Abstract
A rubella virus (RUB) replicon, RUBrep/PAC, was constructed and used to map the 3′ cis-acting elements (3′ CSE) of the RUB genome required for RUB replication. The RUBrep/PAC replicon had the structural protein open reading frame partially replaced by a puromycin acetyltransferase (PAC) gene. Cells transfected with RUBrep/PAC transcripts expressed the PAC gene from the subgenomic RNA, were rendered resistant to puromycin, and thus survived selection with this drug. The relative survival following puromycin selection of cells transfected with transcripts from RUBrep/PAC constructs with mutations in the 3′ CSE varied. The 3′ region necessary for optimal relative survival consisted of the 3′ 305 nucleotides (nt), a region conserved in RUB defective-interfering RNAs, and thus this region constitutes the 3′ CSE. Within the 3′ CSE, deletions in the ∼245 nt that overlap the 3′ end of the E1 gene resulted in reduced relative survivals, ranging from 20 to <1% of the parental replicon survival level while most mutations within the ∼60-nt 3′ untranslated region (UTR) were lethal. None of the 3′ CSE mutations affected in vitro translation of the nonstructural protein open reading frame (which is 5′ proximal in the genome and encodes the enzymes involved in virus RNA replication). In cells transfected with replicons with 3′ CSE mutations that survived antibiotic selection (i.e., those with mutations in the region of the 3′ CSE that overlaps the E1 coding region), the amount of replicon-specific minus-strand RNA was uniform; however, the accumulation of both plus-strand RNA species, genomic and subgenomic, varied widely, indicating that this region of the RUB 3′ CSE affects plus-strand RNA accumulation rather than minus-strand RNA synthesis.
Rubella virus (RUB) is the sole member of the Rubivirus genus within the Togavirus family. The RUB genome consists of a single-stranded, plus-strand RNA molecule 9,762 nucleotides (nt) in length (7) which is 5′ capped and 3′ polyadenylated and serves as an mRNA during infection. The genome contains two open reading frames (ORFs): the 5′-proximal ORF encodes nonstructural proteins (NS-ORF), including the RNA-dependent RNA polymerase, that are necessary for viral RNA replication, while the 3′-proximal ORF encodes the viral structural proteins (SP-ORF), the capsid protein (CP), and two envelope glycoproteins, E1 and E2. The NS-ORF is translated from the genomic RNA, and the SP-ORF is translated from a subgenomic RNA. The untranslated regions (UTRs) within the RUB genome include a 40-nt sequence at the 5′ end (5′ UTR), a 118-nt sequence in the intragenic (junction) region between the ORFs (J-UTR), and a 59-nt sequence at the 3′ end (3′ UTR). Replication of the RUB genome initiates with the synthesis of a genomic-length minus-strand RNA which is then used as a template for the synthesis of both the plus-strand RNA species. The subgenomic RNA is synthesized from an internal promoter within the J-UTR.
cis-Acting elements at the 3′ end of the RUB genome (3′ CSE) are proposed to act as promoters for minus-strand RNA synthesis (8). These sequences may also be important in translation of the NS-ORF (24). Characterization of RUB defective-interfering (DI) RNAs generated during serial undiluted passaging revealed that these DI RNAs maintained the 5′ end of the genome, the NS-ORF, and the 3′ end of the genome but contained deletions in the SP-ORF that could include the subgenomic RNA start site (6). Minimally, the 3′ 305 nt of the genome was maintained, and therefore this region contains the putative 3′ CSE. Included in this region is a prominent stem-loop [3′(+)SL] structure at the end of the SP-ORF, 59 nt from the 3′ terminal poly(A) tract, shown to interact with at least three cellular proteins, one of which was identified as the autoantigen calreticulin (23, 30).
Four stem-loop (SL) structures were thermodynamically predicted to exist within the 3′ terminal 240 nt of the RUB genome (termed SL1 to SL4) (4). Mapping of the elements essential for replication within the 3′ region from SL2 [the 3′ (+)SL which is at the 3′ end of the SP-ORF] through the 3′ end of the genome using the RUB infectious clone, Robo302, has been done previously (4). The existence of SL2, SL3, and SL4 was confirmed by RNase mapping. Most of the 3′ UTR was necessary for viral replication except for the 3′-terminal 5 nt. However, maintenance of SL2 was not required for replication although it was necessary for optimal calreticulin binding, suggesting that the interaction between calreticulin and SL2 was not necessary for replication. Although infectious cDNA clones are extremely useful for studying virus replication, they cannot easily be used to analyze nucleic acid sequences or structures in protein-coding regions, since only site-directed mutagenesis that maintains amino acid sequences can be employed. For this reason, DI cDNA clones are commonly used since they are smaller than infectious clones and deletions can be made without regard to protein-encoding sequences (16). RUB replicons in which reporter genes replace the SP-ORF, thus mimicking RUB DI RNAs, have been developed (32). In this study, we created a RUB replicon in which an antibiotic resistance gene replaced most of the SP-ORF and which is thus capable of generating stable, replicon-containing cell lines following antibiotic selection. This replicon was used for deletion mapping of the 3′ CSE, including the region overlapping the 3′ end of the SP-ORF.
MATERIALS AND METHODS
Cell lines, transfection, and antibiotic selection.
BHK-21 and Vero cells, maintained at 35°C in Dulbecco's minimal essential medium (Invitrogen, Carlsbad, Calif.) supplemented with 5% fetal bovine serum and gentamicin (10 mg/ml) were passaged by trypsinization followed by dilution into fresh medium. Transfection with in vitro RNA transcripts was carried out using Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol. Electroporation followed the standard protocol described in Current Protocols in Molecular Biology (2). For antibiotic selection, at 24 h posttransfection or postelectroporation, the medium was removed from the cells and medium containing puromycin (Sigma, St. Louis, Mo.) at 2 μg/ml was added. Control Vero and BHK cells were killed in about 14 and 5 days of antibiotic treatment, respectively.
After puromycin selection, the cultures were trypsinized and the surviving cells were counted using a hemacytometer. Additionally, 1/30 of the cell suspension was replated in medium containing puromycin. Once cell colonies were visible to the naked eye, the plates were stained with 0.1% crystal violet. This method of directly visualizing colony density was termed a “reverse plaque assay.” Cultures that survived puromycin selection were subcultured up to five times in the presence of puromycin for subsequent experiments.
Replicon construction and site-directed mutagenesis.
RUBrep/PAC was generated from Robo302, an infectious genomic cDNA clone (31) by replacing the majority of the SP-ORF with the antibiotic resistance gene, puromycin acetyltransferase (PAC) (Fig. 1A). Mutations in the 3′ UTR originally created in Robo302 (4) were introduced into RUBrep/PAC by replacing the BamHI-EcoRI fragment with the fragments from Robo302 mutants. To create additional deletions in the 3′ end of the E1 coding region in RUBrep/PAC, oligonucleotide primers containing a unique restriction enzyme site near the deletion site, the nucleotides up to the deletion, and 15 nt beyond the deletion site were designed and used in a PCR with an appropriate upstream or downstream primer. The amplified PCR product was used to replace the corresponding restriction fragment in RUBrep/PAC. Construction of pGEM-GFP220 was done by PCR amplification of a fragment between nt 6463 and 6682 in RUBrep/GFP, a RUB replicon with the green fluorescent protein reporter gene, using primers pGEM-GFP220 (+) and (−) (GGAATTCCTGGTGGGTACCCAA and CCCAAGCTTCGACCAGGATGGGCA), digesting the amplified fragment with HindIII and EcoRI, and cloning it into HindIII-EcoRI-restricted pGEM3Zf(-) vector (Promega, Madison, Wis.). Synthesis of capped in vitro RNA transcripts from EcoRI-linearized Robo302 and RUBrep/PAC constructs using SP6 RNA polymerase was done as previously described (26). After DNase digestion, the reaction mixtures were used directly for transfection and electroporation.
FIG. 1.
Replication of the RUBrep/PAC replicon in BHK and Vero cells. (A) Schematic of RUBrep/PAC compared to RUB genome. RUBrep/PAC has most of SP-ORF replaced by a puromycin resistance (PAC) gene. As indicated, the PAC gene was preceded by an introduced sequence (in lowercase type) that included an XbaI restriction site (underlined), a TAG stop codon in frame with NS-ORF, a Kozak sequence for optimal translation initiation (in italics), and the start codon for the PAC gene. (B) Replication of RUBrep/PAC in BHK and Vero cells by Northern hybridization. Either BHK or Vero cells were transfected with RUBrep/PAC using Lipofectamine 2000 and subjected to puromycin selection, and intracellular RNA from the surviving cells at passage 3 was analyzed by Northern hybridization using a DIG-11-dUTP-labeled, nick-translated, RUBrep/PAC DNA probe. Intracellular RNA preparations from mock-infected and RUB-infected BHK or Vero cells were included for comparison. Viral genomic RNA (vG), viral subgenomic RNA (vSG), rG, and rSG are indicated.
Analysis of virus- and replicon-specific intracellular RNA species.
Intracellular RNA was prepared using Tri-Reagent following the protocols of the manufacturer (Molecular Research Center Inc., Cincinnati, Ohio). To detect RUB-specific RNA species by Northern hybridization, standard protocols supplied with Northern-Hyb kits (Ambion, Austin, Tex.) were used. Northern blots were probed with RUBrep/PAC plasmid DNA that has been random-primed labeled with digoxigenin (DIG)-11-dUTP using a DIG DNA labeling kit (Roche, Branchburg, N.J.). Detection of RNA species that hybridized to DIG-labeled probes was accomplished by enzyme immunoassay with chemiluminescence using DIG luminescent detection kit (Roche) followed by exposure to Kodak X-ray film.
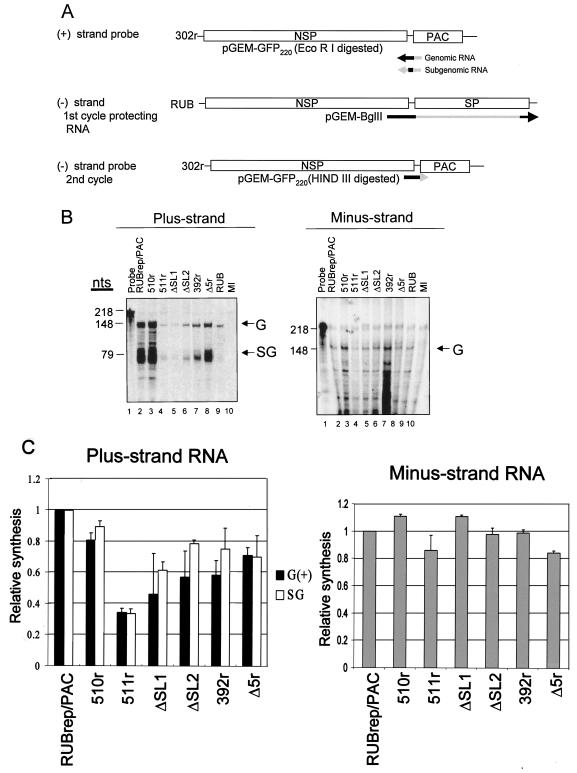
To detect and quantitate strand-specific RNA, a one-cycle (plus-strand) or two-cycle (minus-strand) RNase protection assay (RPA) developed by Liang and Gillam (17) was employed with minor modifications: most importantly, to eliminate DNA contamination, both the intracellular RNA sample and the probe mixture were treated with RNase-free DNase (1 U/μl; Roche) followed by inactivation of DNase by 10 mM EDTA at 65°C for 20 min and ethanol precipitation prior to hybridization. 35S-labeled probes were produced by in vitro runoff transcription of pGEM3Z-GFP220. Plus-strand probe (to detect plus-strand RNA) was synthesized by SP6 RNA polymerase from GFP220 linearized with EcoRI, while minus-strand probe (to detect minus-strand RNA) was synthesized by T7 RNA polymerase and HindIII-linearized template. In both cases, the reaction cocktail contained 25 nM unlabeled CTP and 35S-CTP (10 μCi/ml [1,000 Ci/mmol]; Amersham, Arlington Heights, Ill.). For the two-cycle RPA, the denatured, intracellular RNA was first hybridized with an unlabeled, plus-strand RNA transcript from pGEM-BglII (first cycle protecting RNA), which contains RUB sequences from the unique BglII site at nt 5352 of the RUB genome through the 3′ end of the genome. Equal amounts (in micrograms) of first-cycle protecting RNA and minus-strand probe were used in two-cycle RPA, resulting in an ∼10-fold excess of probe in the final hybridization.
In vitro translation.
In vitro RNA transcripts from RUBrep/PAC constructs synthesized in the presence of cap analog were phenol-chloroform extracted, and the RNA was ethanol precipitated in the presence of 400 mM sodium acetate. The quality of the RNA was examined by electrophoresis in a 0.8% agarose gel and the amount of RNA was determined spectrophotometrically (Beckman DU640 spectrophotometer). Equal amounts of RNA transcripts (250 ng/10 μl) were added to an in vitro-translation cocktail (TNT Quick Master Mix; Promega) containing RNase inhibitor (1 U/μl; Roche) and [35S]methionine (400 μCi/ml [1,000 Ci/mmol]; ICN Pharmaceutical, Inc., Costa Mesa, Calif.). Following a 2 h-incubation at 30°C, one-fifth of the reaction mixture was analyzed on a sodium dodecyl sulfate-8% polyacrylamide gel.
Quantitation of radiolabeled products.
Quantitation of in vitro translation products or Northern gel or RPA bands was done using a FluorChem imaging system (Alpha Innotech Corporation, San Leandro, Calif.).
RESULTS
Construction of RUBrep/PAC and generation of puromycin-resistant cells.
As shown in Fig. 1A, RUBrep/PAC maintained the entire 5′ ORF, but the entire coding regions of C, E2, and part of E1 were deleted and replaced with a dominant selectable marker puromycin N-acetyl transferase (PAC). To optimize translation of the PAC gene and to ensure the correct reading frame was translated, the 5′ junction sequences of the insert were modified to contain a stop codon (UAG) followed by an optimal Kozak sequence prior to the start codon (Fig. 1A). The 3′-terminal 600 nt in the RUB genome were maintained in RUBrep/PAC, and a UAG stop codon was introduced at the 3′ end of the PAC gene to prevent production of a fusion protein with the E1 coding sequences remaining in this region. Because RUBrep/PAC retained the NSP-ORF and CSE required for viral RNA replication, autonomous replication of RUBrep/PAC should occur in the absence of helper virus conferring antibiotic resistance. Following transfection of either BHK or Vero cells with RUBrep/PAC and antibiotic selection, colonies of antibiotic-resistant cells grew (see below). Both replicon-specific genomic RNAs (rG) and replicon-specific subgenomic RNAs (rSG) were detected in puromycin-selected cells (Fig. 1B, lanes 3 and 6). Because BHK cells were more sensitive to puromycin and yielded a better recovery of resistant colonies after transfection with RUBrep/PAC than Vero cells, they were used in the following experiments.
Mapping the 3′ CSE required for RUB genome replication using RUBrep/PAC.
The 3′-terminal 305 nt of the RUB genome, the region conserved in all DI RNAs and thus comprising the putative 3′ CSE (6), was predicted to contains four SL structures: SL1, nt 9527 to 9647, and SL2, nt 9671 to 9702, both of which are at the 3′ end of the SP-ORF; and SL3, nt 9703 to 9730; and SL4, nt 9742 to 9753 (Fig. 2A) (4). The SL3 and SL4 structures are within the 3′ UTR. SL4 is followed by a 7-nt single-stranded leader sequence and the poly(A) tract.
FIG. 2.
Effects of deletions and point mutations in the 3′-terminal 600 nt of RUBrep/PAC on the generation of puromycin-resistant colonies. (A) Deletion analysis in E1 coding region. Four mutations deleting large regions of the 3′ 600 nt of the RUB genome contained in RUBrep/PAC were made as indicated. The regions of predicted RNA secondary structures in the 3′-terminal 240 nt of RUB genome are represented by different patterns of boxes (SL1 [cross-hatch pattern], SL2 [dark shading], SL3 [light shading], and SL4 [black with white dots]), while the regions without significant predicted RNA secondary structure are denoted by very light shading (white boxes with widely spaced black dots). Deletions are represented by dashed lines, and the nucleotides are numbered according to their positions in the RUB genome. Coding sequences for the reporter gene, PAC, are represented as a solid box. BHK cells were electroporated with in vitro transcripts from RUBrep/PAC or one of the RUBrep/PAC mutants. Following antibiotic selection for 7 days, the number of cells surviving antibiotic selection was determined and compared to surviving cells of RUBrep/PAC (relative survival). The relative survival shown is the average of five electroporations with each construct. Approximately 1/30 of the surviving cells from one electroporation was replated, colonies were allowed to grow, and colonies were stained in order to give a direct visualization of the number of cells surviving antibiotic selection (reverse plaque assay). (B) Mutagenic analysis of SL2 and 3′ UTR. The relative survival of various SL2 and 3′ UTR modifications of RUBrep/PAC is compared with previously published results of the same modifications in an infectious clone (Robo302) (4). The names of the RUBrep/PAC and the infectious clone construct (in parentheses) are both given (first column). A description of the construct is given (column 2); negative numbers indicate the position from the 3′ end of the RUB genome. Results from the previous work (4) (columns 3 and 4) and the present study (column 5) with replicons are shown. Replicon results were determined as described for panel A. Relative survival (R.S.) for each replicon construct is shown. Note that the relative infectivity of the infectious clone construct 392 refers to the original mutant, not the revertant.
Mapping using the infectious clone Robo302 showed that the 3′ UTR with the exception of the 3′ 5 nt was necessary for virus replication (4). Site-directed mutagenesis within SL2 revealed that destabilization of the SL decreased calreticulin binding but had no effect on infectivity. However, deletion mapping within the E1 coding region could not be done easily without interfering with the amino acid sequence. Since nt 9170 through 9762, the 3′ ∼600 nt of the RUB genome, was present in RUBrep/PAC following the PAC termination codon, we were able to make a series of deletions across these sequences in this vector (Fig. 2).
Equal amounts of transcripts from the parent and each mutated replicon construct were used to electroporate BHK cells, and puromycin selection began 24 h postelectroporation. The number of surviving cells was determined after 96 to 120 h of puromycin selection; the percentage of surviving cells in comparison with parental RUBrep/PAC was defined as the relative survival. Additionally, approximately 1/30 of cell suspensions used to determine surviving cell numbers were replated for a visual display of relative survival, termed a reverse plaque assay. Interestingly, high relative survival, twice that of RUBrep/PAC, was obtained with 510r, a construct in which the 3′-terminal sequences were reduced to the 3′ 305 nt retained in DI RNAs. Deletions within the 305-nt region were deleterious. Deletion from the 5′ boundary of the 3′ 305-nt element to SL1 (Δ 9452-9526) reduced relative survival to 8% (511r). Deletion of SL1 resulted in a more dramatic reduction in relative survival of 100-fold (in addition, following selection, cells containing ΔSL1 grew more slowly than did cells containing other constructs), and deletion of SL2 reduced relative survival to 8%. The effects of these deletions could be visualized by reverse plaque assay in that the intensity of staining was proportional to relative survival (Fig. 2A). These results showed that deletions of the E1 coding region within the 3′ 305-nt element did not abolish replication, and thus these sequences are not required for replication but are necessary for high relative survival.
Deletions, rearrangements, and single site mutations introduced in the 3′ UTR, shown in Fig. 2B, had previously been made in Robo302 (4), including ΔSL3 (Δ 9703-9730), 393r (Δ −6 to −10), 450r (destabilized SL4 structure), 524r (Δ −8 to −10), 392r (Δ −6 and −7), and Δ5r (Δ −1 to −5). The Δ5 mutation in the 3′ leader had no effect on relative survival, while deletion of nt −6 and −7 reduced relative survival by 100-fold (392r). None of the replicons with deletions in SL3 or SL4 were viable. These results were consistent with the mutagenesis results obtained with the infectious clone (4). The exception was mutation 524r, which failed to yield antibiotic-resistant cells in RUBrep/PAC although in Robo302 viruses were recovered. However, such viruses only replicated to a titer of 1/10 that of Robo302 virus and contained sequence modifications at the 3′ end of the genome that were different than the introduced mutation. Results from reverse plaque assay correlated with observed relative survivals (Fig. 2B): no colonies were obtained from cells transfected with 393r, ΔSL3, 524r, or 450r, while a few colonies can be observed in 392r-transfected cells and a confluent monolayer of Δ5r-transfected cells was achieved. All these data not only confirmed the previous observations on the importance of 3′ UTR to replication but also suggested that the requirement for replicon replication may be more stringent than that for viral replication.
Finally, RUBrep/PAC was used to investigate two SL2 mutations, 430-GAG and 444, which had calreticulin binding affinities similar to those of wild-type (wt) SL2, but yielded virus that grew to ∼10-fold lower titers than did wt virus and had altered plaque morphologies. In these two mutations, a G was substituted for one member of the U-U bulge in SL2, producing a G-U or U-G base pair and thus “sewing up” the bulge (430-GAG changed U9696 to G, while 444 changed U9673 to G). However, both mutations resulted in changes to the E1 coding sequence: 444 changed L1056 (UUG) to W (UGG), while 430-GAG changed the E1 gene stop codon UAG to an R (GAG). Thus, it was not clear whether the reduced-yield phenotype of these mutant viruses was due to defects in viral RNA replication or to the changes in E1 amino acid sequence. When introduced into RUBrep/PAC, GAGr had a relative survival of roughly 2% that of RUBrep/PAC, while the survival of 444r was 10-fold higher than that of GAGr, but still less than 20% that of RUBrep/PAC. These results indicate that the phenotype expressed by these SL2 mutants was due to the change in RNA sequence or structure affecting replication rather than to the amino acid sequence change in E1.
To confirm that the antibiotic resistance of the cells surviving antibiotic selection was due to the replication of the replicon, the presence of replicon was confirmed by Northern hybridization. In each antibiotic-resistant cell line, rG and rSG were detected (data not shown). Because the RUBrep/PAC constructs were 1.4 to 1.6 kb shorter than the RUB genome, these RNAs migrated proportionality faster than did those of the virus; the migration rate of the genomic and subgenomic RNAs of each replicon construct relative to those of RUBrep/PAC corresponded to the size of the deletion within the replicon. The relative amounts of the replicon RNA species varied, and with some constructs—notably, those with low relative survivals (most notably 511r, ΔSL1, and GAGr)—greater amounts of total cell RNA were necessary to visualize the genomic and subgenomic RNAs. This showed that the lower relative survival associated with these replicons was related to the replication efficiency of the replicon. To confirm that the intended mutations were maintained in the replicons, the nucleotide sequence of replicons with point mutations, such as GAGr and 444r, and small deletions, such as 392r, Δ5r, and ΔSL2, were determined; in all cases the intended mutations were present without second-site mutations. In our previous study using infectious clones, reversions and second-site alterations were detected with some mutated constructs such as 524 and 392.
Effect of 3′ CSE mutations on replicon replication: translation of the NS-ORF.
Translation of eukaryotic mRNAs involves interaction of the 5′ and 3′ ends of the RNA molecule (28), and it was shown that inclusion of the 3′ end of the RUB genome in a reporter gene construct initiating from the 5′ end of the RUB genome enhanced both in vitro and in vivo expression by at least 50% (24). To investigate if the difference in relative survival among the RUBrep/PAC constructs with 3′ CSE mutations was due to reduction in translation efficiency of the NS-ORF, the translation of the NSPs from each RUBrep/PAC mutant was examined in vitro. The translation efficiencies among these mutants, based on the amount of P200 synthesized normalized to RUBrep/PAC, are shown in Fig. 3. Most of the mutants synthesized an amount of P200 that was within 20% of that synthesized by RUBrep/PAC. The only exception was ΔSL3, which had a translation efficiency of ∼50% relative to RUBrep/PAC. Thus, although there was some variation in the translation of the NS-ORF, the differences do not simply explain results from antibiotic survival experiments (e.g., ΔSL3 is lethal but has an NS-ORP translation efficiency by 50%).
FIG. 3.
Effects of 3′ modifications on NSP synthesis in replicons. Equal amounts of in vitro transcripts from the indicated RUBrep/PAC constructs were translated in a rabbit reticulocyte lysate containing [35S]methionine. Following resolution by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, P200 polyprotein was quantitated by densitometry and the relative intensity, averaged from three independent experiments, was calculated by normalization to that of RUBrep/PAC. Error bars, standard deviations.
Effect of 3′ CSE mutations on replicon replication: replicon RNA synthesis in transfected cells.
The 3′ CSE of plus-strand RNA viruses may contain both the promoter and the initiation site for the minus-strand RNA synthesis (8). To examine if mutations in the 3′ CSE affected RNA synthesis by RUBrep/PAC replicons, the amount of minus- and plus-strand replicon-specific RNA present in cells transfected by parental and mutant replicon following drug selection was examined using an RPA (17) (Fig. 4). A representative RPA gel is shown in Fig. 4B, and the mean minus- and plus-strand RPA signal from three independent experiments is shown in Fig. 4C. The amount of minus-strand RPA product varied among the 3′ CSE mutants by only ±20% from that produced by RUBrep/PAC; however, the amount of plus-strand RPA product varied more widely. While the amount of plus-strand RPA product from 510r-transfected cells (the construct that maintains the 3′ CSE) was within 10 to 20% of that from RUBrep/PAC-transfected cells, the amount of plus-strand RPA product produced by the 3′ CSE mutants ranged from ∼35 to ∼70% that of RUBrep/PAC. These results were in agreement with Northern analysis in which roughly twice as much RNA from cells transfected with 511r and ΔSL1 was required to generate genomic and subgenomic bands of equal intensity to that for RUBrep/PAC (data not shown); these mutants produced ∼35 and ∼45% of the plus-strand RPA products of RUBrep/PAC. While most of these 3′ CSE mutants exhibited a lower relative survival after antibiotic a selection, a strict correlation between relative survival and strength of plus-strand RPA signal was not observed. For example, while 511r (∼8% relative survival) and ΔSL1 (∼1% relative survival) had plus-strand RPA signals of ∼35 and ∼45%, ΔSL2, 392r, and Δ5r (∼8, ∼1, and 130%) had plus-strand RPA signals of 60 to 70% of RUBrep/PAC.
FIG. 4.
Strand-specific RNA synthesis in BHK cells transfected with various RUBrep/PAC constructs. (A) Schematic representation of probes and protecting RNA used for RPA. To detect plus-strand RNAs, including genomic and subgenomic RNAs, 35S-labeled RNA transcripts of negative polarity with a total of 218 nt were prepared by runoff transcription in vitro using digested pGEM-GFP220 and were used directly in RPA. To detect minus-strand RNA, unlabeled RNA probe transcribed from a construct, pGEM-BglII, encoding 3′ 4,000 nt in the RUB genome, was first used, and the protected products from this first cycle of RPA were hybridized to a 35S-labeled pGEM-GFP220 probe of positive polarity. The regions that would hybridize to replicon genomes are denoted as black lines, while those that would be digested from protecting RNA and probes by RNase are shown in gray. Equal amounts of RNA extracted from cells within five passages following puromycin were used for RPA analysis. (B) Representative gels are shown: left panel, plus strand RNA; right panel, minus strand RNA. The product protected by genomic (G) RNA (both plus and minus strand) was 148 nt, and that protected by the subgenomic RNA (SG) was 79 nt, as indicated on the right margin of the gel. RPA band intensities were quantitated using a FluorChem imaging system. (C) The results of quantitation are the means of three RPA analyses using intracellular RNA from two independent transfections.
DISCUSSION
A RUB replicon, RUBrep/PAC, in which most of the SP-ORF was replaced with a puromycin resistance gene, PAC, was used in this study to analyze the 3′ CSE of the RUB genome. Transfection of BHK or Vero cells with this replicon allowed selection with puromycin of cells harboring the replicating replicon. Cells surviving selection contained both replicon-specific plus-strand RNAs, the genomic and subgenomic RNAs (Fig. 1), as well as minus-strand RNA (Fig. 4). This replicon offered the advantage over replicons which did not contain an antibiotic resistance gene (32) of an easy assay for replication: determining the number of cells surviving antibiotic selection (relative survival). Additionally, cotransfection with helper viruses and amplification by passaging was not necessary (16), and replicon-specific macromolecular synthesis could be studied in the cells that survived selection and were thus enriched for the presence of the replicon. Since replicons such as RUBrep/PAC do not spread between cells, revertants and second-site mutations, which were a problem in some of the mutations introduced into the 3′ CSE in the Robo302 infectious clone (4), were less problematic. Similar useful replicon systems have been developed for other viruses such as alphaviruses (1) and flaviviruses (15, 19).
In an earlier study (4), we made extensive series of mutations in the 3′ UTR using the Robo302 infectious clone, and in the present study we found that similar results were obtained when these mutations were introduced into RUBrep/PAC: namely, the 3′-terminal 5 nt (Δ5) was dispensable for replication while mutations in other regions of the 3′ UTR were not tolerated. Deletions of nt 6 to 7 from the 3′ end (392) dramatically attenuated replication; the 392r replicon construct had a relative survival of 1% of the parental replicon, while the infectious clone with this mutation (392) yielded virus that formed tiny plaques, replicated to 1/10 of the wt titer, and yielded viruses that had acquired sequence alterations. Constructs with SL3 deleted (ΔSL3), as well as two constructs with rearrangements in SL4 (393 and 450), were nonviable in both systems. A rearrangement in SL4 (524) was nonviable in the replicon system, but yielded virus in the infectious clone system, although it replicated to 14% of wt levels and contained modified 3′ terminal sequence. These results also demonstrate that the replicon system is less subject to rearrangements than is the infectious clone system.
Using RUBrep/PAC, we were able to extend deletion analysis to the complete putative 3′ CSE, including the E1 coding region which could not be so analyzed using the Robo302 infectious clone. Interestingly, a RUBrep/PAC construct which contained only the 3′ 305-nt element of the RUB genome which is conserved in DI RNAs (510r) had a higher survival rate than did RUBrep/PAC, which contained the 3′ 600 nt of the genome. However, three RUBrep/PAC constructs with large deletions in the E1 coding region of the 3′ 600-nt element had greatly lowered survival rates: 511r, which lacked the ∼5′ 75 nt of the 3′ 305-nt element preceding SL1, ΔSL1 and ΔSL2, which deleted SL1 and SL2, respectively. Taken together, these results demonstrated that the 3′ 305-nt element retained in DI RNAs constitutes the authentic 3′ CSE required for efficient replication. The 3′ CSE consists of ∼245 nt of the 3′ end of the SP-ORF followed by the ∼60-nt 3′ UTR, and while the majority of the 3′ UTR is necessary for replication, the region of the 3′ CSE containing E1 coding sequences is not since all of the replicon constructs that deleted part of the 3′ CSE within the E1 coding sequences yielded surviving cells following selection. The presence of RNA sequence or secondary structure elements within the protein coding regions that function in viral replication has been observed in other viruses. For example, in tobacco etch virus, a potyvirus, an internal RNA secondary structure in the capsid protein gene, located at the 3′ end of the genome, was found in conjunction with the 3′ UTR to be important for viral genome amplification (12, 20). In picornaviruses, an internal RNA structure located in capsid protein coding near the 5′ end of the genome (termed CRE) was found to function in recovery of replication of nonreplicating replicons (21, 22). RNA templates containing the CRE are preferentially used as templates for VPg uridylylation in vitro (27).
Of the landmarks within the RUB 3′ CSE, SL2 has attracted attention because of its ability to bind calreticulin in vitro and in vivo (23, 30). Using the infectious cDNA clone, single-base substitutions were made to change the structure of SL2 while avoiding interruption of the E1 coding frame, and an inverse correlation between calreticulin binding affinity and relative survival was found (4). Two of the mutations that were key in this conclusion were 444 and 430-GAG, each of which replaced one of the members of the U-U bulge in SL2 with a G, resulting in SL2 structures without a bulge. SL2 transcripts with either mutation bound calreticulin in vitro with equal affinity, as did the wt SL2 with the bulge, but viruses containing these mutations formed tiny plaques and produced titers ∼1.5 to 2.0 logs lower than did wt Robo302 virus. However, since both 444 and 430-GAG introduced amino acid changes into the E1 protein, it was not clear if the effect of these mutations was due to changes in RNA sequence or structure or changes in the E1 protein. In RUBrep/PAC, the 444r mutation reduced the survival rate by 5-fold, while the GAGr mutation reduced the survival rate by 50-fold, and thus the effect of both mutations is due to their effect on RNA sequence or structure, confirming the inverse correlation between calreticulin-binding efficiency and replication found in the earlier study (4). Since the SL2 structure formed by both of these mutants has the same calculated thermodynamic stability, the reason for the 10-fold difference in survival rate of the mutant replicons is puzzling. In RNase mapping studies, SL2 and SL3 were found to form two alternate structures that appeared to be in an equilibrium (4), and this equilibrium may be disrupted by 430-GAG. Deletion of SL2 entirely yielded a replicon that was viable, but with reduced relative survival (about 10% of wt). This finding also indicates that calreticulin binding to this SL is not necessary for replication, although the presence of this SL enhances replication. This finding indicates that calreticulin binding to SL2 may be necessary for optimal replication, however, since SL2 deletion was large, it is more likely that ΔSL2 impacts the overall structure of the 3′ CSE, reducing survival, rather than having a specific effect on calreticulin binding (see below).
We also used RUBrep/PAC to examine the effects of 3′ CSE mutations on specific steps in the virus replication cycle. The first step which the 3′ CSE could play a role is in translation of the replicase proteins from the NS-ORF. Although the 3′ end of eukaryotic mRNAs is involved in translation (29) and Pogue et al. (24) found that the RUB 3′ CSE increased expression of a reporter gene placed downstream from the RUB 5′ CSE, we found that none of the mutations introduced into the 3′ CSE of RUBrep/PAC significantly reduced in vitro translation of the NS-ORF. Secondly, the 3′ CSE potentially serves as the promoter for minus-strand RNA synthesis (8); however, we were unable to detect differences in minus-strand RNA synthesis in cells that survived antibiotic selection, including all of the mutants in the region of the 3′ CSE overlapping the E1 coding region and the viable mutants in the 3′UTR. Studies of other plus-strand RNA viruses have shown that the 3′ CSE is not necessarily required for minus-strand RNA accumulation (3, 18) or virus replication (31). In a study using Sindbis virus, a member of the Alphavirus genus of the Togavirus family, the 5′ CSE was found to be the critical element in directing minus-strand RNA synthesis (9). We also discovered that replicons with mutations in the 3′ CSE/E1 coding overlap region varied in the amount of plus-strand RNA present in cells following antibiotic selection. To our knowledge, this is the first report that the 3′ CSE of a plus-strand virus plays such a role in virus replication. We also attempted to analyze replicon-specific RNA synthesis in cells following transfection and before selection to gain information on RNA synthesis by nonviable replicon mutants, but both our RPA and Northern gels were not sensitive enough to detect plus-strand RNA synthesis (data not shown). We have recently produced a cell line that expresses the RUB capsid protein and exhibits enhanced replication of transfected RNA of both wt and mutant replicons; in this cell line we were able to detect plus-strand RNA before selection in cells transfected with replicon constructs which imparted high or intermediate relative survivals (e.g., 510r and ΔSL2), but not replicon constructs which imparted low relative survivals (e.g., 511r and ΔSL1), consistent with results on relative levels of RNA accumulation by these constructs in cells following selection.
The effect of the 3′ CSE in plus-strand RNA accumulation could be due to participation of the 3′ CSE in plus-strand RNA synthesis, presumably through an interaction between the 5′ CSE and 3′ CSE (albeit in the minus strand). Direct interactions of the 3′ CSE with the 5′ CSE have been reported for other plus-strand RNA viruses. In the case of flaviviruses, this interaction is mediated by complementary base pairing (14, 25), while in picornaviruses, the 5′ and 3′ termini may be bridged by cellular proteins (13). Circularization of the genome RNA of Sindbis virus has been observed previously (9); however, the sequences which mediated this interaction have not been identified. It is not known if the RUB genome is capable of circularizing, and thus the effect that mutations in the 3′ CSE would have on circularizing is hypothetical. Alternatively, the 3′ CSE could function in determining the stability of the plus-strand RNA species after synthesis. In cell mRNAs, determinants of differential stability commonly reside within the 3′ UTR (5, 28), and we found that mutations in the 3′ CSE affected both genomic and subgenomic RNA synthesis equally. The lack of strict correlation between plus-strand RNA accumulation and relative survival also indicates that the 3′ CSE could play other roles in replicon replication, e.g., translation efficiency of the subgenomic RNA, which is critical in production of the antibiotic resistance factor. Note that mutations introduced into the 3′ CSE may disturb the global structure of the entire RNA molecule, resulting in alteration or concealment of RNA sequences or structures that interact with the viral RNA replication complex and/or other trans-acting cellular factors essential for viral replication. For example, deletion of SL2 altered the structure of both SL1 and SL3 when analyzed using the Mfold program in the Genetics Computer Group package (35). Such large changes could also render the RNA more susceptible to intracellular degradation (11) or affect subgenomic RNA translation efficiency.
Finally, as discussed previously (32), the development of RUB replicons makes possible the generation of suicide vectors which cannot spread from cell to cell for delivery and expression of foreign genes. Such a system will require an efficient packaging systems for the replicons. In this study, the use of RUB replicons has been extended by manipulating them to express an antibiotic resistance gene such that stably transfected cell lines can be selected. Similar replicon-transfected stable cell lines have been developed for a number of plus-strand RNA viruses, and these replicons have been manipulated to express foreign genes in addition to the antibiotic resistance gene (1, 19, 33, 34). A problem encountered with some of these systems is the inherent cytopathogenicity of the parent virus, leading to the necessity of selection of noncytopathic variants, an attribute which may be restricted to the cell line in which the variants were selected (10). RUB offers the advantage of being noncytopathic in most cell lines. As with other replicon systems, we have also constructed replicons capable of expressing both the antibiotic resistance gene and additional foreign genes by incorporating additional subgenomic promoters or internal ribosome entry sequence elements (M.-H. Chen and W.-P. Tzeng, unpublished results).
Acknowledgments
We thank Ping Chiang at Georgia State University for oligonucleotide synthesis and automated sequencing.
This research was funded by grant AI21789 from NIH. M.-H.C. was supported by Georgia State University Research Program Enhancement and by an IPA between the Centers for Disease Control and Prevention and Georgia State University.
REFERENCES
- 1.Agapov, E. V., I. Frolov, B. D. Lindenbach, B. M. Pragai, S. Schlesinger, and C. M. Rice. 1998. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent., R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1998. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 3.Boccard, F., and D. Baulcombe. 1993. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology 193:563-578. [DOI] [PubMed] [Google Scholar]
- 4.Chen, M.-H., and T. K. Frey. 1999. Mutagenic analysis of the 3′ cis-acting elements of the rubella virus genome. J. Virol. 73:3386-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decker, C. J., and R. Parker. 1995. Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr. Opin. Cell Biol. 7:386-392. [DOI] [PubMed] [Google Scholar]
- 6.Derdeyn, C. A., and T. K. Frey. 1995. Characterization of defective-interfering RNAs of rubella virus generated during serial undiluted passage. Virology 206:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez, G., C. Y. Wang, and T. K. Frey. 1990. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during togavirus evolution. Virology 177:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 9.Frolov. I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolov, I., E. Agapov, T. A. Jr. Hoffman, B. M. Pragai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 12.Haldeman-Cahill, R., J. A. Daros, and J. C. Carrington. 1998. Secondary structures in the capsid protein coding sequence and 3′ nontranslated region involved in amplification of the tobacco etch virus genome. J. Virol. 72:4072-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levis, R., B. G. Weiss, M. Tsiang, H. Huang, and S. Schlesinger. 1986. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell 44:137-145. [DOI] [PubMed] [Google Scholar]
- 17.Liang, Y., and S. Gillam. 2000. Mutational analysis of the rubella virus nonstructural polyprotein and its cleavage products in virus replication and RNA synthesis. J. Virol. 74:5133-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, Y. J., C. L. Liao, and M. M. Lai. 1994. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 68:8131-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 20.Mahajan, S., V. V. Dolja, and J. C. Carrington. 1996. Roles of the sequence encoding tobacco etch virus capsid protein in genome amplification: requirements for the translation process and a cis-active element. J. Virol. 70:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakhasi, H. L., N. K. Singh, G. P. Pogue, X. Q. Cao, and T. A. Rouault. 1994. Identification and characterization of host factor interactions with cis-acting elements of rubella virus RNA. Arch. Virol. Suppl. 9:255-267. [DOI] [PubMed] [Google Scholar]
- 24.Pogue, G. P., X. Q. Cao, N. K. Singh, and H. L. Nakhasi. 1993. 5′ sequences of rubella virus RNA stimulate translation of chimeric RNAs and specifically interact with two host-encoded proteins. J. Virol. 67:7106-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Proutski, V., E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pugachev, K. V., E. S. Abernathy, and T. K. Frey. 1997. Improvement of the specific infectivity of the rubella virus (RUB) infectious clone: determinants of cytopathogenicity induced by RUB map to the nonstructural proteins. J. Virol. 71:562-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross, J. 1995. M-RNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sachs, A. B., P. Sarnow, and M. W. Hentze. 1997. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89:831-838. [DOI] [PubMed] [Google Scholar]
- 30.Singh, N. K., C. D. Atreya, and H. L. Nakhasi. 1994. Identification of calreticulin as a rubella virus RNA binding protein. Proc. Natl. Acad. Sci. USA 91:12770-12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzeng, W. P., M. H. Chen, C. A. Derdeyn, and T. K. Frey. 2001. Rubella virus DI RNAs and replicons: requirement for nonstructural proteins acting in cis for amplification by helper virus. Virology 289:63-73. [DOI] [PubMed] [Google Scholar]
- 33.Xiong, C., R. Levis, P. Shen, S. Schlesinger, C. M. Rice, and H. V. Huang. 1989. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science 243:1188-1191. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, X., D. R. Hinton, D. J. Cua, S. A. Stohlman, and M. M. Lai. 1997. Expression of interferon-gamma by a coronavirus defective-interfering RNA vector and its effect on viral replication, spread, and pathogenicity. Virology 233:327-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker, M., and P. Stiegler. 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 9:133-148. [DOI] [PMC free article] [PubMed] [Google Scholar]