Abstract
RNAstructure is a software package for RNA secondary structure prediction and analysis. This contribution describes a new set of web servers to provide its functionality. The web server offers RNA secondary structure prediction, including free energy minimization, maximum expected accuracy structure prediction and pseudoknot prediction. Bimolecular secondary structure prediction is also provided. Additionally, the server can predict secondary structures conserved in either two homologs or more than two homologs. Folding free energy changes can be predicted for a given RNA structure using nearest neighbor rules. Secondary structures can be compared using circular plots or the scoring methods, sensitivity and positive predictive value. Additionally, structure drawings can be rendered as SVG, postscript, jpeg or pdf. The web server is freely available for public use at: http://rna.urmc.rochester.edu/RNAstructureWeb.
INTRODUCTION
RNA is an important biomolecule, as a catalyst (1), a director of post-transcriptional modification (2) and gene regulation (3), a target of drugs (4,5) and also a pharmaceutical (6,7). Its structure is generally hierarchical (8). Primary structure is the sequence of nucleotides, and is a covalent bond structure. Secondary structure is the set of the canonical base pairs, and tertiary structure is the set of additional contacts and the complete three-dimensional structure.
Because RNA folding is generally hierarchical, secondary structure can often be predicted and analyzed without predicting tertiary structure. Secondary structure prediction can provide a framework for understanding the mechanism of action of RNA. Secondary structure prediction is also an important consideration in the design of siRNA and antisense DNA oligonucleotides both to avoid structure in the mRNA target of these agents and in avoiding self-structure in agents, either of which prevents them from hybridizing with their targets (9–14). RNA structure prediction can also make predictions about which regions of sequence are accessible for interacting with proteins (15). Finally, secondary structure prediction can be used to identify novel functional RNA sequences encoded in genomes (16–18).
RNAstructure is a software package for RNA secondary structure prediction and analysis (19). It was first reported in 1998 as a free energy minimization program for Microsoft Windows (20). It has been expanded to include methods for predicting bimolecular structure (12), conserved structures in multiple homologs (21–24) and siRNA design (9). Several methods are available for predicting structures for a single sequence, including maximum expected accuracy (25), stochastic sampling (26), exhaustive traceback (27) and pseudoknot prediction (28). Graphical user interfaces are provided for Microsoft Windows, Macintosh OS-X and Linux. Command line interfaces are also available for these operating systems. Finally, the set of underlying C++ classes are available as a library for use by programmers.
This report describes a new set of web servers that provide the functionality of RNAstructure. These web servers are open to the public and can be found at http://rna.urmc.rochester.edu/RNAstructureWeb.
ORGANIZATION
The RNAstructure web servers are organized around two schemes. The first scheme provides specific programs for web users. Table 1 shows a list of programs that are available. The second scheme is a set of themes defined by the biological problem. For example, to predict a secondary structure for a single sequence, the server called Predict a Secondary Structure can be used to run calculations by multiple programs. The list of themes and the exact programs included are listed in Table 2. This approach is designed to be user-friendly because it bundles all the available methods for addressing a specific problem, and for this reason most users will want to use these servers.
Table 1.
List of individual programs provided by the RNAstructure web server
| Servers | References | Description |
|---|---|---|
| AllSub | (27,29) | Generate all possible low free energy structures for a nucleic acid sequence. |
| bifold | (12) | Predict the lowest free energy structure for two interacting sequences, allowing intramolecular base pairs. |
| bipartition | (30) | Perform a partition function calculation for two interacting nucleic acid sequences. Intramolecular pairs are not allowed. |
| CircleCompare | (19) | Compare two structures using overlaid circular plots to emphasize pairing differences and pseudoknots. |
| ct2dot | (19) | Convert a CT-formatted structure into a dot bracket file. |
| dot2ct | (19) | Convert a dot bracket file into a CT file. |
| draw | (19) | Draw the secondary structure of a nucleic acid strand, with or without color annotation. |
| DuplexFold | (31) | Predict the lowest free energy structure for two interacting sequences, not allowing intramolecular base pairs. |
| Dynalign | (17,21,32) | Calculate the lowest free energy secondary structures common to two unaligned sequences. |
| efn2 | (33) | Calculate the folding free energy of structures in a CT file. |
| EnsembleEnergy | (30) | Calculate the ensemble folding free energy change for a sequence. |
| Fold | (33,34) | Predict the lowest free energy structure and a set of low free energy structures for a sequence. |
| MaxExpect | (25) | Generate a structure or structures composed of highly probable base pairs. |
| Multilign | (22) | Predict low free energy secondary structures common to three or more sequences using progressive iterations of Dynalign. |
| oligoscreen | (35) | Predict stability of hybridization and self-structure for a list of oligonucleotides. |
| OligoWalk for siRNA Design | (9,11) | Design efficient siRNAs for a given miRNA target. |
| partition | (30) | Perform a partition function calculation on a single sequence to calculate base pair probabilities. |
| PARTS | (23,36) | Predict the common secondary structure, including base pair probabilities, for two unaligned sequences. |
| ProbablePair | (30) | Generate secondary structures composed of base pairs with probabilities that exceed a specified threshold. |
| ProbKnot | (28) | Predict a secondary structure composed of probable base pairs, which might include pseudoknots. |
| RemovePseudoknots | (19,37) | Remove pseudoknots from a structure. |
| scorer | (31,30) | Calculate the sensitivity and positive predictive value (PPV) for a predicted as compared to the accepted structure. |
| stochastic | (26,36) | Generate a representative sample of structures using stochastic sampling. |
| TurboFold/TurboKnot | (24,38) | Calculate the conserved structures of three or more unaligned sequences using iteratively refined partition functions. |
Table 2.
List of RNAstructure web servers organized around a problem
| Servers | Programs run (see Table 1 for description) |
|---|---|
| Predict a secondary structure | Fold, MaxExpect, ProbKnot, partition |
| Predict a secondary structure common to two sequences | Dynalign, Parts |
| Predict a secondary structure common to three or more sequences | Multilign, TurboFold/TurboKnot |
| Predict a bimolecular secondary structure | bifold, DuplexFold |
INPUT
The structure prediction servers work using either bare sequences pasted into a window, or FASTA-formatted files uploaded to the server. The exception to this is the set of servers that predict a structure common to three or more sequences. For these servers, including Multilign and TurboFold, the sequence window requires multiple FASTA-formatted sequences. Alternatively, a FASTA-formatted file with multiple sequences can be uploaded.
Servers that act on structures, such as CircleCompare, draw, efn2, RemovePseudoknots or scorer, require an upload of a CT file. These can be generated manually or by a structure prediction server. Another alternative is to use a dot-bracket formatted structure and convert it to CT format using the dot2ct component of the RNAstructure server.
For each individual server, sample input data can be generated automatically to illustrate format and to provide a test case for the server. For structure prediction servers, clicking a link pastes sample sequences into the sequence windows. For the servers that act on structures, a link is provided to download a sample CT file, and this can be uploaded back to the server as sample data.
For Fold, partition and Predict a Secondary Structure, structure prediction can be restrained using SHAPE mapping data, when available, to improve the accuracy of structure prediction (39). AllSub, Dynalign, Fold, partition and Predict a Secondary Structure accept folding constraints, including the ability for force specific base pairs, forbid specific pairs, force a nucleotide paired, force a nucleotide unpaired, and specify that a nucleotide is accessible to chemical modification (33,34). These constraints and restraints are mediated by file uploads.
OUTPUT FORMATS
Once a calculation is submitted, a notification page is displayed. This page continues to refresh until the calculation output is available. At that time, the page is replaced with a page that contains the output. This page can be bookmarked and returned to at a later time. Alternatively, an e-mail address can be provided as part of the input data. If this is done, an e-mail is sent to the address when the calculation is complete and the results are available.
The structure prediction servers display the predicted structure using an SVG drawing, which can be rendered by web browsers with an SVG plugin (Figure 1). If more than one structure is predicted, i.e. suboptimal structures or a structure sample is present, ‘previous’ and ‘next’ buttons are displayed to enable scrolling between structures. For each predicted structure, the structure can be downloaded as a jpeg, svg, pdf, postscript or CT file.
Figure 1.
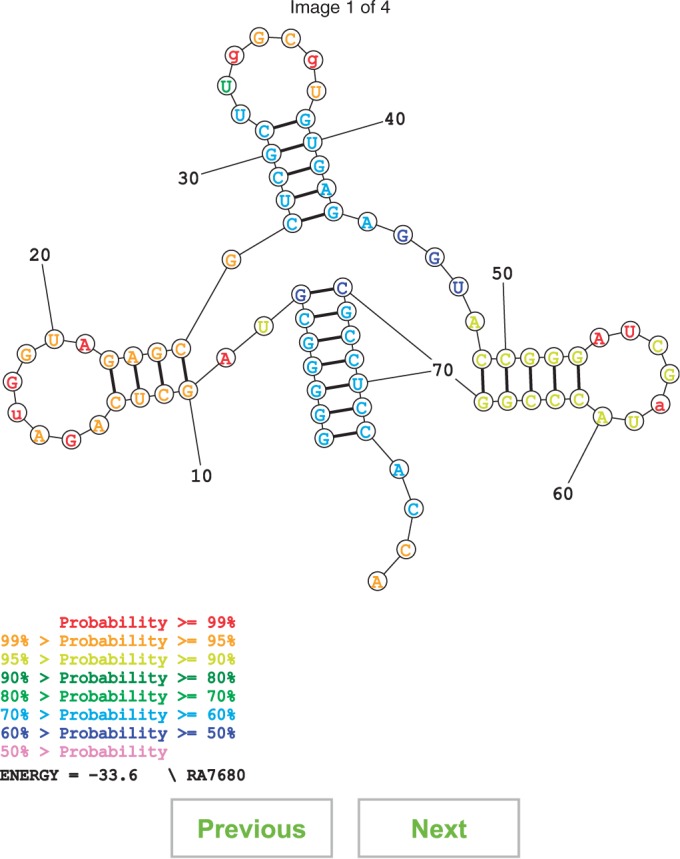
Sample structure prediction. This is the predicted lowest free energy structure for the sample sequence for the ‘Predict a Secondary Structure’ server. This sequence is the tRNA RA7680 from the Sprinzl database (41). It is color annotated according to base pairing probability; more probable pairs are more likely to be correctly predicted than less probable pairs (30).
Each output page displays the RNAstructure executable and the command line used to generate the results (40). Each input file is a link, allowing download of the processed-form data. This makes clear exactly what calculation is performed, and facilitates a user learning the capabilities of the RNAstructure software.
IMPLEMENTATION
The web servers use PHP to process queries. The server does not rely on JavaScript running on the client side, although a single exception to this is the previously available OligoWalk web server (11). The server itself is a computer cluster with a head node and five compute nodes, each with two dual core processors. A queuing system prioritizes the order at which queries are executed, which is generally in the order in which queries were made. The exceptions to this rule are calculations submitted for Dynalign, Multilign or TurboFold, which are submitted to the server as multiple-core calculations. These may need to wait in the queue at times when a number of other time-consuming calculations are already running.
The RNAstructure web servers have limitations on the calculations that can be done. This ensures that the resource will be available to the broader community with reasonable wait times. A list of limitations is available with the online help. Currently, for example, the single sequence structure prediction methods are limited to sequences of 2500 nucleotides or less. TurboFold is limited to a maximum of 12 sequences up to 1600 nucleotides in length. If the required calculation exceeds the web server requirements, the software can be downloaded (http://rna.urmc.rochester.edu/RNAstructure.html) and run locally.
HELP
Extensive online help is available for using the RNAstructure web server. The help documents each of the input and output fields, and is organized by individual server. For each server help page, a link is provided to the underlying RNAstructure executable (19), which provides additional details about the program. A separate help page details the server limitations, as explained above.
CONCLUSION
RNAstructure is designed to be a user-friendly software package, accessible to the community of investigators studying RNA (19). These new web servers continue this tradition, and provide the software to a wider user base. The accuracy of the algorithms has been extensively tested in prior publications. As the algorithms in RNAstructure are improved, and new algorithms developed, the web servers will be updated to continue to make RNAstructure available to the community.
FUNDING
National Institutes of Health (NIH) [R01 GM076485 to D.H.M.]. Funding for open access charge: NIH [R01 GM076485].
Conflict of interest statement. None declared.
REFERENCES
- 1.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 2.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 3.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sucheck SJ, Wong CH. RNA as a target for small molecules. Curr. Opin. Chem. Biol. 2000;4:678–686. doi: 10.1016/s1367-5931(00)00142-3. [DOI] [PubMed] [Google Scholar]
- 5.Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem. Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 6.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 7.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 9.Lu ZJ, Mathews DH. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res. 2007;36:640–647. doi: 10.1093/nar/gkm920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu ZJ, Mathews DH. Fundamental differences in the equilibrium considerations for siRNA and antisense oligodeoxynucleotide design. Nucleic Acids Res. 2008;36:3738–3745. doi: 10.1093/nar/gkn266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu ZJ, Mathews DH. OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008;36:W104–W108. doi: 10.1093/nar/gkn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathews DH, Burkard ME, Freier SM, Wyatt JR, Turner DH. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- 14.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA. 2010;16:1096–1107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uzilov AV, Keegan JM, Mathews DH. Detection of non-coding RNAs on the basis of predicted secondary structure formation free energy change. BMC Bioinformatics. 2006;7:173. doi: 10.1186/1471-2105-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 2006;16:885–889. doi: 10.1101/gr.5226606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathews DH, Andre TC, Kim J, Turner DH, Zuker M. An updated recursive algorithm for RNA secondary structure prediction with improved thermodynamic parameters. In: Leontis NB, SantaLucia J Jr, editors. Molecular Modeling of Nucleic Acids. Washington, D.C.: American Chemical Society; 1998. pp. 246–257. [Google Scholar]
- 21.Mathews DH, Turner DH. Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 2002;317:191–203. doi: 10.1006/jmbi.2001.5351. [DOI] [PubMed] [Google Scholar]
- 22.Xu Z, Mathews DH. Multilign: an algorithm to predict secondary structures conserved in multiple RNA sequences. Bioinformatics. 2011;27:626–632. doi: 10.1093/bioinformatics/btq726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harmanci AO, Sharma G, Mathews DH. PARTS: probabilistic alignment for RNA joint secondary structure prediction. Nucleic Acids Res. 2008;36:2406–2417. doi: 10.1093/nar/gkn043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmanci AO, Sharma G, Mathews DH. TurboFold: iterative probabilistic estimation of secondary structures for multiple RNA sequences. BMC Bioinformatics. 2011;12:108. doi: 10.1186/1471-2105-12-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu ZJ, Gloor JW, Mathews DH. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA. 2009;15:1805–1813. doi: 10.1261/rna.1643609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan S, Mathews DH, Turner DH. Interpreting oligonucleotide microarray data to determine RNA secondary structure: application to the 3' end of Bombyx mori R2 RNA. Biochemistry. 2006;45:9819–9832. doi: 10.1021/bi052618x. [DOI] [PubMed] [Google Scholar]
- 28.Bellaousov S, Mathews DH. ProbKnot: fast prediction of RNA secondary structure including pseudoknots. RNA. 2010;16:1870–1880. doi: 10.1261/rna.2125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wuchty S, Fontana W, Hofacker IL, Schuster P. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999;49:145–165. doi: 10.1002/(SICI)1097-0282(199902)49:2<145::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 30.Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piekna-Przybylska D, DiChiacchio L, Mathews DH, Bambara RA. A sequence similar to tRNA3Lys gene is embedded in HIV-1 U3/R and promotes minus strand transfer Nat. Struct. Mol. Biol. 2009;17:83–89. doi: 10.1038/nsmb.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmanci AO, Sharma G, Mathews DH. Efficient pairwise RNA structure prediction using probabilistic alignment constraints in dynalign. BMC Bioinformatics. 2007;8:130. doi: 10.1186/1471-2105-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 34.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matveeva OV, Mathews DH, Tsodikov AD, Shabalina SA, Gesteland RF, Atkins JF, Freier SM. Thermodynamic criteria for high hit rate antisense oligonucleotide design. Nucleic Acids Res. 2003;31:4989–4994. doi: 10.1093/nar/gkg710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harmanci AO, Sharma G, Mathews DH. Stochastic sampling of the RNA structural alignment space. Nucleic Acids Res. 2009;37:4063–4075. doi: 10.1093/nar/gkp276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smit S, Rother K, Heringa J, Knight R. From knotted to nested RNA structures: a variety of computational methods for pseudoknot removal. RNA. 2008;14:410–416. doi: 10.1261/rna.881308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seetin MG, Mathews DH. TurboKnot: rapid prediction of conserved RNA secondary structures including pseudoknots. Bioinformatics. 2012;28:792–798. doi: 10.1093/bioinformatics/bts044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]


