbohydrate-binding sites better on larger nucleotides, FAD, NAD and NAP, compared with the other nucleotides; a similar behavior can be observed with phosphate-binding sites in proteins binding guanine-containing nucleotides.
Performance on Apo and Holo protein structure
We tested the method on a set of apo/holo structure pairs of 64 proteins collected from LigASite (20) to see whether the conformational change between the apo and holo states of the same nucleotide-binding proteins could affect the method performance. We observed on average a 7% difference between the performance on apo and holo structures when considering the top-ranked prediction, while the performance difference decreased to an average value of 2% when considering the top five predictions. We can conclude that the method is only slightly affected by conformational changes between apo and holo structures, thus representing a valid tool for the analysis of protein structures crystallized without a nucleotide.
Evaluation of predicted nucleotide-binding sites
To predict complete nucleotide-binding sites, the method links the predicted binding site for each nucleotide module. Nucleotide modules are linked following distance thresholds that have been empirically derived by analyzing all the nucleotide–protein complexes in the PDB. These thresholds are composed of a maximum and minimum distance for a particular pair of modules, say nucleobase–carbohydrate, to be linked. We evaluated a predicted nucleotide-binding site to be correct if the RMSD of the modules in the predicted binding site with the corresponding modules in the crystallized ligand is ≤5 Å. The method placed a correct prediction in the first rank in 59% of the analyzed proteins.
In 98% of the cases, all the modules composing a correct prediction are predicted from a template-binding site belonging to non-homologous proteins (30% sequence identity threshold). This shows that complete nucleotide-binding sites can be assembled from their components, which can belong to unrelated and non-homologous proteins.
DISCUSSION
Nucleos is a web server for the identification of nucleotide-binding sites in protein structures. Nucleos is based on the concept of nucleotide and binding-site modularity and represents a valuable resource for structural biologists. It offers the possibility of immediately visualizing the results of the prediction on a query PDB structure or on a user-uploaded PDB structure directly on the web with a graphical interface. Predicted nucleotide modules, and the information about their prediction, can be inspected both independently and combined into nucleotide-binding sites. The prediction regards not only the position in space of the nucleotide modules, but also the protein amino acids involved in their binding. All together these information represent a valuable resource for drug-design and docking-guided experiments. The performance of the method has been tested on a set of 924 non-redundant nucleotide-binding protein structures and on apo-holo pairs of nucleotide-binding protein structures. We observed no significant variation in the method performances when analyzing apo and holo structures. Moreover, in the majority of the cases, the method was able to identify the correct nucleotide-binding site with the top-ranked prediction using template-binding sites from non-homologous proteins. This enables the prediction of unknown or not annotated nucleotide-binding sites in newly discovered protein structures.
FUNDING
PRIN 2010 [prot. 20108XYHJS_006 to M.H.C.]; FIRB ‘Futuro in ricerca’ Project [RBFR08ZSXY to G.A.]. Funding for open access charge: AIRC [IG 10298 to M.H.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ji HF, Kong DX, Shen L, Chen LL, Ma BG, Zhang HY. Distribution patterns of small-molecule ligands in the protein universe and implications for origin of life and drug discovery. Genome Biol. 2007;8:R176. doi: 10.1186/gb-2007-8-8-r176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;I:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fry DC. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc. Natl Acad. Sci. 1986;83:907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spiegel A, Carter A, Brann M, Collins R, Goldsmith P, Simonds W, Vinitsky R, Eide B, Rossiter K, Weinstein L. Signal transduction by guanine nucleotide-binding proteins. Recent Prog. Horm. Res. 1988;44:337–375. doi: 10.1016/b978-0-12-571144-9.50015-6. [DOI] [PubMed] [Google Scholar]
- 5.Denessiouk KA, Johnson MS. “Acceptor–Donor–Acceptor” motifs recognize the Watson–Crick, Hoogsteen and Sugar “Donor–Acceptor–Donor” edges of adenine and adenosine-containing ligands. J. Mol. Biol. 2003;333:1025–1043. doi: 10.1016/j.jmb.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita K, Sadanami K, Kidera A, Go N. Structural motif of phosphate-binding site common to various protein superfamilies: all-against-all structural comparison of protein mononucleotide complexes. Protein Eng. 1999;12:11–14. doi: 10.1093/protein/12.1.11. [DOI] [PubMed] [Google Scholar]
- 7.Ausiello G, Gherardini PF, Gatti E, Incani O, Helmer-Citterich M. Structural motifs recurring in different folds recognize the same ligand fragments. BMC Bioinformatics. 2009;10:182. doi: 10.1186/1471-2105-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stockwell GR, Thornton JM. Conformational diversity of ligands bound to proteins. J. Mol. Biol. 2006;356:928–944. doi: 10.1016/j.jmb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Chauhan JS, Mishra NK, Raghava GPS. Identification of ATP binding residues of a protein from its primary sequence. BMC Bioinformatics. 2009;10:434. doi: 10.1186/1471-2105-10-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chauhan JS, Mishra NK, Raghava GPS. Prediction of GTP interacting residues, dipeptides and tripeptides in a protein from its evolutionary information. BMC Bioinformatics. 2010;11:301. doi: 10.1186/1471-2105-11-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansari HR, Raghava GPS. Identification of NAD interacting residues in proteins. BMC Bioinformatics. 2010;11:160. doi: 10.1186/1471-2105-11-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen K, Mizianty MJ, Kurgan L. Prediction and analysis of nucleotide-binding residues using sequence and sequence-derived structural descriptors. Bioinformatics (Oxford, England) 2012;28:331–341. doi: 10.1093/bioinformatics/btr657. [DOI] [PubMed] [Google Scholar]
- 13.Taroni C, Jones S, Thornton JM. Analysis and prediction of carbohydrate binding sites. Protein Eng. 2000;13:89–98. doi: 10.1093/protein/13.2.89. [DOI] [PubMed] [Google Scholar]
- 14.Saito M, Go M, Shirai T. An empirical approach for detecting nucleotide-binding sites on proteins. Protein Eng. 2006;19:67–75. doi: 10.1093/protein/gzj002. [DOI] [PubMed] [Google Scholar]
- 15.Parca L, Gherardini PF, Helmer-Citterich M, Ausiello G. Phosphate binding sites identification in protein structures. Nucleic Acids Res. 2011;39:1231–1242. doi: 10.1093/nar/gkq987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parca L, Mangone I, Gherardini PF, Ausiello G, Helmer-Citterich M. Phosfinder: a web server for the identification of phosphate-binding sites on protein structures. Nucleic Acids Res. 2011;39:W278–W282. doi: 10.1093/nar/gkr389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gherardini PF, Ausiello G, Russell RB, Helmer-Citterich M. Modular architecture of nucleotide-binding pockets. Nucleic Acids Res. 2010;38:3809–3816. doi: 10.1093/nar/gkq090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parca L, Gherardini PF, Truglio M, Mangone I, Ferrè F, Helmer-Citterich M, Ausiello G. Identification of nucleotide-binding sites in protein structures: a novel approach based on nucleotide modularity. PloS One. 2012;7:e50240. doi: 10.1371/journal.pone.0050240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gherardini PF, Ausiello G, Helmer-Citterich M. Superpose3D: a local structural comparison program that allows for user-defined structure representations. PloS One. 2010;5:e11988. doi: 10.1371/journal.pone.0011988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dessailly BH, Lensink MF, Orengo CA, Wodak SJ. LigASite—a database of biologically relevant binding sites in proteins with known apo-structures. Nucleic Acids Res. 2008;36:D667–D673. doi: 10.1093/nar/gkm839. [DOI] [PMC free article] [PubMed] [Google Scholar]