Abstract
Mitochondria and plastids (chloroplasts) are cell organelles of endosymbiotic origin that possess their own genetic information. Most organellar DNAs map as circular double-stranded genomes. Across the eukaryotic kingdom, organellar genomes display great size variation, ranging from ∼15 to 20 kb (the size of the mitochondrial genome in most animals) to >10 Mb (the size of the mitochondrial genome in some lineages of flowering plants). We have developed OrganellarGenomeDraw (OGDRAW), a suite of software tools that enable users to create high-quality visual representations of both circular and linear annotated genome sequences provided as GenBank files or accession numbers. Although all types of DNA sequences are accepted as input, the software has been specifically optimized to properly depict features of organellar genomes. A recent extension facilitates the plotting of quantitative gene expression data, such as transcript or protein abundance data, directly onto the genome map. OGDRAW has already become widely used and is available as a free web tool (http://ogdraw.mpimp-golm.mpg.de/). The core processing components can be downloaded as a Perl module, thus also allowing for convenient integration into custom processing pipelines.
INTRODUCTION
Mitochondria and plastids (chloroplasts) are organelles in eukaryotic cells that originated from the endosymbiotic uptake of an α-proteobacterium and a cyanobacterium, respectively. Both mitochondria and plastids have retained genomes of double-stranded DNA that are replicated and expressed within the organelles and are usually present in high copy numbers per cell. Compared with the genomes of their bacterial ancestors, present-day plastid and mitochondrial genomes are much reduced in gene content. This is mainly due to the massive transfer of genetic information from the genome of the endosymbionts to the (nuclear) genome of the host cell in the course of evolution (1–3). The plastid genomes of seed plants, for example, contain a relatively conserved set of only ∼130 genes. These include many photosynthesis-related genes (encoding, e.g. subunits of the two photosystems, the cytochrome b6f complex, the chloroplast ATP synthase and an NAD(P)H dehydrogenase), a large set of so-called genetic system genes whose gene products are involved in the expression of the plastid genome (e.g. rRNAs, tRNAs, ribosomal proteins and a bacterial-type RNA polymerase) and a few other genes and open reading frames (4–6). Similarly, mitochondrial genomes mainly encode gene functions involved in the expression of the mitochondrial genome or in respiration, the main metabolic function of mitochondria in all aerobic organisms (7–9). Despite their relatively low coding capacity, organellar genomes can display enormous variations in genome size. Although, for example, the human mitochondrial genome is only 16.6 kb and contains 37 densely packed genes (7), the genomes of some flowering plants reach sizes of >11 Mb (10), even though their gene content is similar or even smaller than that of the human mitochondrial DNA. Thus, the largest plant mitochondrial genomes are much larger than most bacterial genomes and even larger than some eukaryotic nuclear genomes, but to a large extent consist of DNA that presumably is non-coding. In addition to the enormous size variation, there is also structural variation in that most organellar genomes map as circular DNAs, but some are known to be linear [e.g. the mitochondrial genomes in some lineages of protists and green algae (8–10)].
The specifics of organellar genomes and their extraordinary variability in structure, size and gene density make it difficult to create high-quality visual maps from sequenced genomes using standard molecular biology software. To date, there are several software tools available that can be used for drawing graphical maps of DNA sequences. Commercial molecular biology software suites like VectorNTI (14) and DNAStar (15) provide excellent options for processing sequence data, but usually do not provide sophisticated functions that would allow the generation of high-quality physical maps. In particular, these software packages are not suited for drawing maps of comparably large organellar genomes because they were designed for small plasmid-type vector sequences. There are also a few freeware or shareware software packages for manipulating DNA sequences that provide map drawing functions, such as pDRAW32 (http://www.acaclone.com/), Plasm (http://en.bio-soft.net/plasmid/Plasm.html) and PlasmaDNA (16). However, their functionality is again mainly optimized for small plasmid-like vector sequences (e.g. PlasMapper, wishart.biology.ualberta.ca/PlasMapper/, rejects sequences >20 kb). CGView (17) provides a more flexible framework for creating circular genome maps, and the CGView server also has a web browser interface. However, the program cannot be used anonymously and has not been optimized for organellar genomes and gene functions. In addition to functional restrictions and licensing requirements, many of the currently available tools are operating system specific and cannot be conveniently accessed via a web browser interface.
To provide software tools that are suitable for generating high-quality graphical maps of plastid and mitochondrial genomes, we launched OrganellarGenomeDRAW (OGDRAW) in 2007 (18). The initial version was mainly designed for displaying publication-quality physical maps using a consensus color code for the gene classes commonly present in plastid and mitochondrial genomes (18). In OGDRAW, all computational processing is done on the server (http://ogdraw.mpimp-golm.mpg.de/), and the input and output parameters can be conveniently defined by the user via a web browser interface that is independent of the underlying operating system. Since its initial release, OGDRAW has become widely used and has helped researchers with producing physical maps for numerous published chloroplast and mitochondrial genome sequences. In the new version presented here, we have reworked and extended the web browser interface to improve stability and user friendliness. In addition, we have included a new workflow that allows the visualization of quantitative expression data directly on the physical map of the genome.
RESULTS AND DISCUSSION
Drawing physical maps of organellar genomes
OGDRAW uses annotated DNA sequences in the widely used GenBank format as input (Figure 1). If the genome sequence to be drawn as physical map has already been deposited in the GenBank repository, it is sufficient to enter the accession number. Alternatively, users can upload GenBank files directly to OGDRAW—the data will only be used for generating the maps and are deleted subsequently to ensure confidentiality.
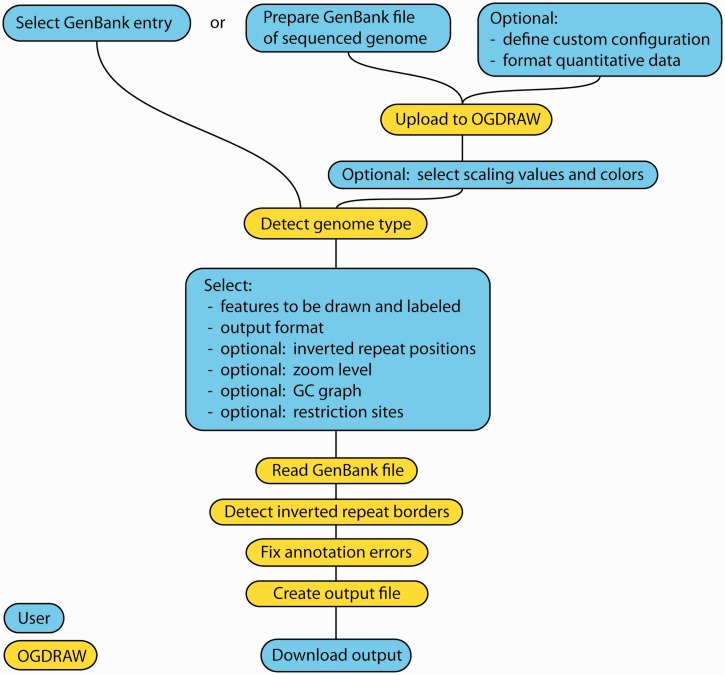
Figure 1.
Chart illustrating the workflow of OGDRAW. Cyan fields indicate user input, yellow fields denote steps performed by OGDRAW. See text for details.
To draw a map, OGDRAW first extracts the annotations of all sequence features from the GenBank file and executes some pre-processing steps that enhance the visual consistency of the final map (e.g. by removing inconsistent feature annotation entries). Briefly, duplicated entries and entries with identical start and end positions but different names will be merged into one feature. Unusually large features (e.g. trans-spliced genes) are decomposed into their subfeatures, and only the subfeatures are subsequently drawn to prevent these features from visually covering a large section of the map. For each feature, a name and the feature type (e.g. protein-coding gene, tRNA, etc.) is extracted and subsequently used to determine the (functional) category the gene belongs to. As DNA sequences stored in GenBank or provided by users do not always adhere to the nomenclature conventions for plastid and mitochondrial genes and/or the standard rules for annotating features, some further (optional) pre-processing steps were implemented that help to improve the map. When activating the ‘tidy-up’ option, apparent genes that are >10 kb and cannot be dissected into exons or sublocations will be ignored. (No known organellar gene is >7 kb, and such giant ‘genes’ usually represent annotation errors.) The ‘tidy-up’ function will also reformat gene names to meet the nomenclature conventions laid out in the ‘naming guidelines’ section of the OGDRAW web site.
By default, all sequence features will be drawn using a pre-set color scheme that chooses the color based on the name and the type of the feature. The default color scheme will color all genes according to their function in the organelle and/or their association with conserved organellar multiprotein complexes. For example, all plastid gene names starting with the letters ‘psa’ will be recognized as components of photosystem I and drawn in the same (photosystem I-specific) color (Figures 1 and 2). OGDRAW provides default color schemes for the gene classes present in plastid and mitochondrial genomes (18) (Figure 2). However, these color sets are freely configurable and can be customized by supplying a configuration file in which each category of genes to be drawn in a distinct color is defined using a simple XML-like format. To aid users in creating or editing configuration files, we provide several sample configuration files and a comfortable graphical Java-based tool that enables users to generate custom configuration files without having to modify the XML file directly. Both the sample files and the configuration tool (OGDrawConfig) are available for download from the web site.
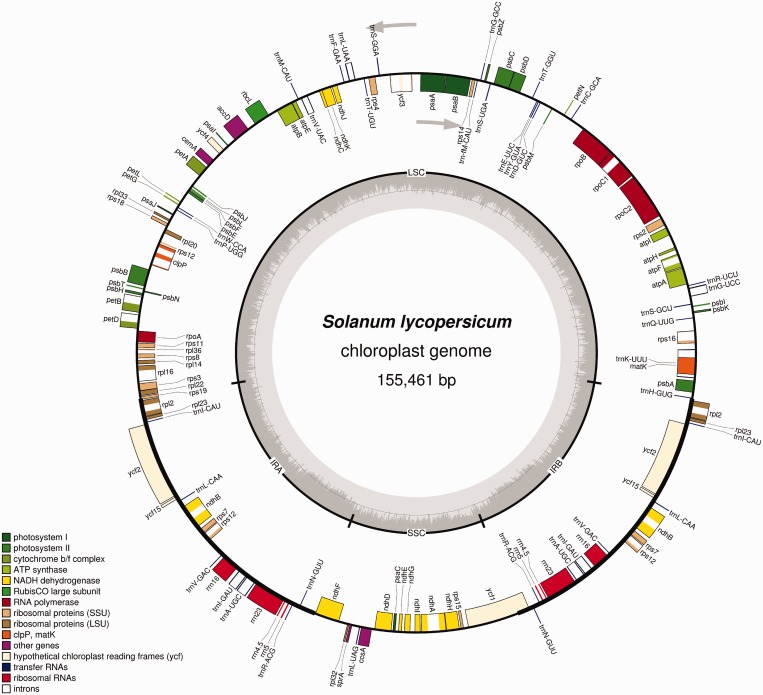
Figure 2.
Map of the tomato (Solanum lycopersicum) chloroplast genome. The map was drawn using the complete genome sequence as input ((23); GenBank accession number NC_007898.2). The gray arrows indicate the direction of transcription for the two DNA strands. The circle inside the GC content graph marks the 50% threshold.
Following submission of the sequence data, the map drawing process can be customized (Figure 1). By default, all classes of gene functions defined in the configuration file will be included in the map, but the user can choose to exclude individual gene classes (or other features) by unchecking them in the list. OGDRAW also offers the possibility to perform in silico restriction mapping and include selected restriction sites in the physical map generated. The genome conformation (linear versus circular) is detected automatically, and, accordingly, the map is drawn as either circular or linear genome. Most plastid DNAs map as circular genomes and contain characteristic inverted repeat regions (IRA/IRB) that are separated by a large and a small single copy region. When working with plastid genome data, OGDRAW can automatically detect these regions and indicate them in the final map (Figure 2). Alternatively, users can manually specify the extent of the repeat and single copy regions (Figure 1) in case automatic detection failed (e.g. because the repeat regions are <2 kb or not fully identical copies of each other). The criterion for automatic IR detection is complete sequence identity because plastid genomes exhibit high gene conversion activity, which usually leads to fast copy correction of sequence deviations between the two IRs (19).
After pre-processing, the customized feature list is used to generate a map image in the graphics format and resolution chosen on the option page. Available formats are TIFF, PNG, JPEG and GIF in selectable resolutions ranging from 72 dpi (quick view) to 600 dpi (publication-quality image) and PostScript. Internally, OGDRAW always generates a PostScript image first, which is then converted to the graphics format specified by the user. Users who want to modify the map should use PostScript as output format, since PostScript images can be conveniently edited using common software tools, such as Inkscape (http://inkscape.org), Adobe Illustrator (http://www.adobe.com/products/illustrator.html) or CorelDraw (http://www.corel.com/).
The maps generated by OGDRAW use inner/outer circle depiction to visualize the orientation of genes and features. Features transcribed clockwise will be drawn on the inside of the circle, whereas features annotated on the complementary strand (and transcribed counter-clockwise) will be drawn on the outside of the circle. Irrespective of the genome conformation, OGDRAW also offers the option to zoom into a region of the sequence and draw a linear map of the specified genomic region and the specific features contained in it. Features transcribed from left to right will then be shown above the baseline, and features transcribed in the opposite direction will be shown below the baseline of the map. As an additional option, users can choose to include a graph representation of the GC content of the sequence in the circular map (Figure 2). In the highly AT-rich chloroplast genomes, the GC content graph usually shows two distinct humps of higher GC content. They correspond to the conserved ribosomal RNA-encoding regions located within the IRs (Figure 2).
Visualizing expression data sets
The workflow for displaying quantitative data from transcript profiling, polysome profiling (20,21) or proteomics experiments is largely identical to the above-described standard procedure for drawing physical maps (Figure 1). First, the data sets (e.g. from genome-wide expression profiling experiments) are uploaded as a simple tab-separated text file. The first column of the text file should contain the gene identifiers, which must be identical to the identifiers used in the sequence file (i.e. the GenBank entry). The actual numerical data for each gene should be provided in the second column. It is recommended to adhere to the nomenclature conventions in both the sequence file and the expression data file, as the ‘tidy-up’ contingency option is only applicable to the GenBank file.
On the subsequent screen, the sequence file to be uploaded (or the accession number to be used) should be provided, and the color scale for displaying the numerical data can be configured. The colors for depicting upregulated, downregulated and non-responding genes can be freely chosen. However, the data set is expected to contain values that are centered around zero, as for example, is the case with gene expression studies in which the log2-fold changes of expression between two conditions are computed. In such a data set, upregulated genes have positive values, whereas downregulated genes have negative log2-fold changes. Non-responding genes have a value of zero. OGDRAW automatically scales the legend and links the highest and lowest expression values in the data set to maximum color intensities (Figure 3). However, the user can manually override these settings by choosing different maximum intensity values. Values between the maxima will be drawn in a color intensity that is scaled to the gradient (Figure 3). After accepting the settings for the color scale, the workflow proceeds to the drawing options where the user can specify all options as described earlier in the text for the standard drawing workflow. Instead of presenting a choice of functional gene classes, each individual gene is now listed and can be excluded from the final map by unchecking the corresponding box. The color corresponding to the quantitative value for a given gene is then used as fill color for the respective gene (feature) in the map.
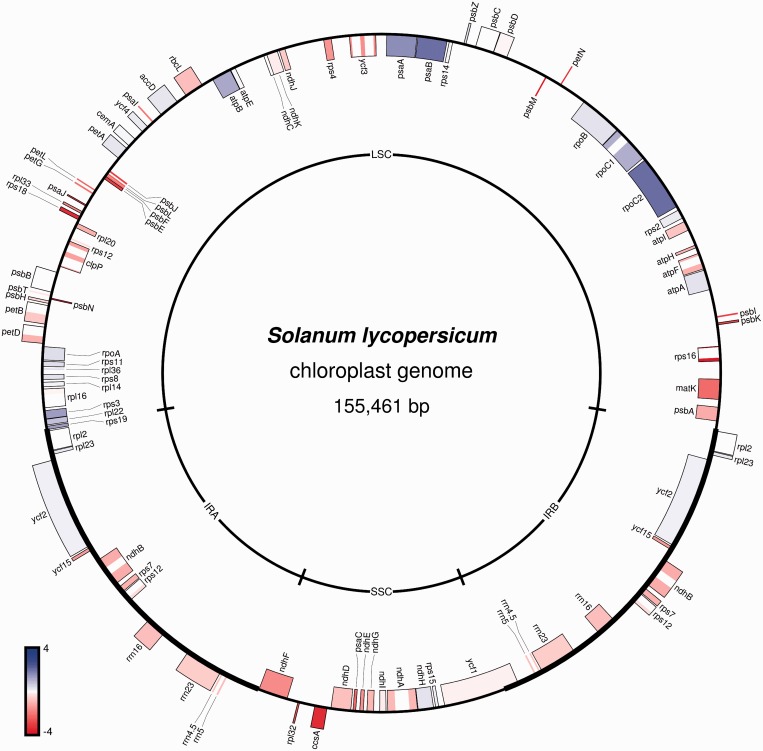
Figure 3.
Visualization of a gene expression data set on the physical map of the tomato (S. lycopersicum) chloroplast genome. The expression data are from a polysome profiling experiment (see text for details).
As an example, Figure 3 shows the changes in plastid translation rates determined by a polysome profiling experiment (translatomics) (20,22) and plotted onto the physical map of the tomato chloroplast DNA (23). In plastids, the regulation of gene expression is highly complex, with changes in transcription and mRNA abundance often not correlating with changes in protein synthesis or abundance. This is because there is extensive translational regulation of plastid gene expression, which can largely override changes in transcript abundance (24). To determine the fraction of mRNAs that are actively translated in tomato leaf chloroplasts, polysomal RNA (mRNAs loaded with ribosomes and therefore assumed to be translated) was purified by sucrose density gradient centrifugation. A total of 10 µg of leaf polysomal RNA and the identical amount of total leaf RNA were hybridized to a custom-made microarray containing specific oligonucleotide probes for all plastid-encoded genes and open reading frames (20). In Figure 3, the ratios of these two samples are presented on a log2 scale. The data visualize the differences in relative transcript abundance in the translated mRNA population compared with the pool of total RNA. Although some transcripts are underrepresented in the actively translated RNA pool (e.g. ccsA, rps18, matK), other mRNAs are highly enriched (e.g. rpoC2, psaA, psaB). This result reflects the high degree of translational control in chloroplasts and shows that there is no direct correlation between mRNA abundance and the rate of protein synthesis (Figure 3). Consequently, transcriptome data are an insufficient basis to draw solid conclusions about the regulation of gene expression at the level of protein accumulation.
By mapping gene expression data to the physical map of the genome, additional layers of information are revealed, which would not become apparent if the same data set was displayed as a simple heat map. Most importantly, the expression map allows the user to correlate changes in expression with the operon structure of the plastid genome. Adjacent genes in plastid (and mitochondrial) genomes often form a bacterial-type operon and are co-transcribed giving rise to a polycistronic primary transcript. Visualizing expression data on the physical map of the genome can thus uncover co-regulation patterns and potentially relate them to changes in transcription rates, RNA processing defects or altered stability of polycistronic mRNAs (e.g. in mutants or specific environmental conditions) (21).
IMPLEMENTATION
OGDRAW was developed in Perl, and D and is available as a set of Perl modules (GeneMap) that provide a flexible object-oriented interface for easy integration into custom scripts. The Perl modules can be downloaded freely from the OGDRAW web site. For general parsing and manipulation of GenBank files, OGDRAW makes use of software provided by the Bioperl project (25). Image generation and output functions in OGDRAW use the PostScript-Simple (http://search.cpan.org/∼mcnewton/PostScript-Simple-0.07/lib/PostScript/Simple.pm) and ImageMagick/PerlMagick (http://www.imagemagick.org) modules. Active elements providing additional information on the web site require a JavaScript-enabled web browser. The web page has been tested on all common browsers (including Firefox, Internet Explorer, Safari and Chrome), has been continuously available since the publication of the first version of OGDRAW (18) and individual user support has been provided.
CONCLUSIONS
OGDRAW provides the community with a user-friendly flexible toolbox for drawing high-quality maps of organellar genomes and for visualizing expression data sets. OGDRAW is freely accessible via a cross-platform web browser interface (http://ogdraw.mpimp-golm.mpg.de/). The input data can be provided in the form of standard GenBank files, which contain both the annotation and the raw sequence in a convenient format. As GenBank flat files represent a standard sequence storage format and, moreover, can be read and edited with most commonly used molecular biology software packages, it is simple and straightforward for users to generate input files for OGDRAW and quickly obtain physical maps of newly sequenced genomes. OGDRAW has been specifically designed for drawing organellar genome maps and provides a number of options to properly capture the specific structural and functional characteristics of mitochondrial and plastid genomes, such as color-coded conserved gene classes present in organellar genomes, IRs and polycistronic transcripts. However, OGDRAW can also be used for plasmids, cloning vectors and any other small-to-medium-size genetic element or genome. There is no upper limit to the size of the genome submitted to OGDRAW (and the current limit of the file size of 2 MB can be increased on request), but gene-dense and/or feature-rich genomes that are substantially >1 Mb will produce rather crowded maps on printing on standard-size paper. The detailed maps generated by OGDRAW can be obtained in a range of output formats that (especially when choosing the vector graphics format PostScript) can easily be further customized using standard graphics editing software. Maps from sequences deposited in the GenBank database can be obtained by just a few mouse clicks using the accession number as input.
The newly implemented module for visualization of quantitative gene expression data sets by drawing expression maps extends the application range of OGDRAW beyond the display of structural features by enabling users to view genome structure and gene expression activities at the same time. This offers the unique possibility to uncover relationships between an expression response and the position of genes in the genome or their location within operons.
Finally, the quality of the map obtained depends on the consistency and accuracy of the annotation data provided in the input data file. Wrongly or non-uniformly annotated (or named) features will predictably lead to incorrect maps. In the frequently asked questions section of the OGDRAW web site, common types of annotation errors that can cause problems with map drawing are listed along with instructions how these can be rectified by correcting the input data.
To our knowledge, OGDRAW is currently the only web-based tool that, owing to its specific design, is suitable to draw complete publication-quality physical maps and expression maps of organellar genomes (as well as of other types of input sequences).
FUNDING
Funding for open access charge: Max Planck Society (MPG).
Conflict of interest statement. None declared.
REFERENCES
- 1.Gray MW. Origin and evolution of organelle genomes. Curr. Opin. Genet. Dev. 1993;3:884–890. doi: 10.1016/0959-437x(93)90009-e. [DOI] [PubMed] [Google Scholar]
- 2.Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phyl. Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 3.Bock R, Timmis JN. Reconstructing evolution: gene transfer from plastids to the nucleus. BioEssays. 2008;30:556–566. doi: 10.1002/bies.20761. [DOI] [PubMed] [Google Scholar]
- 4.Shimada H, Sugiura M. Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res. 1991;19:983–995. doi: 10.1093/nar/19.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakasugi T, Tsudzuki T, Sugiura M. The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynthesis Res. 2001;70:107–118. doi: 10.1023/A:1013892009589. [DOI] [PubMed] [Google Scholar]
- 6.Bock R. Structure, function, and inheritance of plastid genomes. Top. Curr. Genet. 2007;19:29–63. [Google Scholar]
- 7.Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–464. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 8.Gray MW, Lang BF, Cedergren R, Golding GB, Lemieux C, Sankoff D, Tumel M, Brossard N, Delage E, Littlejohn TG, et al. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nosek J, Tomáska L, Fukuhara H, Suyama Y, Kovác L. Linear mitochondrial genomes: 30 years down the line. Trends Genet. 1998;14:184–188. doi: 10.1016/s0168-9525(98)01443-7. [DOI] [PubMed] [Google Scholar]
- 10.Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr. Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 12.Kubo T, Mikami T. Organization and variation of angiosperm mitochondrial genome. Physiol. Plant. 2007;129:6–13. [Google Scholar]
- 13.Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10:e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu G, Moriyama EN. Vector NTI, a balanced all-in-one sequence analysis suite. Brief. Bioinform. 2004;5:378–388. doi: 10.1093/bib/5.4.378. [DOI] [PubMed] [Google Scholar]
- 15.Burland TG. DNASTAR's Lasergene sequence analysis software. Methods Mol. Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- 16.Angers-Loustau A, Rainy J, Wartiovaara K. PlasmaDNA: a free, cross-platform plasmid manipulation program for molecular biology laboratories. BMC Mol. Biol. 2007;8:77. doi: 10.1186/1471-2199-8-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 19.Khakhlova O, Bock R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006;46:85–94. doi: 10.1111/j.1365-313X.2006.02673.x. [DOI] [PubMed] [Google Scholar]
- 20.Kahlau S, Bock R. Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: Chromoplast gene expression largely serves the production of a single protein. Plant Cell. 2008;20:856–874. doi: 10.1105/tpc.107.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter M, Piepenburg K, Schöttler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R. Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 2010;64:851–863. doi: 10.1111/j.1365-313X.2010.04377.x. [DOI] [PubMed] [Google Scholar]
- 22.Valkov VT, Scotti N, Kahlau S, MacLean D, Grillo S, Gray JC, Bock R, Cardi T. Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control. Plant Physiol. 2009;150:2030–2044. doi: 10.1104/pp.109.140483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahlau S, Aspinall S, Gray JC, Bock R. Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J. Mol. Evol. 2006;63:194–207. doi: 10.1007/s00239-005-0254-5. [DOI] [PubMed] [Google Scholar]
- 24.Eberhard S, Drapier D, Wollman F-A. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2002;31:149–160. doi: 10.1046/j.1365-313x.2002.01340.x. [DOI] [PubMed] [Google Scholar]
- 25.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]