Abstract
The variable domain of an immunoglobulin (IG) sequence is encoded by multiple genes, including the variable (V) gene, the diversity (D) gene and the joining (J) gene. Analysis of IG sequences typically requires identification of each gene, as well as a comparison of sequence variations in the context of defined regions. General purpose tools, such as the BLAST program, have only limited use for such tasks, as the rearranged nature of an IG sequence and the variable length of each gene requires multiple rounds of BLAST searches for a single IG sequence. Additionally, manual assembly of different genes is difficult and error-prone. To address these issues and to facilitate other common tasks in analysing IG sequences, we have developed the sequence analysis tool IgBLAST (http://www.ncbi.nlm.nih.gov/igblast/). With this tool, users can view the matches to the germline V, D and J genes, details at rearrangement junctions, the delineation of IG V domain framework regions and complementarity determining regions. IgBLAST has the capability to analyse nucleotide and protein sequences and can process sequences in batches. Furthermore, IgBLAST allows searches against the germline gene databases and other sequence databases simultaneously to minimize the chance of missing possibly the best matching germline V gene.
INTRODUCTION
The immunoglobulins (IG) are a group of antigen-binding proteins produced by the B lymphocytes. They serve as critical defensive components that protect our bodies against invading pathogens. An IG consists of two heavy (H) chains and two light (L) chains. Structurally, each chain can be divided into the variable (V) domain and the constant (C) domain. The V domain is responsible for binding the antigens and can be further divided into the framework regions (FR) and the complementarity determining regions (CDR) (1).
To counter a vast repertoire of antigens, the immune system has devised multi-layer mechanisms to produce an extraordinarily diverse pool of IG proteins (2). One critical mechanism is that the actual antigen-binding domain (i.e. the V domain) is jointly encoded by multiple genes (the VH domain is encoded by V, D and J genes, whereas the VL domain is encoded by V and J genes). These genes are initially separated in the germline genome but are subsequently joined by a process called the V-(D)-J rearrangement in the precursor B cells. As there are multiple genes for each gene type, and any of these genes can combine into an IG V domain, the resulting repertoire of V domain is very large. Other mechanisms contributing to the V domain diversity include imprecise joining between any of the recombining genes, nucleotide trimming of the V, D and/or J genes, addition of P nucleotides and random addition of nucleotides (N regions) at rearranging junctions, pairing of different H and L chains, as well as somatic mutations that occur in the V domains and are then selected when B lymphocytes encounter antigens. Thus, the total diversity in IG molecules is virtually unlimited.
Studying IG proteins often requires a detailed analysis of their gene sequences. This includes, but is not limited to, identifying the contributing germline V, D and J genes, analysing the V-(D)-J junction details, finding the boundaries for FR and CDR and comparing with other IG sequences in a database. Although the popular BLAST program (3) can be used to search against various databases of nucleotide and protein sequences at the National Center for Biotechnology Information (NCBI), it has only limited capability for IG sequences. Different IG genes have different characteristic lengths, with the D genes being as short as ∼10 bases and V genes being ∼290 bases long. BLAST needs special parameters to identify short matches, but these parameters are not optimal for longer matches. Therefore, it is necessary to perform multiple searches for a single IG sequence. In addition, manual assembly of different genes together from BLAST results is difficult and error-prone. Finally, because BLAST was developed as a general purpose sequence similarity search program, it does not provide information specific for IG sequences.
There are several software tools that have been developed for IG sequence analysis. Notably, IMGT®, the international ImMunoGeneTics information system®, offers IMGT/JunctionAnalysis (4), IMGT/V-QUEST (5,6) and its version for next-generation sequencing, IMGT/HighV-QUEST (7). It also maintains many widely used reference germline gene databases. Other tools include VBASE2 (8), iHMMune-align (9) and JoinSolver (10). Although these tools provide valuable analysis capabilities, such as germline gene identification, FR and CDR delineation and mutational analysis, they have various limitations. For example, they all lack the ability to search against more comprehensive databases like the NCBI nr or genomic databases, as well as the ability to search protein sequences. In addition, these tools either are slow to process a large batch of query sequences or lack that ability altogether. Other limitations include the inability to analyse short sequences and no support for FR/CDR delineation in the Kabat (11) system. To address these issues, we have developed a more flexible IG V domain sequence analysis tool named IgBLAST. This tool uses the well-known BLAST algorithm (3) to perform sequence similarity search and provides commonly sought information for IG sequences.
SEARCH STRATEGY AND IMPLEMENTATION
IgBLAST consists of several components that are responsible for finding matches to the individual V, D and J genes, finding IG V region annotation information and combining all information to produce the final result. IgBLAST is implemented using the NCBI C++ toolkit. Default BLAST search parameters are used unless indicated otherwise. The implementation details are described below.
Identifying the FR/CDR boundaries
A query is searched with BLAST against the IMGT or NCBI germline V gene database (the sequences in such databases have been pre-annotated for the FR/CDR boundaries). The top database sequence hit is used to map the pre-annotated FR/CDR boundary information to the query sequence. The BLAST search parameters are Expect cut-off, 20; word size 9; mismatch penalty, −1; Dust filtering, off.
Identifying the V, D and J gene hits
Multiple BLAST searches are performed to identify all genes. To identify the germline V gene hits, a query is searched against a user-selected germline V gene database (search parameters: Expect cut-off, 20; word size 9; mismatch penalty, −1; Dust filtering, off). To avoid irrelevant BLAST hits when searching the D and J gene databases, the query region matching the top germline V gene is masked. A BLAST search is then performed with the masked query against the user-selected J gene database (search parameters: Expect cut-off 1000, word size 7, mismatch penalty, −3; Dust filtering, off) and against the user-selected D gene database if the query is a heavy chain (search parameters: Expect cut-off 100 000; Dust filtering, off). As a D gene is short, its identification is more likely subject to spurious matches that are caused by random nucleotide additions, somatic mutations, as well as other homologous D genes. Therefore, the word size and the mismatch penalty for the D gene search are adjustable by users who have different requirements for the match stringency for D genes. The default word size is 5, which requires a minimum of five consecutive nucleotide matches for a D gene to be found. The default mismatch penalty is conservatively set to a relatively high value (−4) to minimize the chance of spurious matches. However, this setting inevitably favours finding D gene alignments that have few mismatches rather than those that have more mismatches but are longer.
Some rearrangement constraints are assumed when searching for the V, D and J genes. These assumptions include that a valid D gene must be positioned between the V and J gene, and that only genes from the same locus [i.e. the heavy locus (IGH), the κ locus (IGK) or the λ locus (IGL)] are allowed in a rearrangement.
Determination of rearrangement frame
A rearrangement frame (or V-J frame) is tagged in-frame if the last complete coding triplet for the V gene in the query is in-frame with the first complete coding triplet for the J genes. Otherwise it is tagged out-of-frame. A rearrangement is tagged productive only when it is in-frame and contains no stop codon.
RESULTS AND DISCUSSION
Program input
IgBLAST is a robust tool for IG sequence analysis. A query sequence can be a full length or a partial IG V domain sequence and does not need to contain a D or J gene (although a V gene sequence containing at least nine bases of a germline V gene is required). Deletions and insertions in the V, D or J genes of a query sequence are allowed, as the underlying BLAST algorithm has the capability to handle such cases (3). IgBLAST accepts several different query formats, including raw sequences, FASTA sequences, GenBank accessions or GI numbers.
IgBLAST offers a few options for custom parameter adjustment. These include setting the stringency for D gene detection, choosing the Kabat or IMGT V region delineation system and selecting different views of alignments. IgBLAST has the flexibility to search against germline V, D and J gene databases separately. A few germline gene databases are available from different sources, including the IMGT/V-QUEST reference directory sets (5), UNSWIg human heavy chain repertoire (12) and the NCBI germline gene collections. Currently supported organisms are human, mouse, rat and rabbit. IgBLAST also provides an option to search a custom database, which is useful when a user believes there is discrepancy in the germline gene composition between the study subjects and available databases. For example, one study suggests that some entries in the human germline gene collection may represent errors and should be removed (12). On the other hand, a user may wish to include a germline gene sequence that is not present in available germline gene databases. To make a custom database, a user need only save sequences in FASTA format in a text file.
IgBLAST offers the unique capability to search sequence databases such as the NCBI nr or genomic database. In particular, the NCBI nr database is a large collection of the annotated nucleotide sequences submitted to GenBank. Its content is updated daily to reflect the latest submissions. Although searching the germline V gene database is fast and is sufficient for many users, it should be pointed out that, as it takes time and efforts to collect germline gene sequences, there might be a delay before a new germline gene is added to a germline gene database. Therefore, users should consider adding additional database, such as the NCBI nr database if it is absolutely essential to include any potential new germline V genes in the search.
For users who are interested in analysing IG V domain protein sequences (for example, antibody modelling and peptide mapping), IgBLAST can be used to identify the V gene with FR/CDR delineation.
IgBLAST is capable of processing multiple queries (we recommend not exceeding 1000 sequences per batch). For users desiring maximal flexibility in high-throughput searches, we provide a stand-alone version of IgBLAST (user instruction can be found by following the ‘Stand-alone IgBLAST’ link on the IgBLAST web page).
Program output
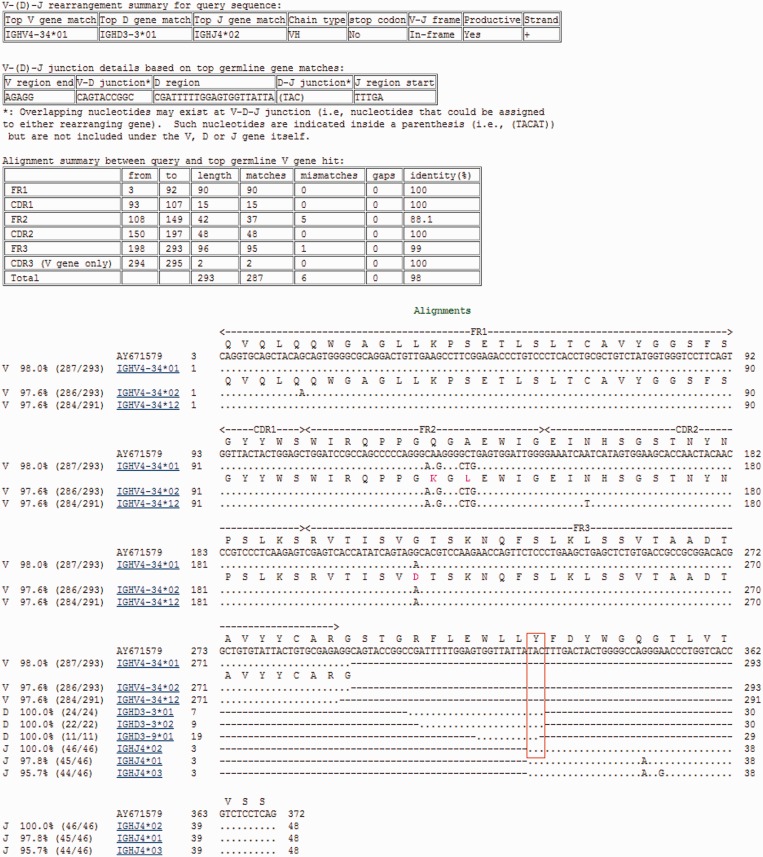
IgBLAST output presents a clear and informative view of the search result. Figure 1 shows the result of searching a human IG sequence against the IMGT germline gene databases. In addition to showing the actual detailed alignment between the query and various hits, the report includes a tabulated summary of results based on the alignments between the query and the top matched germline V, D and J gene. The summary information includes the identifiers of the best matched V, D and J gene, the relationship between the coding frames of the V and J genes, the details of the V-(D)-J junctions and the match statistics for various FR/CDR. All fields in the summary table should be self-evident.
Figure 1.
IgBLAST result page. This example used a human IG sequence (GenBank accession AY671579) to search against the default germline gene databases [IMGT human V genes (F + ORF + in-frame P), IMGT human D genes (F + ORF) and IMGT human J genes (F + ORF)]. The search used default values for all parameters. A red box was added to indicate the overlapping nucleotides TAC at the D–J junction. The search was performed on 25 February 2013.
It is worth noting that the IgBLAST report provides information on overlapping nucleotides at a rearrangement junction that might have been contributed by either of the rearranging genes because of homology-directed recombination events (13). Such nucleotides are listed inside a parenthesis under the relevant junction in the summary table (i.e. the bases TAC under the D–J junction field in Figure 1) and are also evident by examining the alignment details (as highlighted by the red box in Figure 1).
The alignment section uses a familiar multiple alignment view with the hits aligned to the query (shown in the first row). The alignments show the three top hits from the V, D and J gene matches by default, but this is user adjustable. The far left column indicates the gene category (i.e. V, D or J) for the germline gene hits. The second column shows the percent identity between the query and each hit (the number of matches and the alignment length are indicated in parenthesis in the third column). Each line is preceded by a number indicating the starting nucleotide position for the line and ends with a number indicating the ending nucleotide position. Users can choose the format with a dot, indicating that the hit is identical to the query (Figure 1) or the format showing the original letters for the hit. A dash in the alignment indicates a gap in the relevant sequence. FR/CDR boundaries are directly annotated on top of the query sequence.
To make it easier to view the effect of a nucleotide substitution on a protein sequence, IgBLAST offers the option to show translations for a nucleotide query. If there is a difference in the amino acid between the query and the germline V gene, the corresponding amino acid in the germline V gene is coloured purple (Figure 1).
As indicated previously, IgBLAST has the capability to search against the germline gene databases, as well as other sequence databases at the same time. Figure 2 shows one such example. Similar to the result of searching against the germline gene databases only, the alignment section first lists the hits from germline gene databases where one can see the top matched germline V gene hit is IGHV1-9*01 (97.9% similarity to the query over 290 bases). Below the germline gene database hits, the result page lists hits from the NCBI nr database, including the 16 hits (excluding the self-hit AF104468) that show a 100% match to the query over 290 bases. IgBLAST offers a convenient feature that displays the sequence titles when a user mouses over the sequence identifiers (for example, the accession AF021857). A quick examination of sequence titles suggests that many of these 16 hits come from different sources [for example, M17723 is from an anti-dextran hybridoma, whereas BC018315 is a cDNA clone from the Mammalian Gene Collection project (http://mgc.nci.nih.gov/)]. As independent isolation of identical IG V gene sequences is often used as an indication of a germline sequence (14), these 16 hits may represent a possible new germline V gene. In fact, sequence titles from three of these 16 sequences (AF021857, AF021859 and AF021861) indicate that these are un-mutated (i.e. germline) sequences. Obviously, whether these hits definitively represent a germline V gene remains to be investigated and is out of scope for this article. Thus, the IgBLAST results from searching against the germline gene databases and the NCBI nr database together alert a user about a possible germline V gene that is a better match than the one from the germline V gene database.
Figure 2.
Example IgBLAST result of searching against the germline gene databases and the NCBI nr database simultaneously. A mouse IG sequence (GenBank accession AF104468) was searched against the default mouse germline gene databases [IMGT mouse V genes (F + ORF + in-frame P), IMGT mouse D genes (F + ORF + in-frame P) and IMGT mouse J genes (F + ORF + in-frame P)]. The ‘organism’ field was set to mouse, and the nr database was selected for the ‘additional database’ field. Default values are used for all other parameters, except the ‘number of alignments for additional database’ was 25. The light blue pop-up message box is a feature that displays the sequence title when the mouse pointer is moved over the sequence identifier (i.e. the accession AF021857 in the example). Only part of the result page is shown because of space limitation. A red box was added to indicate the hits from the nr databases that have 100% matches to the query over the 290 bases. The search was performed on 25 February 2013.
Performance
Searching a single IG sequence against a germline gene database typically generates the result page instantly. For batch submission, a test search of 1000 human IG heavy chain sequences (between 300 and 600 bases) takes ∼44 s to return the results.
Program evaluation
Identifying original germline genes from a rearranged sequence with certainty (particularly for short D genes) is a difficult task, as there are multiple germline genes that share high similarity. This task is further complicated by the random nucleotide additions at rearrangement junctions, as well as somatic mutations. As a result, a rearranged sequence is often similar to multiple germline gene sequences. The quality of IG sequence analysis is typically judged by expert visual examination of the assigned V, D and J genes, but this can be subjective. Hence, Gaeta et al. (9) propose some tests using objective criteria, and the results from that study suggest that iHMMune-align performs best for germline gene identification among several IG sequence analysis tools. Although IgBLAST results have been subject to numerous visual examinations during development, it is also important to test IgBLAST objectively. Thus, we use the same strategy as Gaeta et al. and compare the results with iHMMune-align.
The first test data set includes 100 randomly chosen IG heavy chain sequences without V gene mutations. Gaeta et al. (9) argue that the D and J elements in these sequences should contain few or no mutations, as mutation rates drops rapidly from 5′ side of the V gene. The test results are summarized in Table 1. IgBLAST reports that the average length of assigned D and J genes are 16 and 44 bases, respectively, with 0.04 and 0.08 nucleotide mismatches (per sequence) on average to germline D and J genes, respectively. Thus, IgBLAST indeed reports very low mutations in D and J genes for sequences that have no mutations in V genes. The test with iHMMune-align shows similar results.
Table 1.
Characteristics of the D and J genes identified in 100 random IG heavy chain sequencesa
| IgBLAST | iHMMune-align | |
|---|---|---|
| Average D gene mutations per sequence (average D gene length) | 0.04 (16.29) | 0.056 (17.43) |
| Average J gene mutations per sequence (average J gene length) | 0.08 (44.29) | 0.23 (44.52) |
aOne hundred IG heavy chain sequences are randomly selected from NCBI nr database (available in Supplementary File S1). The selection is based on 100% identity match to any heavy chain germline gene from IMGT database as determined by BLAST program with default parameters; therefore, there is no previous knowledge about their D and J gene compositions. Tests were performed using web IgBLAST and stand-alone iHMMune-align (version iHMMune-align_26-11-2007.zip) with default search parameters. iHMMune-align did not return a D gene match for 11 sequences that were excluded from D gene analysis.
We next use clonally related sequences to test IgBLAST. The rational for this test (9) is that these sequences originate from the same rearrangement but are then divergent because of somatic mutations (most sequences carry 20+ mutations in these data sets); therefore, a good sequence analysis tool should report the same V, D and J genes for most or all sequences. Data set 1 contains 57 sequences with the IGHV4-34*01-IGHD7-27*01-IGHJ3*02 rearrangement identified as the dominant alignment by iHMMune-align (9). Table 2 presents results for this test. IgBLAST reports that 52 sequences have IGHV4-34*01 and IGHJ3*02 as the closest-matched germline V and J genes, respectively, and 54 sequences have IGHD7-27*01 as the closest-matched germline D gene. The iHMMune-align results are similar. IgBLAST and iHMMune-align both find the dominant IGHV4-34*01-IGHD7-27*01-IGHJ3*02 rearrangement in 47 sequences.
Table 2.
Number of sequences with correctly identified V, D and J genes or rearrangements in clonally related sequence data setsa
| IgBLAST | iHMMune-align | |
|---|---|---|
| Data set 1 (57 sequences) | ||
| IGHV4-34*01 | 52 | 51 |
| IGHD7-27*01 | 54 | 55 |
| IGHJ3*02 | 52 | 52 |
| IGHV4-34*01-IGHD7-27*01- IGHJ3*02 rearrangement | 47 | 47 |
| Data set 2 (101 sequences) | ||
| IGHV4-34*01 | 96 (96)b | 95 |
| IGHD6-6*01 | 87 (48)b | 86 |
| IGHJ6*02 | 101 (101)b | 97 |
| IGHV4-34*01- IGHD6-6*01- IGHJ6*02 rearrangement | 82 | 80 |
aThe clonally related sequences were obtained from Wilson and co-workers (15). Tests were performed using web IgBLAST and stand-alone iHMMune-align (version iHMMune-align_26-11-2007.zip) with default search parameters, except that the mismatch penalty for D gene is set to −1 (instead of default −4) for IgBLAST test with data set 2. The identified germline genes are the top hits (or one of the top equivalent hits that have identical match scores, as well as identical per cent identity) from IgBLAST or iHMMune-align searches. iHMMune-align did not return a D gene match for 1 and 6 sequences for data set 1 and data set 2, respectively. IHMMune-align also did not return any germline gene matches for one sequence in both data sets because of presence of deletions in V gene.
bResults from IgBLAST using default mismatch penalty for D genes.
A second set of clonally related sequences (data set 2) with more mutations in the V-(D)-J junction regions are also analysed. The dominant rearrangement was previously identified as IGHV4-34*01 IGHD6-6*01- IGHJ6*02 by iHMMune-align (9). As shown in Table 2, IgBLAST reports 96 sequences use IGHV4-34*01 and all use IGHJ6*02, whereas iHMMune-align finds similar number for IGHV4-34*01 (95) but slightly lower number for IGHJ6*02 (97). For D genes, although iHMMune-align finds 86 sequences that use IGHD6-6*01, IgBLAST only finds 48 sequences with this D gene. As discussed in the ‘Search Strategy and Implementation’ section, IgBLAST uses a high mismatch penalty (−4) for D genes by default that is not optimal for identifying D genes with more mismatches (as is the case for data set 2). Therefore, we reduced the D gene mismatch penalty to −1 for this test. Indeed, reducing the D gene mismatch penalty results in identification of IGHD6-6*01 in 87 sequences (which is similar to iHMMune-align result) while not affecting the V and J gene findings. IgBLAST also finds the previously identified dominant IGHV4-34*01 IGHD6-6*01- IGHJ6*02 rearrangement in 82 sequences, whereas iHMMune-align finds such rearrangement in 80 sequences.
Overall, IgBLAST produces expected results for all three test data sets involving IG heavy chain sequences with and without mutations. iHMMune-align generates similar results, although it has an advantage that no search parameter adjustment is needed, at least for our test cases.
CONCLUSIONS
IgBLAST is a web tool that we have developed for analysis of IG V domain sequences. It is robustly implemented to handle a variety of query sequences in different formats and addresses common analysis tasks, such as identifying the V, D and J genes, viewing rearrangement junction details and delineation of FR/CDR for the V gene. IgBLAST also offers the unique capability to search against germline gene databases, as well as other sequence databases (such as the NCBI nr database) simultaneously to minimize the chance of missing possibly the best matching germline V gene. IgBLAST is a free public tool with no login requirement.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary File 1.
FUNDING
Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge members of the BLAST group, the user help group and the C++ toolkit group at the NCBI for their work that has made this tool possible.
REFERENCES
- 1.Lefranc M-P, Lefranc G. The Immunoglobulin Factsbook. San Diego: Academic Press; 2001. [Google Scholar]
- 2.Schatz DG, Oettinger MA, Schlissel MS. V(D)J recombination: molecular biology and regulation. Annu. Rev. Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 3.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yousfi Monod M, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004;20(Suppl. 1):i379–i385. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
- 5.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for Ig and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giudicelli V, Brochet X, Lefranc MP. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (Ig) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb. Protoc. 2011;2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 7.Alamyar E, Giudicelli V, Li S, Duroux P, Lefranc MP. IMGT/HighV-QUEST: the IMGT(R) web portal for immunoglobulin (Ig) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012;8:26. [Google Scholar]
- 8.Retter I, Althaus HH, Munch R, Muller W. VBASE2, an integrative V gene database. Nucleic Acids Res. 2005;33:D671–D674. doi: 10.1093/nar/gki088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaeta BA, Malming HR, Jackson KJ, Bain ME, Wilson P, Collins AM. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 10.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J. Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 11.Kabat EA. Sequences of Proteins of Immunological Interest, National Institutes of Health Publication. 5th edn. Bethesda: United States Department of Health and Human Services; 1991. [Google Scholar]
- 12.Wang Y, Jackson KJ, Sewell WA, Collins AM. Many human immunoglobulin heavy-chain IGHV gene polymorphisms have been reported in error. Immunol. Cell. Biol. 2008;86:111–115. doi: 10.1038/sj.icb.7100144. [DOI] [PubMed] [Google Scholar]
- 13.Gu H, Forster I, Rajewsky K. Sequence homologies, N sequence insertion and JH gene utilization in VHDJH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO J. 1990;9:2133–2140. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu H, Tarlinton D, Muller W, Rajewsky K, Forster I. Most peripheral B cells in mice are ligand selected. J. Exp. Med. 1991;173:1357–1371. doi: 10.1084/jem.173.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng NY, Wilson K, Wang X, Boston A, Kolar G, Jackson SM, Liu YJ, Pascual V, Capra JD, Wilson PC. Human immunoglobulin selection associated with class switch and possible tolerogenic origins for C delta class-switched B cells. J. Clin. Invest. 2004;113:1188–1201. doi: 10.1172/JCI20255. [DOI] [PMC free article] [PubMed] [Google Scholar]