Abstract
Tissue-specific transcription is advantageously investigated by using viral promoters, which are selected for compact regulatory elements. Mouse mammary tumor virus (MMTV) has adapted to specialized cell types and targets initially B lymphocytes. We previously showed that, in B-cell lines, glucocorticoid-induced MMTV transcription requires an ETS family factor, GA-binding protein (GABP), bound in tandem to the MMTV DNA next to the glucocorticoid receptor (GR). We now report that transforming growth factor β (TGF-β) superinduces this response up to 10-fold through binding of its effectors, Smads, between the GABP-binding motifs. The basal level was unaffected. The TGF-β-glucocorticoid cooperation also depended on GR and GABP binding, was transferable to another promoter, and occurred both with transiently transfected and with integrated templates. Smad3 associated in vitro with GR, with GABPα (via the MH2 domain), and with GABPβ, Smad4 only with GABPα. Interactions of Smad3 with GABP (when coexpressed or endogenous to B cells) were shown by coprecipitation and by mammalian two-hybrid assay. This composite DNA element integrates three signaling pathways deriving from TGF-β, glucocorticoid hormones, and a unique ETS factor, and may allow MMTV to exploit factors from the milk. It may as well indicate novel possibilities for cellular regulatory networks.
Transforming growth factor β (TGF-β) is a member of a superfamily of structurally related proteins that regulate cell differentiation and proliferation in a wide variety of organisms (reviewed in references 49 and 50). Through serine/threonine kinase receptors, it activates transcription factors called Smads along a unique signaling pathway that has been the focus of intense research efforts in recent years (reviewed in references 4, 51, and 53). Receptor-regulated Smad2 or Smad3 become phosphorylated and associate with a co-Smad (Smad4). They activate transcription by binding to DNA elements and/or by associating with DNA-bound factors. Among these, some were newly discovered, such as FAST-1 (14) and FAST-2 (43), whereas others were already known, such as AP-1 (70), Sp1 (54), TFE3 (35), or AML-1 (55). Smad proteins contain highly conserved domains (see Fig. 6C): an N-terminal, DNA-binding MH1 and a C-terminal MH2 mediating most protein-protein interactions. Only rather degenerate common binding sequences could be defined by comparing TGF-β-responsive elements of various genes (18; reference 63 and references therein). The biological effects of TGF-β are extremely varied, according to the type and environment of the target cell (reviewed in references 19 and 49). In the immune system, it functions as a modulator of differentiation and as an immunosuppressor, e.g., in regulating cytokine production by T cells and immunoglobulin expression by B cells (13, 56). Identifying novel Smad partners and regulators is crucial for understanding TGF-β function.
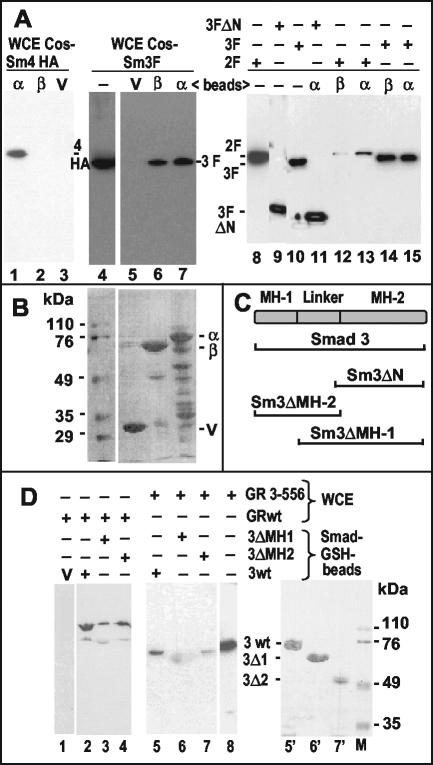
FIG. 6.
Association of Smads with GABP and GR in vitro. (A) GST pull-down assays were performed as described in Materials and Methods. GST-GABPα (α), GST-GABPβ (β), or GST vector alone (V) bound to glutathione-Sepharose beads were mixed with whole-cell lysates (WCE) of transfected COS cells expressing HA-Smad4 (lanes 1 to 3), Flag-Smad2 (lanes 12 and 13), or Flag-Smad3 (full-length in lanes 5 to 7 and lanes 14 and 15 or N terminally deleted in lane 11). The eluted complexes were analyzed by Western immunoblot with antibodies directed to the respective epitope tags. Lanes without added beads (−) contain ca. 5% of the whole-cell lysate inputs as a control. (B) Ponceau-S-stained membrane corresponding to lanes 5 to 7 of panel A showing the GST proteins. Numbers on the left identify the molecular mass markers on the same gel. (C) Scheme of the Smad3 protein and its domains. Horizontal brackets below denote the fractions of the protein present in the mutants used. (D) Beads carrying GST fusions with the indicated parts of Smad3 (see scheme in panel C) or the GST vector (V) were mixed with whole-cell lysates (WCE) of transfected COS cells expressing full-length GR (GRwt) or an N-terminal fragment (GR 3-556; see scheme Fig. 4C). The eluted complexes were analyzed by Western immunoblot with antibodies against GR that recognize its N-terminal domain. The diffuse spot in lane 6 is due to nonspecific sticking of the secondary antibody to GST-Smad3ΔMH1 (see lane 6′). Lane 8 shows the input lysate (∼5%). Lanes 5′ to 7′, Ponceau-S-stained membrane corresponding to lanes 5 to 7; lane M, molecular size markers (numbers on the right in kilodaltons).
Mouse mammary tumor virus (MMTV) is a B-type retrovirus that causes carcinomas of the mammary gland in females of susceptible mouse strains (11) through insertional activation of cellular int genes (reviewed in reference 66). The major site of MMTV replication is the mammary gland, under the stimulation of pregnancy-related hormones. MMTV DNA is the prototype gene for studying the genomic effects of glucocorticoids, i.e., effects that are mediated by binding of the hormone-activated glucocorticoid receptor (GR) to the glucocorticoid regulatory element (GRE), upstream of the promoter in the long terminal repeat (LTR) (9, 10; for a review, see reference 7). The GR contains domains for transactivation, DNA binding, and hormone binding (see scheme in Fig. 4B) (8, 48). MMTV has a distinctive tissue specificity of expression and biological adaptation that make it an unmatched model system. For primary infection, MMTV targets B lymphocytes in the intestine, eliciting a critical immune reaction involving a viral superantigen (reviewed in reference 47). Lymphocytes are required for virus transportation to the mammary gland (65), and B cells are essential for virus propagation in the infected animal (32). Both B and T cells get infected and are able to transmit virus (3, 25). We were therefore interested in the regulation of MMTV expression in B cells. In a recent study we identified a novel DNA regulatory region, adjacent to the distal GRE, that contains a tandem of motifs interacting with the heterodimeric ETS factor GA-binding protein (GABP) present in mature B-cell lines (5) (see scheme in Fig. 1A). In these cells the GABP-binding sites are essential for stimulation by glucocorticoids, and GABP functionally cooperates with the GR. GABP is composed of a DNA-binding α subunit and a transactivating β subunit linked to α through ankyrin repeats. Interactions between β subunits of two neighboring dimers greatly stabilize the complex on DNA (reviewed in reference 27) (Fig. 1A). Since our study was the first to show cooperation between the GR and GABP, we were interested in defining signaling pathways that could act through GAPB and possibly modify the hormone response.
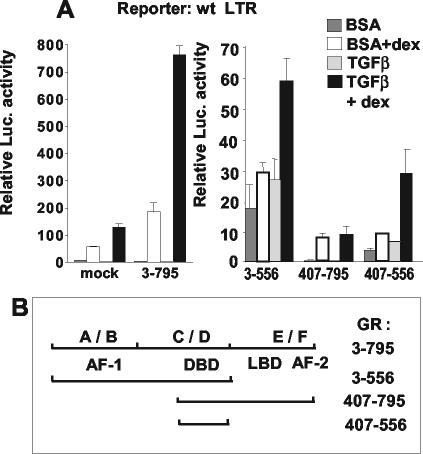
FIG. 4.
Differential contributions of GR domains to the TGF-β superinduction. Transient transfection and treatment of M12 cells were as described for Fig. 1B. (A) Cotransfection of 3 μg of pCI vector (mock) or of pSTC-TK encoding various parts of the rat GR (see scheme in panel B) with 3 μg of pGL3-basic/wt LTR. Normalization was done with the protein content, and error bars denote the standard deviations. Note the difference in scale between the left and right portions of panel A. (B) Scheme of the GR and its segments used in panel A. Numbers to the right indicate amino acids. Positions 3 to 795 is the full-length GR. A to F above the line denote conventional divisions of the GR. Below the line are functional domains for transactivation (AF-1 and AF-2), DNA binding (DBD), and ligand binding (LBD). BSA, bovine serum albumin.
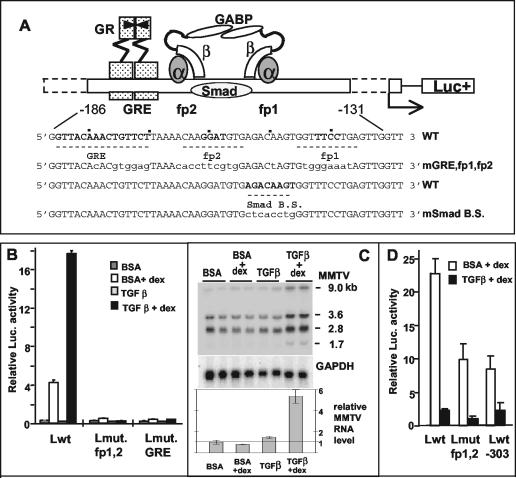
FIG. 1.
Opposite effects of TGF-β1 in a B-lymphoma cell line (B and C) versus a mammary tumor cell line (D). (A) Scheme of the arrangement of factors binding to the regulatory elements upstream of the MMTV promoter that are relevant for the present study. In the wild-type sequence (WT), the numbering refers to the transcription initiation site, and fp1 and fp2 denote the DNase I footprints with M12-cell nuclear extracts (5), with the core sequences for GABP binding in boldface. The putative Smad binding site is also marked. Mutated bases in the different reporter constructs are in lowercase letters (mGRE, fp1, and fp2 are mutated in all three sites; mSmad B.S. is mutated in the putative Smad binding site [modified from references 27 and 48]). (B and D) pGL-3 plasmids (B, 3 μg; D, 2 μg) with various LTRs were transiently transfected into M12 B cells (B) or GR mouse mammary tumor cells (D) that were kept thereafter in serum-free medium plus 5 ng of TGF-β1/ml or the carrier solution containing bovine serum albumin for 24 h (B) or 46 h (D). The “+dex” samples received Dex (50 [B] or 300 [D] nM) for the last 4 h (B) or 8 h (D) before cell lysis for determination of luciferase activity. The LTRs used were as follows: wild type (Lwt), LTR mutated in the fp1/fp2 GABP-binding sites (Lmut fp1,2), LTR mutated in the GRE (Lmut. GRE [LS−193/−162] [10]), and LTR 5′ truncated at position −303 (Lwt-303). The values for firefly luciferase activity were normalized against the Renilla luciferase activity produced by cotransfected plasmids (50 ng of pRL-SV40Δ enhancer [B] or 200 ng of pRL-SV40 [D]). Error bars denote the standard deviations in duplicate samples. The experiment in panel D was performed twice; the experiment in panel B was performed many times. (C) Cooperative effect of TGF-β1 and Dex on the endogenous MMTV RNA levels of M12 cells. Northern blot analysis of total RNA from M12 cells treated with TGF-β1 (5 ng/ml) or bovine serum albumin buffer for 24 h in serum-free medium in the presence or absence of 50 nM Dex for the last 4 h before RNA extraction. Total RNA was separated in a 1% agarose gel containing formaldehyde and blotted onto a nylon membrane, which was hybridized with a 32P-labeled probe of the MMTV env region (upper panel) and then with a probe of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA as a loading control (lower panel). RNA samples are in duplicate. On the right, the sizes in kilobases of the viral mRNAs are given (see the text). The total intensity of the MMTV bands relative to the GAPDH control was quantified and plotted (relative MMTV RNA; values ± the standard deviation are indicated).
An involvement of TGF-β in the biology of MMTV was previously described only in mouse mammary tumor cell lines as an indirect repression of the LTR transcriptional activity (12). Concerning the site of viral entry, the gut-associated lymphoid tissue, nothing is known about possible effects of TGF-β on infection by MMTV. In the present study we show that the LTR element composed of the GRE and two GABP-binding sites also mediates a transcriptional superinduction by TGF-β of the glucocorticoid effect through the binding of Smads to DNA located between the GABP sites and protein-protein interactions involving Smads, GABP, and GR. This superinduction, observed in B lymphocytes, may play a positive role in the early phase of viral infection in the gut. On the other hand, it constitutes a novel combination of sequences and transcription factors activated by TGF-β in a naturally occurring promoter.
MATERIALS AND METHODS
Cells, reagents, and plasmids.
M12.4.1 (23), an immunoglobulin G2a-expressing BALB/c mouse B-lymphoma cell line representative of mature B cells was obtained from H. R. MacDonald (Ludwig Institute for Cancer Research, Lausanne, Switzerland) and cultured in Dulbecco modified Eagle medium containing 5% heat-inactivated fetal calf serum (FCS; both from Gibco-BRL), 50 μM 2-mercaptoethanol, 2 mM l-glutamine, and antibiotics. The GR mouse mammary tumor cell line and the COS-7 cell line (24) were maintained in Dulbecco modified Eagle medium, 10% FCS, and antibiotics. Cultures were kept at 37°C in a humidified 5% CO2 atmosphere. Dexamethasone (Dex) was from Sigma, and recombinant human TGF-β1 was from Sigma, BioPharmacy Ltd. (Japan), or R&D Systems. Rabbit polyclonal antibodies against GR (M-20, sc-1004) and against glutathione S-transferase (GST; Z-5, sc-459) were purchased from Santa Cruz Biotechnology; rabbit polyclonal antibodies against GABPα and GABPβ were a generous gift from E. Flory (22). The mouse monoclonal antibodies used were purchased from Roche (M2 against the Flag peptide and 12CA5 against the hemagglutinin[HA] peptide), from BD-Transduction Laboratories (against Smad2/3), and from Santa Cruz Biotechnology (B8 against Smad4). The plasmids pCMV5-DPC4-HA (encoding Smad4 with a 3′-end HA tag [29]), pCMV/DPC4(1-514) encoding a truncated form of Smad4 acting as a dominant-negative mutant, and pCMV5/TβR-I (T204D) encoding an activated TGF-β receptor were provided by J. Massagué (Memorial Sloan-Kettering Cancer Center, New York, N.Y.). Cytomegalovirus (CMV)-based plasmids expressing the rat GR and subsets of it—pSTC-TK-GR3-795, pSTC-TK-GR3-556, pSTC-TK-GR407-795, and pSTC-TK-GR407-556 (41) were provided by S. Rusconi (University of Fribourg, Fribourg, Switzerland); a plasmid expressing an activated TGF-β receptor R(II-I) (17) was provided by the laboratory of R. Derynck (University of California, San Francisco). The pGex4-Ti plasmids expressing GST fusions with Smad4, Smad3, Smad3-ΔMH1, and Smad3-ΔMH2 (36) were from C. H. Heldin (Ludwig Institute for Cancer Research, Uppsala, Sweden). The plasmid pUC-In-wt/K1 containing the simian virus 40 (SV40) large-T antigen and a mutated SV40 origin (21) was from the laboratory of Ellen Fanning via P. Beard (ISREC, Lausanne, Switzerland). pGL3-Basic vectors (Promega) containing the wild-type MMTV LTR (GR strain, from positions −1196 to +133 from the RNA start site), the LTR truncated at position −303, LTRs (full-size or truncated) mutated in the GABP-binding sites fp1 and/or fp2, LTRs with mutations in the GRE (LS[−193/−162] and LS[−175/−166], carrying an octameric HindIII linker inserted between the indicated positions [39]), and the negative control plasmid ΔP deleted from positions −105 to +133 have been previously described (5). Bacterial expression plasmids for GABPα or GABPβ cDNAs fused to GST at their N termini were made with the pGEX-KG vector (Amersham Biosciences). The equivalent GST fusions for expression in mammalian cells were constructed in the pCMV-GST vector received from R. R. Reed, Johns Hopkins University, Baltimore, Md. (64). The cDNAs of GABPα and GABPβ (22) were subcloned by PCR starting from the second amino acid to their natural stop codon. Mammalian expression plasmids for Smad3-Flag and Smad2-Flag were prepared from pRK5-vector constructs (received from the laboratory of R. Derynck, University of California at San Francisco) by subcloning the coding region plus the Flag epitope and the SV40 origin into the pCI vector (Promega). For Smad3, an EcoRI/HpaI fragment was ligated into pCI cut with EcoRI and BamHI (filled in with Klenow enzyme). For Smad2, since it contains an HpaI site, a three-way ligation of an EcoRI/SalI fragment containing the coding region, a separately obtained SalI/HpaI fragment containing the Flag epitope and the SV40 sequences, and a pCI vector cut with EcoRI and BamHI (filled in with Klenow enzyme) was performed. Smad3F-ΔN was made from the pCI/Smad3F clone by digestion with BamHI and PflMI, filling in with T4 polymerase, and self-ligation. The first AUG in this construct corresponds to amino acid 212, which is roughly at the start of the MH2 domain. Mutations of the fp1/fp2+GRE sites and of the Smad binding site in the LTR were carried out by the method of splicing-by-overlap extension of PCR products (33) and verified by sequencing. For constructs with the enhancer-less SV40 promoter, a region from positions −209 to −116 was amplified by PCR with linked BamHI and BglII sites. Purified fragments were ligated with BglII-digested pGL3-promoter (Promega) in the presence of BglII and BamHI to get oriented inserts. For the mammalian two-hybrid analysis, we used plasmids of the CheckMate system (Promega). The reporter pG5luc expresses the firefly luciferase under the control of a minimal TATA box and five GAL4-binding sites. The coding sequences of GABPα and of GABPβ were each cloned by PCR as EcoRV-XbaI fragments into the pBIND vector in frame with the yeast GAL4 DNA-binding domain. The coding sequences of GABPβ and of Smad3 were each cloned by PCR as EcoRV-XbaI fragments into the pACT vector in frame with the herpes simplex virus VP16 activation domain.
Cell transfections.
M12 cells were transfected by the DEAE-dextran method as described previously (5). GR mammary tumor cells were transfected by a different DEAE-dextran method (200 ng of DEAE-dextran/ml for 8 h at 37°C), followed by a dimethyl sulfoxide shock (10%, 2 min at room temperature). COS-7 cells were incubated with 5 μg of DNA in 2.5-ml/10-cm dish of 500 ng of DEAE-dextran/ml in phosphate-buffered saline at 37°C for 30 min. After the addition (12 ml/dish) of medium containing 3% FCS and 0.1 mM chloroquine, the incubation was continued for 2.5 h at 37°C. The cells were then treated with 10% dimethyl sulfoxide in complete medium for 1.5 min at room temperature, washed, and incubated overnight. After the medium was changed, the cells were incubated for another 24 to 30 h. Transfection mixtures also contained Renilla luciferase expression vectors as internal standards (pRL-SV40 or pRL-TK [Promega]). A novel control plasmid was constructed (pRL-SV40Δ enhancer) by cloning the SV40 promoter from the pGL3-promoter plasmid (digested with BglII/HindIII) into pRL-null (Promega). Luciferase assays were performed with the Dual-Luciferase kit (Promega), and measurements of luminescence were made with a Berthold Lumat luminometer. The results were expressed as the ratio of the firefly to the Renilla luciferase activities or, in some cases, as the firefly activity per constant amount of protein (as determined by the BCA assay [Pierce]) after having verified beforehand the reproducibility of the transfection conditions (5).
Protein interaction assays.
GST fusion proteins were produced and purified as described previously (36), but bacteria expressing GST-GABPα were grown at 16°C for 18 to 24 h after induction due to its instability at higher temperatures. Whole-cell extracts from COS cells were prepared as described by Liv et al. (44). GST pull-down assays with bacterial proteins bound to GSH-Sepharose beads were performed as described by Hittelman et al. (34), with a 4°C incubation and rotary motion. Isolation of GST-fused proteins made in mammalian cells was done as described by Tsai and Reed (64). Immunoprecipitation from cell extracts was done in a lysis buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 8), and 1% NP-40, plus protease and phosphatase inhibitors (28), with 2 μg of rabbit antiserum/ml at 4°C. After the addition of protein A-Sepharose (Sigma) and rotation at 4°C for several hours, the beads were washed five times with lysis buffer. Bound proteins were eluted from Sepharose beads, separated by electrophoresis in sodium dodecyl sulfate-10% polyacrylamide gels, and transferred to a nitrocellulose membrane (28). Western immunoblotting and detection with horseradish peroxidase-coupled goat anti-mouse or anti-rabbit antibodies (Pierce) and chemiluminescence (SuperSignal West Pico [Pierce]) were performed according to standard protocols (28).
Electrophoretic mobility shift assays (EMSAs).
They were performed as described previously (5). Double-stranded probes were made by annealing an oligonucleotide that was 5′ end labeled with [32P]ATP and T4 polynucleotide kinase with its unlabeled complementary strand. Their sequences were as follows: for the wild-type Smad binding site (Smad B.S.wt), positions −143 to −160 plus three nucleotides on each side (lowercase) with mutated parts of fp1/fp2, 5′-cttATGTGAGACAAGTGGTTTatg-3′ and 5′-catAAACCACTTGTCTCACATaag-3′; for the mutant (m, lowercase) Smad binding site, 5′-cttATGTGctcacctgGGTTTatg-3′ and 5′-catAAACCcaggtgagCACATaag-3′; and for fp1,2, containing both fp1/fp2 binding sites with the Smad box in the middle, positions −172 to −133, 5′-CCAACTCAGGAAACCACTTGTCTCACATCCTTGTTTTAAG-3′ and 5′-CTTAAAACAAGGATGTGAGACAAGTGGTTTCCTGAGTTGG-3′. The unlabeled, double-stranded competitor oligonucleotides with a different sequence were fp2 (−175 to −152), 5′-ggaCTTAAAACAAGGATGTGAGAC-3′ and 5′ GTCTCACATCCTTGTTTTAAGtcc-3′. The nucleotides in lowercase letters were added to avoid an overlap with the GRE. The unrelated, hepatic nuclear factor 3 binding site of the transthyretin gene (−109 to −84) was 5′-TGACTAAGTCAATAATCAGAATCAGC-3′ and 5′-GCTGATTCTGATTATTGACTTAGTCA-3′.
Biotinylated oligonucleotide precipitation.
Wild-type and mutant Smad-binding site oligonucleotides were synthesized with the addition of biotin to the 5′ end of one strand and annealed with the nonmodified complementary strand. For each binding reaction, 50 pmol of annealed oligonucleotides was coupled to 60 μl of streptavidin paramagnetic beads (Promega) in 10 mM HEPES (pH 7.6), 1 mM EDTA, and 2 M NaCl at 25°C for 40 min. The beads were washed four times in the same buffer by using a magnetic stand. Whole-cell extracts (400 ng) (44) were precleared with 30 μl of streptavidin beads for 3 h on ice and then incubated with the oligonucleotide-bound beads and 4 μg of poly(dI-dC)-poly(dI-dC) with a final concentration of 10 mM HEPES (pH 7.6), 100 mM NaCl, 0.1 mM EGTA, 6.6% glycerol, and 0.2% NP-40 for 2 h at 4°C. The beads were washed four times with 10 mM HEPES-150 mM NaCl-1 mM EDTA-0.1% NP-40, and bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
Isolation of total RNA and Northern blot analysis were performed by standard methods (16, 58).
RESULTS
Identification of a TGF-β- and glucocorticoid-responsive DNA sequence in the MMTV LTR.
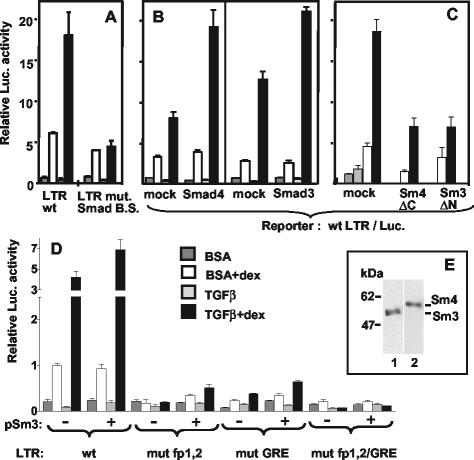
TGF-β plays an important role in cells of the immune system, and we were therefore interested in examining the possible effects of this cytokine on MMTV gene expression in the mature B-cell line we are using as model: the murine M12 B-cell lymphoma. The cells were transiently transfected with reporter plasmids containing the firefly luciferase gene under the control of the MMTV LTR (5) from the mouse strain called GR (the name having no connection to the common abbreviation used for the glucocorticoid receptor) in its wild-type form or with various mutations (Fig. 1A). Transfected M12 cells were treated with TGF-β in absence of serum, with or without the synthetic glucocorticoid, Dex. The results are shown in Fig. 1B. Whereas the basal level was slightly reduced, the Dex-induced level of luciferase activity was increased fivefold by TGF-β. The effect appeared as a superinduction of the glucocorticoid stimulation, as it was abolished, along with the hormonal induction, in an LTR mutated in the GRE or in both GABP-binding sites (fp1/fp2, Fig. 1B). The superinduction was not restricted to transfected templates. The level of MMTV RNA transcribed from endogenous proviral copies of M12 cells was also enhanced by TGF-β plus Dex, compared to Dex alone, as shown by Northern blot analysis of M12 cell RNA (Fig. 1C). We noticed that endogenous transcripts were present at a relatively high basal level, which could not be increased by Dex. However, only the combination of Dex and TGF-β produced an increase (fivefold) of steady-state endogenous MMTV RNA, suggesting that glucocorticoids are involved in the effect from integrated templates as well. All forms of endogenous transcripts of B cells were increased, i.e., the 9-kb full-length, the 3.6-kb env, and the 1.7-kb superantigen-encoding RNAs, as well as a previously described 2.8-kb RNA (46). A twofold increase of endogenous MMTV transcripts with Dex plus TGF-β was also observed in primary BALB/c spleen cells cultured for 4 days in the presence of lipopolysaccharide, a B-cell activator (data not shown). The cooperative effect of TGF-β and Dex observed in B cells was in striking contrast to the repression observed in the mouse mammary tumor GR cell line transfected with the same wild-type LTR reporter. The results with GR cells (Fig. 1D) showed a drastic reduction of reporter activity in TGF-β-treated cells, a finding in agreement with a published report on endogenous (Mtv-2) transcripts (12). Only the Dex-induced level could be measured: with this assay, the basal level in mammary cells, in contrast to B cells, is exceedingly low and below detection due to negative regulatory factors (71). A strong negative effect of TGF-β was also observed with an LTR truncated at position −303 or mutated in both GABP-binding sites. With these two templates, the absolute levels of Dex-induced activity were lower than with a wild-type LTR due to the removal of weakly positive elements, as observed previously (9, 10). These results show that TGF-β acts in opposite ways on the MMTV LTR promoter depending on the cell type (mature B cells versus mammary tumor cells), suggesting a tissue specificity of the effect. To start mapping the LTR sequences required for the TGF-β superinduction in B cells, we compared the activities of a complete LTR (Fig. 2A) and of an LTR truncated at position −303 (Fig. 2B). The latter showed a higher basal level due to the removal of negative regulatory elements (45) but retained a TGF-β induction that was completely dependent on the GABP-binding sites. With LTRs carrying only one mutated site (either fp1 or fp2; Fig. 2C), the residual activities of each mutant showed additivity with respect to glucocorticoid induction, in agreement with our previous results (5). However, they showed a synergism with respect to the TGF-β effect (Fig. 2C), suggesting a requirement for the occupancy of both GABP-binding sites, presumably by an α2-β2 tetramer (27) for maximal TGF-β response.
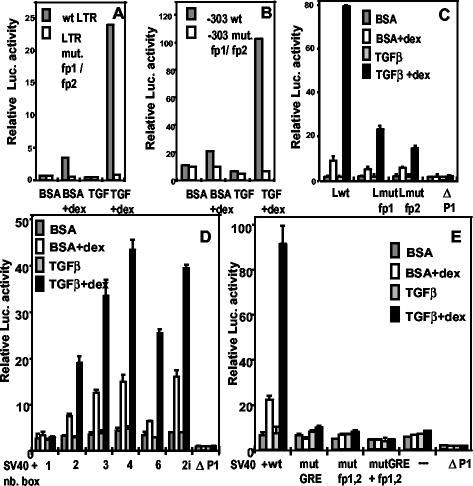
FIG. 2.
The GABP-binding sites are required, together with the GRE, for the TGF-β superinduction of the MMTV promoter (A, B, and C) and of the SV40 promoter (D and E). Transient transfection and treatment of M12 cells were as described in the legend to Fig. 1B. (A to C) pGL3-basic containing the following LTRs: wild type (wt LTR or Lwt); LTR mutated in fp1/fp2 (LTR mut. fp1/fp2 [see Fig. 1A]); LTR 5′ truncated at −303, wild type (-303 wt), or LTR mutated in fp1/fp2 (-303 mut. fp1/fp2); LTR mutated only in fp1 (Lmut fp1) or in fp2 (Lmut fp2); a promoter-negative LTR deleted from positions −105 to +133 (ΔP1), as a negative control. (D and E) pGL3-(SV40)-promoter containing the indicated number of head-to-tail boxes (nb. box) with the wild-type sequence from positions −116 to −209 (2i = two in inverted orientation) (D) or none (−), or else two boxes, either wild type (wt) or mutated in the GRE (mutGRE = LS−175/−166 [10]), in fp1/fp2 (mut fp1,2), or in both GRE and fp1/fp2 (mutGRE + fp1,2) (E). The negative control was as in panel C. Cotransfected internal standards (50 ng) were pRL-SV40 in panels A and B and pRL-SV40Δ enhancer in panels C, D, and E. Error bars indicate the standard deviations. Experiments were repeated at least twice.
A minimal construct is active as a dimer with a heterologous promoter.
We next sought to define a minimal effector sequence and to assess any contribution by the MMTV promoter. The DNA fragment from positions −209 to −116, comprising the GRE and the GABP-binding sites with only short flanking sequences, was cloned in front of the SV40 enhancer-less promoter linked to the firefly luciferase gene, either as a single copy or as head-to-tail oligomers. The results of transient assays in M12 cells (Fig. 2D) show that (i) a single copy was inactive, both in Dex alone and in Dex+TGF-β stimulation; (ii) a dimer was competent for both effects, to an extent similar to that of an MMTV-promoter construct; (iii) three or four copies, but not six, produced a further boost; (iv) a dimer in reverse orientation with respect to the promoter was more active than one in a direct orientation. We conclude that the combination of GRE and GABP-binding sites behaves as an enhancer sequence similarly to a simple GRE with respect to glucocorticoid stimulation (67), whereby the superinduction by TGF-β appears as an amplification of the hormone effect. The pattern of reporter activity of the dimer is reminiscent of that of the MMTV promoter, which indeed contains two GREs (10). The essential role of the GABP recognition sequences was conserved in the heterologous construct, since their mutation abolished the responsiveness of a dimer-containing plasmid (Fig. 2E), suggesting that the GRE/fp1/fp2 sequence constitutes an element that mediates the glucocorticoid stimulation and the superimposed TGF-β response in B cells.
A putative Smad-recognition sequence in the MMTV LTR is involved in the superinduction by TGF-β of the glucocorticoid response in B cells.
We noticed that the DNA sequence linking fp1 and fp2 (position −155/−149) resembles some of the proposed binding sites for Smad3/4 (reviewed in references 20 and 51). We mutated this putative AGACA “Smad box” plus three nucleotides on the 3′ side (Fig. 1A) in the context of the LTR/luciferase reporter plasmid and tested its activity in transiently transfected M12 cells. Two types of results were obtained that depended on the physiological state of the cells at the time of transfection. On one hand, exponentially growing cultures showed little reduction of the TGF-β effect in presence of Dex compared to a wild-type LTR (not shown). On the other hand, cultures that were transfected at a high cell density showed an impairment of the TGF-β superinduction but not of stimulation by Dex alone (Fig. 3A). This inhibition was not due to exhaustion of nutrients, since it was not found in cultures that had been prestarved in absence of serum for 24 h before transfection (not shown). This difference may suggest the existence of redundant signaling pathways for TGF-β (50); alternatively, the mutation may be partially compensated for in actively growing cells by protein-protein contacts of Smads with GR and GABP (see below). To see whether one could affect the process, we overexpressed either Smad4 or Smad3 (Fig. 3B) with the wild-type LTR reporter construct. An increase (∼2-fold) of the TGF-β superinduction was detected in several instances, but not always, reflecting a difficulty often encountered in this type of experiments with cells endowed with endogenous levels of factors (Fig. 3E shows a Western immunoblot for Smad3 and Smad4 of TGF-β+Dex-treated M12 cell extracts). The role of Smad proteins in the TGF-β superinduction was confirmed when dominant-negative mutants of Smads were used (Fig. 3C). Overexpression of truncated forms of Smads lacking essential domains for mutual interaction (Smad4ΔC [68]) or for nuclear translocation (Smad3ΔN [69]) reduced the extent of TGF-β superinduction (Fig. 3C). Smad4ΔC also inhibited the Dex response by an unidentified mechanism that might involve protein-protein interactions (see below) and will require further analysis. The peculiar position of the Smad recognition sequence, which is intercalated between the GABP-binding sites next to the GRE suggested that it may depend for its function on their occupancy by the cognate factors. The DNA-binding ability of Smads is generally low, and it relies in many instances on cooperation with other DNA-binding factors. We therefore tested the ability of overexpressed Smad3 to further increase the TGF-β superinduction by using LTR templates mutated in the GABP-binding sites, or in the GRE, or in all sites. As shown in Fig. 3D, mutations in either the GRE or the GABP sites still allowed some TGF-β effect, whereas an LTR mutated in both the GABP and the GRE sites was unable to respond, despite the presence of the intact Smad binding site. This result suggests a participation of GABP and GR to the enhancement. We next sought to define more precisely the role of the GR in the TGF-β response. The partial agonist RU486, which can bind to the receptor and induce its nuclear translocation but does not efficiently activate transcription, did not cooperate with TGF-β (data not shown), suggesting a requirement for the transactivation function(s) of the receptor. We showed previously that in M12 cells the Dex induction due to endogenous receptor could be significantly improved by overexpressing a full-length GR (amino acids 3 to 795) exogenously (5) (Fig. 4A). Figure 4A also shows that addition of TGF-β superinduced this (already increased) level by fourfold. To start identifying the domains of the receptor participating in the TGF-β response, we introduced mutant GRs lacking various domains (Fig. 4B) in a cotransfection assay, and compared the superinduction factors with that of the full-length protein (Fig. 4A). As expected, the results in the absence of TGF-β showed variable absolute levels of activity depending on the forms of truncated GR. Their respective functions have been elucidated previously (41) and were verified in the present context, indicating that the overexpression levels were sufficient to overcome the action of endogenous GR. The constitutively active mutant GR(3-556) was still stimulated twofold in the presence of TGF-β, whereas GR(407-795), a dominant-negative mutant with respect to Dex induction, was unable to cooperate with TGF-β. Surprisingly, GR(407-556), including only the DNA-binding and a weak transactivation function (52) mediated a threefold increase by TGF-β. These data suggest that the GR C-terminal domain exerts an inhibitory effect on the TGF-β response, whereas a minimal DNA-binding domain of GR is able to mediate a TGF-β induction.
FIG. 3.
Smad proteins and a putative Smad-binding site in the MMTV LTR between fp1 and fp2 play a role in TGF-β superinduction. Transient transfection and treatment of M12 cells were as described for Fig. 1B. The internal control was pRL-SV40Δ enhancer (50 ng in panels A, B, and D and 25 ng in panel C). (A) pGL3-Basic (3 μg) with a wild-type LTR (LTR wt) or an LTR with an 8-bp mutation between fp1 and fp2 (LTR mut. Smad B.S. [see Fig. 1A]) were transfected into M12 cells grown to high density (see the text). The result is representative of two independent experiments with duplicate samples. (B) Three micrograms of pGL3-basic/wt-LTR was cotransfected with 3 μg of either empty pCI vector (mock), pCMV5-DPC4-HA expressing the Smad4 protein or pCI-Smad3F. (C) One hundred nanograms of pGL3-basic/wt-LTR were cotransfected with 8 μg of either empty pCI vector (mock), pCMV/DPC4(1-514) (Sm4 ΔC), or pCI-Smad3F ΔN (Sm3 ΔN). (D) pGL3-basic (3 μg) with a wild-type LTR (wt), or an LTR mutated in fp1 and fp2 (mut fp1,2; see Fig. 1A), or in GRE (mut GRE = LS −175/−166 [10]), or in both GABP and GR binding sites (mut fp1,2/GRE) was cotransfected with 6 μg of either pCI (−) or pCI-Smad3F (+). Error bars indicate the standard deviations. (E) Western immunoblot of a whole-cell lysate from M12 cells treated with TGF-β plus Dex. Replicated slots were blotted and assayed with antibodies to Smad2/3 (lane 1) or Smad4 (lane 2). The position of molecular size markers is indicated on the left in kilodaltons.
Smad3 and Smad4 bind to an AGACA box in the LTR at position −155.
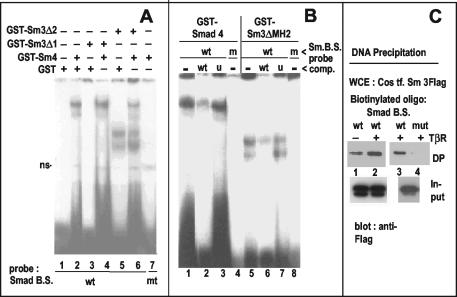
To verify the role of the putative Smad box defined above by mutation, we used it as a probe for EMSAs with bacterially produced GST-Smads (see scheme in Fig. 6C). Full-length Smad4 and the DNA-binding moiety of Smad3 (Smad3ΔMH2 [29]) formed retarded complexes (Fig. 5A and B), whereas GST alone did not (Fig. 5A, lane 1) nor, as expected, did a Smad3 lacking the MH1 domain (Smad3ΔMH1; Fig. 5A, lane 3). The presence of two retarded complexes is likely due to homodimer formation (38). Because Smads can form heterodimers, we tested the combination of GST-Smad4 with the GST-tagged subsets of Smad3 in EMSAs by using the GST protein as a control (Fig. 5A). We found no evidence for higher-order complexes but rather an inhibition of Smad4 binding to the probe in the presence of Smad3-ΔMH2 (lane 6 versus lane 4). The complexes of Smad4 and of Smad3ΔMH2 were competed for by unlabeled homologous oligonucleotides (Fig. 5B, lanes 2 and 6) but not by a heterologous one (lanes 3 and 7). No complexes were detected (Fig. 5A, lane 7; Fig. 5B, lanes 4 and 8) with a probe that included the same base mutations (Fig. 1A) that abolished the TGF-β response in the luciferase assay (Fig. 3A). To demonstrate the binding of full-length Smad3 to the MMTV AGACA box, we used a DNA precipitation assay. A biotinylated double-stranded oligonucleotide containing the wild-type Smad-binding site, but not one containing the mutation in the relevant bases, was able to precipitate Flag-tagged Smad3 from transfected COS cell lysates (Fig. 5C). The coexpression of an activated TGF-β receptor improved the binding only slightly. We conclude that Smad proteins, purified from bacteria or present in a mammalian cell lysate, are able to bind the MMTV LTR sequence between the GABP-binding sites shown in Fig. 1A.
FIG. 5.
Direct binding of Smad3 and Smad4 to the MMTV LTR sequence −160/−143 containing an AGACA (Smad) box. (A) EMSA with E. coli-expressed GST-Smads or GST control alone (lane 1) incubated with 32P-labeled probes encompassing the putative Smad box (without fp1/fp2 [Smad B.S. wt]) or a mutated one (mt, lane 7; see Fig. 1A). ns, nonspecific. (B) EMSA with E.coli-expressed GST-Smads and the wild-type Smad box probe (wt, lanes 1 to 3 and 5 to 7) or the mutated box (m, lanes 4 and 8). Unlabeled, double-stranded oligonucleotides were added in ∼300-fold excess over the probe (lanes 2 and 6). Their sequences were as follows: wild type (wt) or an unrelated (u) hepatic nuclear factor 3 binding site. (C) DNA precipitation assay. Lysates of COS cells expressing Flag-Smad3 with (lanes 2 to 4) or without (lane 1) an activated TGF-β receptor (T204D) were incubated with biotinylated, double-stranded Smad-binding site probes, either wild type (wt, lanes 1 to 3) or mutated (mut, lane 4), and precipitated with streptavidin magnetic beads. DNA-bound precipitates (DP; upper panels) or aliquots of input lysates (lower panels) were subjected to immunoblot analysis with an anti-Flag antibody. Lanes 1 and 2 and lanes 3 and 4 are from two independent experiments.
In vitro association between Smads 3/4 and GABP and between Smad3 and GR.
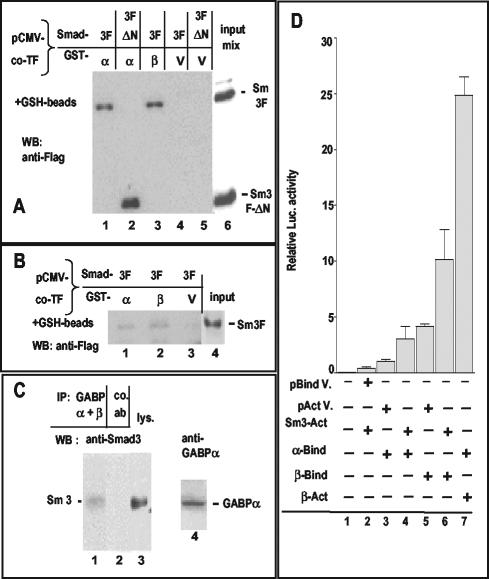
GST fusion proteins with GABPα or GABPβ were produced in bacteria (Fig. 6B) and incubated with extracts of COS cells overexpressing epitope-tagged Smads in GST pull-down assays (Fig. 6A). HA-Smad4 interacted with GST-GABPα (Fig. 6A, lane 1) but not with GABPβ or with GST alone (lanes 2 and 3). Flag-Smad3 interacted with GABPα (lanes 7 and 15) and GABPβ (lanes 6 and 14) but not with GST (lane 5). Flag-Smad2 (another receptor-regulated Smad) interacted more weakly than Smad3 with GABPα (lane 13) and even less with GABPβ (lane 12). In some experiments, Smad3 also bound less well to β than to α (lane 7 versus lane 6 and data not shown). Whether the interaction is direct or requires an intermediate protein from the cell lysate will have to be determined. An N-terminally truncated Smad3 (Sm3ΔN) containing essentially the MH2 domain (see scheme in Fig. 6C) was also able to interact with GST-GABPα (Fig. 6A, lane 11). GST pull-down assays were also performed by using bacterially produced GST-Smad3 or truncated forms of it (lacking either the MH1 or the MH2 domains; Fig. 6D, lanes 5′ to 7′) linked to GSH-beads and incubated with extracts of COS cells overexpressing untagged GR (either full-length or truncated at the C terminus; see scheme in Fig. 4B). GR was subsequently detected on a Western immunoblot with an antibody recognizing its N-terminal portion. Full-length GR was pulled down by GST-Smad3 (Fig. 6D, lane 2), pulled down more weakly by Smad3-ΔMH1 (lane 3) and Smad3-ΔMH2 (lane 4), and not pulled down by GST alone (lane 1). The GR subfragment GR(3-556) also reacted with GST-Smad3 (lane5) and weakly with Smad3-ΔMH2 (lane 7) but not with Smad3-ΔMH1 (lane 6). In contrast, no interactions were detected between any of the GST-Smads (3 or 4) constructs mentioned above and an epitope-tagged GR fragment containing essentially the DNA-binding domain (positions 407 to 556 [see scheme in Fig. 4B; data not shown]).
In vivo association between Smad 3 and GABP.
To assess whether interactions observed by in vitro GST pull-down assays also occurred intracellularly, we applied various experimental approaches to the instance of Smad3-GABP. (i) We used a mammalian CMV-GST vector to coexpress GST-fused GABPα or GABPβ with tagged Smad3 in the same cells, namely, COS-7 (Fig. 7A) or M12 (Fig. 7B). In both cell types, isolation of the GST-carrying proteins on glutathione beads caused the copurification of Smad3: with GABPα (Fig. 7A and B, lane 1) and with GABPβ (Fig. 7A, lane 3, and 7B, lane 2). The C-terminal MH2 portion of Smad3 (ΔN) was also found in association with GABPα in COS cells, similar to the in vitro observation (Fig. 7A, lane 2). As a control, no Smad3 or Smad3ΔN was pulled down by the coexpressed GST protein alone (Fig. 7A, lanes 4 and 5, and B, lane 3). (ii) Anti-GABP immunoprecipitates of endogenous GABP from TGF-β-treated M12 cells contained endogenous Smad3 (Fig. 7C, lane 1), which was not immunoprecipitated by a control antiserum (Fig. 7C, lane 2). (iii) Finally, intracellular interactions between Smad3 and GABP were assayed by using a mammalian two-hybrid system in COS cells, which lack endogenous GABP (Fig. 7D). Luciferase activity from a promoter preceded by five GAL-binding sites was measured in presence of cotransfected GABPα fused to the GAL4 DNA-binding domain and of Smad3 fused to the VP16 activation domain. An enhancement of ∼3-fold was observed, compared to VP16 alone (Fig. 7D, lane 4 versus lane 3). GABPβ fused to the GAL4 DNA-binding domain displayed, as expected, an intrinsic transactivation (Fig. 7D, lane 5), which was increased threefold by the coexpression of Smad3-VP16 (Fig. 7D, lane 6 versus lane 5). These results suggest that Smad3 associates with each of the GABP proteins in the nucleus of transfected COS cells, albeit not as strongly as GABPα with GABPβ, which were used as positive control (Fig. 7D, compare lane 7 to lane 3).
FIG. 7.
Intracellular interactions of Smads with GABP in COS cells (A and D) and M12 B cells (B and C). (A) COS cells were cotransfected with CMV-based plasmids expressing Flag-tagged Smad3 (full length or N terminally deleted, as depicted in Fig. 6C), a constitutively active TGF-β receptor, and a mammalian CMV-GST vector encoding GST-fused GABPα or GABPβ or GST alone (vector, lanes marked V). Whole-cell lysates were incubated with glutathione-Sepharose beads, and bound proteins were analyzed by Western immunoblotting with an anti-Flag antiserum. (B) M12 cells were cotransfected with CMV-based plasmids expressing Flag-tagged Smad3, GST-GABPα, GST-GABPβ, or vector (V); a constitutively active TGF-β receptor; and a plasmid encoding the SV40 large-T antigen. The subsequent steps were as in panel A. (C) Interaction between endogenous factors of M12 cells treated with TGF-β1 (5 ng/ml) for 30 min after a 15-h incubation in serum-free medium. GABP was immunoprecipitated from one-half of the soluble cell extract with rabbit polyclonal antibodies against GABPα and GABPβ (lane 1). Control rabbit serum against an unrelated antigen was added to the other half (lane 2). After addition of protein A-Sepharose, bound proteins were analyzed by Western immunoblot with a monoclonal anti-Smad3 antibody. Lanes 3 and 4 contain aliquots of the cell lysate blotted with anti-Smad3 (lane 3) or anti-GABPα (lane 4, from a different gel). (D) Mammalian two-hybrid assay in COS cells transfected with a luciferase reporter plasmid driven by a promoter with five GAL-binding sites, in combination with the plasmids indicated by the “+” signs. The suffix “Bind” denotes constructs with the GAL4-DNA-binding domain; “Act” denotes constructs with the VP16 activation domain. Control plasmids were the respective vectors (V, first two lines). Statistically significant differences were observed between columns 3 and 4 (P value < 0.0001), columns 5 and 6 (P = 0.0009), and columns 3 and 7 (P < 0.0001). For more details, see Results.
DISCUSSION
For a specific transcriptional TGF-β response, it is essential that Smads interact with DNA-binding partners. Of particular interest are combinations with signal-activated factors, leading to convergence of pathways (reviewed in references 4 and 51). We identified here a novel configuration of Smad partners, cooperating on the natural MMTV promoter in a cell type-restricted manner. TGF-β exerts here a positive modulation (up to 10-fold) on the activated GR, itself requiring an ETS family factor, GABP, for transcriptional stimulation in B cells (5). The inhibition observed in mammary cells confirms previous observations (12) and underscores the differential modes of action of TGF-β in epithelial versus B cells (50), as well as the tissue specificity of the MMTV LTR promoter. Importantly, the TGF-β-GR cooperation was also found with an integrated MMTV template, suggesting that cellular genes may respond similarly if endowed with the appropriate DNA elements. MMTV displays an extreme compression of regulatory sequences, with a Smad box tightly inserted between two GABP-binding sites, themselves adjacent to a GRE (see scheme in Fig. 1A). We showed in vitro binding of Smads, produced in bacteria or mammalian cells, to the MMTV Smad box (Fig. 5), the mutation of which reduced or abolished the TGF-β superinduction in the B-cell reporter assay (Fig. 3A). A dimer of the fragment, including the GR-, GABP-, and Smad-binding sites was functional in front of a heterologous promoter, showing the same dependence on the GRE and GABP sites as in MMTV (Fig. 2D and E). Overexpression of Smads could moderately increase the TGF-β superinduction and partially compensate for the absence of one (but not both) neighboring sites, suggesting interactions between Smads and GR/GABP (Fig. 3). These interactions were verified for the GR by cotransfection in the reporter assay (Fig. 4) and for both GABP and GR by GST pull-down assays in vitro (Fig. 6). Moreover, we could show an intracellular association between Smad3 and GABP: (i) in COS cells, by cotransfection of GST-GABP and Smad3 and by a mammalian two-hybrid assay, and (ii) in M12 cells (where the functional synergy takes place), by GST pull-down of transfected factors and by coimmunoprecipitation of the endogenous factors (Fig. 7). The fact that the interaction was specific but not quantitative might point to a role for the specific DNA sequences as a stabilizing element. This question will be studied in further investigations.
Due to their low DNA-binding sequence specificity, Smad boxes frequently function only as multimers (60), and Smads often act in concert with other transcription factors bound nearby to the DNA (reviewed in reference 63). Of the ETS factor family, only Ets1 was shown, in a recent study, to participate in the TGF-β response of a gene in breast cancer cells, along with Sp1 (42). The situation of the MMTV LTR in B cells differs from many described previously in that TGF-β does not affect the basal transcription level (itself not requiring GABP) but acts to amplify the glucocorticoid-mediated stimulation (which requires GABP, in addition to GR [5]). The peculiarly dense arrangement of DNA elements in MMTV may raise the question of steric hindrance. In vitro, the MH1 domain of Smad3 binds to a single Smad box through a β hairpin embedded in the DNA major groove, allowing simultaneous binding of factors close by on the DNA (59). Since the structure of GABP bound to a direct repeat of ETS sites is known (6), it will be interesting to model the situation on the inverted repeat of MMTV DNA. An interaction between factors binding so closely to each other was first suggested by the strong dependence of the TGF-β superinduction on the integrity of the neighboring sites and by the synergistic contribution of each GABP site (Fig. 2C). The results of GST pull-down assays revealed GABP as a novel partner for Smads (Fig. 6). We found an interaction of GABPα with Smad4 and with Smad3 (in its MH2 domain) and of GABPβ with Smad3 but not Smad4. Since the GABP subunits contain a number of characterized domains (6), it will be of interest to analyze the contacts in more detail.
Concerning the interactions between Smad3 and GR, the indications from GST pull-down assays (Fig. 6D) are apparently at variance with those of transient-transfection assays (Fig. 4A), but the parameters involved are quite different. A GR fragment from amino acids 407 to 556 (containing essentially the DNA-binding domain; Fig. 4B) was able to cooperate in the TGF-β superinduction; however, it did not show any in vitro interactions with Smads (data not shown). Interestingly, we observed an in vitro interaction between GR(407-556) and GABPα (E. Buetti, unpublished observations). When the DNA-binding domain was extended to the C terminus [in GR(407-795)], it suppressed the TGF-β superinduction, suggesting an inhibitory function for this domain. This was not the case for a GR extended on the N-terminal side [i.e., GR(3-556); Fig. 4A]. These results point to distinct interactions of GR domains with DNA-bound Smads. In GST pull-down assays (Fig. 6D), full-size GR was able to bind full-size GST-Smad3 and also, albeit less well, Smad3 with MH1 or MH2 (Fig. 6C) deleted. However, GR(3-556) could only bind Smad3-ΔMH2 and not Smad3-ΔMH1, suggesting contacts between the GR-N terminus and the MH1+linker region of Smad3 and, by default, between the GR-C terminus and the MH2+linker region. Further studies will be performed to better define the interacting domains.
Of the steroid receptor family, the activated vitamin D receptor was shown to be potentiated by Smad3 through the MH1 domain (70). Whether Smad boxes on the same DNA are also required (62) is a matter of debate. Contrasting results have been reported also on the androgen receptor, either repressed (30) or enhanced (37) by Smad3 in different cells. Conversely, the activated androgen receptor repressed the TGF-β induction of a gene in prostate cancer cells by preventing Smad3 binding to DNA (15). The GR was shown previously to interact with Smad3 and to inhibit the TGF-β response of the plasminogen activator inhibitor-1 promoter. Interestingly, the repression was not reciprocal (61). In contrast to the above examples, the region of the MMTV promoter analyzed here contains a combination of regulatory sequences that possibly evolved to accommodate viral expression in cell types equipped with particular sets of signaling pathways and transcription factors. From the standpoint of the virus, a transcriptional upregulation of the MMTV promoter by TGF-β in B cells may play a role during the primary infection process, which is remarkably inefficient (31). TGF-β is present at a high concentration in maternal milk and is locally produced in the postnatal small intestine (26, 57); TGF-β, secreted by intestinal epithelial cells (26), plays a major role in the homeostasis of B cells in the Peyer's patches and is critical for immunoglobulin A production (13). Glucocorticoids are present in milk (1) and can be taken up by suckling mice, where they remain biologically active (2); they were shown to variously affect B-cell populations (40). Although the biological implications of TGF-β superinduction for the virus life cycle are still speculative, the identification of GABP and GR as functional partners of TGF-β signaling in B lymphocytes opens the way to further investigations of protein-protein and protein-DNA interactions taking place on this composite enhancer sequence and of the connected pathways. The results may reveal novel aspects of regulatory networks involving TGF-β, and further studies will determine whether they operate in the regulation of cellular genes as well.
Acknowledgments
We thank Joan Massagué, Rik Derynck, and Carl-Henryk Heldin for Smad and the TGF-β receptor plasmids, Sandro Rusconi for GR plasmids; Randall Reed for the the pCMV-GST vector; Peter Beard for the SV40 large-T antigen plasmid; and Egbert Flory for the anti-GABP antisera. We thank Julien Ackermann for assistance with the RNA analysis and Pierre Devaud and Salina Arope for assistance with the two-hybrid analysis. We also thank Françoise Stutz for critical reading of the manuscript and Heidi Diggelmann for support.
This study was supported by the Swiss National Science Fund.
REFERENCES
- 1.Alexandrova, M., and L. Macho. 1983. Glucocorticoids in human, cow, and rat milk. Endocrinol. Exp. 17:183-189. [PubMed] [Google Scholar]
- 2.Appel, R. J., and V. P. Eroschenko. 1992. Passage of methoxychlor in milk and reproductive organs of nursing female mice. I. Light and scanning electron microscopic observations. Reprod. Toxicol. 6:223-231. [DOI] [PubMed] [Google Scholar]
- 3.Ardavin, C., P. Martin, I. Ferrero, I. Azcoitia, F. Anjuere, H. Diggelmann, F. Luthi, S. Luther, and H. Acha-Orbea. 1999. B-cell response after MMTV infection: extrafollicular plasmablasts represent the main infected population and can transmit viral infection. J. Immunol. 162:2538-2545. [PubMed] [Google Scholar]
- 4.Attisano, L., and J. L. Wrana. 2000. Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 12:235-243. [DOI] [PubMed] [Google Scholar]
- 5.Aurrekoetxea-Hernández, K., and E. Buetti. 2000. Synergistic action of GA-binding protein and glucocorticoid receptor for the transcription of mouse mammary tumor virus in B cells. J. Virol. 74:4988-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batchelor, A. H., D. E. Piper, F. C. de la Brousse, S. L. McKnight, and C. Wolberger. 1998. The structure of GABPα/β: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science 279:1037-1041. [DOI] [PubMed] [Google Scholar]
- 7.Beato, M. 1989. Gene regulation by steroid hormones. Cell 56:335-344. [DOI] [PubMed] [Google Scholar]
- 8.Beato, M., P. Herrlich, and G. Schütz. 1995. Steroid hormone receptors: many actors in search of a plot. Cell 83:851-857. [DOI] [PubMed] [Google Scholar]
- 9.Buetti, E., and H. Diggelmann. 1983. Glucocorticoid regulation of mouse mammary tumor virus: identification of a short essential DNA region. EMBO J. 2:1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buetti, E., and B. Kühnel. 1986. Distinct sequence elements involved in the glucocorticoid regulation of the mouse mammary tumor virus promoter identified by linker scanning mutagenesis. J. Mol. Biol. 190:379-389. [DOI] [PubMed] [Google Scholar]
- 11.Cardiff, R. D., and L. J. T. Young. 1980. Mouse mammary tumor biology: a new synthesis, p. 1105-1114. In M. Essex, G. Todaro, and H. Zur Hausen (ed.), Viruses in naturally occurring cancers. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 12.Cato, A. C., S. Mink, B. Nierlich, H. Ponta, D. Schaap, E. Schuuring, and A. Sonnenberg. 1990. Transforming growth factor-beta represses transcription of the mouse mammary tumour virus DNA in cultured mouse mammary cells. Oncogene 5:103-110. [PubMed] [Google Scholar]
- 13.Cazac, B. B., and J. Roes. 2000. TGF-β receptor controls B-cell responsiveness and induction of IgA in vivo. Immunity. 13:443-451. [DOI] [PubMed] [Google Scholar]
- 14.Chen, X., M. J. Rubock, and M. Whitman. 1996. A transcriptional partner for MAD proteins in TGF-β signalling. Nature 383:691-696. [DOI] [PubMed] [Google Scholar]
- 15.Chipuk, J. E., S. C. Cornelius, N. J. Pultz, J. S. Jorgensen, M. J. Bonham, S. J. Kim, and D. Danielpour. 2002. The androgen receptor represses transforming growth factor-beta signaling through interaction with Smad3. J. Biol. Chem. 277:1240-1248. [DOI] [PubMed] [Google Scholar]
- 16.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 17.Choy, L., and R. Derynck. 1998. The type II transforming growth factor (TGF)-β receptor-interacting protein TRIP-1 acts as a modulator of the TGF-β response. J. Biol. Chem. 273:31455-31462. [DOI] [PubMed] [Google Scholar]
- 18.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J. M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF-β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derynck, R., and L. Choy. 1998. Transforming growth factor-β and its receptors, p. 593-636. The cytokine handbook. Academic Press, Inc., New York, N.Y.
- 20.Derynck, R., Y. Zhang, and X. Feng. 1998. Smads: transcriptional activators of TGF-β responses. Cell 95:737-740. [DOI] [PubMed] [Google Scholar]
- 21.Dickmanns, A., A. Zeitvogel, F. Simmersbach, R. Weber, A. K. Arthur, S. Dehde, A. G. Wildeman, and E. Fanning. 1994. The kinetics of simian virus 40-induced progression of quiescent cells into S phase depend on four independent functions of large T antigen. J. Virol. 68:5496-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flory, E., A. Hoffmeyer, U. Smola, U. R. Rapp, and J. T. Bruder. 1996. Raf-1 kinase targets GA-binding protein in transcriptional regulation of the human immunodeficiency virus type 1 promoter. J. Virol. 70:2260-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glimcher, L. H., T. Hamano, R. Asofsky, E. Herber-Katz, S. Hedrick, R. H. Schwartz, and W. E. Paul. 1982. I region-restricted antigen presentation by B cell-B lymphoma hybridomas. Nature 298:283-284. [DOI] [PubMed] [Google Scholar]
- 24.Gluzman, Y. 1981. SV40-transformed simian cells support the replication of early SV40 mutants. Cell 23:175-182. [DOI] [PubMed] [Google Scholar]
- 25.Golovkina, T. V., J. P. Dudley, and S. R. Ross. 1998. B and T cells are required for mouse mammary tumor virus spread within the mammary gland. J. Immunol. 161:2375-2382. [PubMed] [Google Scholar]
- 26.Goodrich, M. E., and D. W. McGee. 1999. Preferential enhancement of B-cell IgA secretion by intestinal epithelial cell-derived cytokines and interleukin-2. Immunol. Investig. 28:67-75. [DOI] [PubMed] [Google Scholar]
- 27.Graves, B. J. 1998. Inner workings of a transcription factor partnership. Science 279:1000-1002. [DOI] [PubMed] [Google Scholar]
- 28.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Hata, A., R. S. Lo, D. Wotton, G. Lagna, and J. Massagué. 1997. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature 388:82-87. [DOI] [PubMed] [Google Scholar]
- 30.Hayes, S. A., M. Zarnegar, M. Sharma, F. Yang, D. M. Peehl, P. ten Dijke, and Z. Sun. 2001. SMAD3 represses androgen receptor-mediated transcription. Cancer Res. 61:2112-2118. [PubMed] [Google Scholar]
- 31.Held, W., G. A. Waanders, H. Acha-Orbea, and H. R. MacDonald. 1994. Reverse transcriptase-dependent and -independent phases of infection with mouse mammary tumor virus: implications for superantigen function. J. Exp. Med. 180:2347-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Held, W., G. A. Waanders, A. N. Shakhov, L. Scarpellino, H. Acha-Orbea, and H. R. MacDonald. 1993. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell 74:529-540. [DOI] [PubMed] [Google Scholar]
- 33.Higuchi, T. 1991. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols. Academic Press, Inc., San Diego, Calif.
- 34.Hittelman, A. B., D. Burakov, J. A. Iñiguez-Lluhí, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hua, X., X. Liu, D. O. Ansari, and H. F. Lodish. 1998. Synergistic cooperation of TFE3 and smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 12:3084-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonk, L. J., S. Itoh, C. H. Heldin, P. ten Dijke, and W. Kruijer. 1998. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273:21145-21152. [DOI] [PubMed] [Google Scholar]
- 37.Kang, H. Y., H. K. Lin, Y. C. Hu, S. Yeh, K. E. Huang, and C. Chang. 2001. From transforming growth factor-beta signaling to androgen action: identification of Smad3 as an androgen receptor coregulator in prostate cancer cells. Proc. Natl. Acad. Sci. USA 98:3018-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawabata, M., H. Inoue, A. Hanyu, T. Imamura, and K. Miyazono. 1998. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 17:4056-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kühnel, B., E. Buetti, and H. Diggelmann. 1986. Functional analysis of the glucocorticoid regulatory elements present in the mouse mammary tumor virus long terminal repeat: a synthetic distal binding site can replace the proximal binding domain. J. Mol. Biol. 190:367-378. [DOI] [PubMed] [Google Scholar]
- 40.Laakko, T., R. C. Schwartz, and P. J. Fraker. 2002. IL-7-mediated protection of pro and pre-B cells from the adverse effects of corticosterone. Cell. Immunol. 220:39-50. [DOI] [PubMed] [Google Scholar]
- 41.Lanz, R. B., S. Wieland, M. Hug, and S. Rusconi. 1995. A transcriptional repressor obtained by alternative translation of a trinucleotide repeat. Nucleic Acids Res. 23:138-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindemann, R. K., P. Ballschmieter, A. Nordheim, and J. Dittmer. 2001. Transforming growth factor beta regulates parathyroid hormone-related protein expression in MDA-MB-231 breast cancer cells through a novel Smad/Ets synergism. J. Biol. Chem. 276:46661-46670. [DOI] [PubMed] [Google Scholar]
- 43.Liu, B., C. Dou, l. Prabhu, and E. Lai. 1999. FAST-2 is a mammalian winged-helix protein which mediates transforming growth factor β signals. Mol. Cell. Biol. 19:424-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, F., C. Pouponnot, and J. Massagué. 1997. Dual role of the Smad4/DPC4 tumor suppressor in TGF-β-inducible transcriptional complexes. Genes Dev. 11:3157-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, J., D. Bramblett, Q. Zhu, M. Lozano, R. Kobayashi, S. R. Ross, and J. P. Dudley. 1997. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol. Cell. Biol. 17:5275-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lund, F. E., and R. B. Corley. 1991. Regulated expression of mouse mammary tumor proviral genes in cells of the B lineage. J. Exp. Med. 174:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luther, S. A., and H. Acha-Orbea. 1997. Mouse mammary tumor virus: immunological interplays between virus and host. Adv. Immunol. 65:139-243. [PubMed] [Google Scholar]
- 48.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schütz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massagué, J. 1998. TGF-β signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 50.Massagué, J. 2000. How cells read TGF-β signals. Nat. Rev. Mol. Cell. Biol. 1:169-178. [DOI] [PubMed] [Google Scholar]
- 51.Massagué, J., and D. Wotton. 2000. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miesfeld, R., P. J. Godowski, B. A. Maler, and K. R. Yamamoto. 1987. Glucocorticoid receptor mutants that define a small region sufficient for enhancer activation. Science 236:423-427. [DOI] [PubMed] [Google Scholar]
- 53.Miyazono, K., P. ten Dijke, and C. H. Heldin. 2000. TGF-beta signaling by Smad proteins. Adv. Immunol. 75:115-157. [DOI] [PubMed] [Google Scholar]
- 54.Moustakas, A., and D. Kardassis. 1998. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc. Natl. Acad. Sci. USA 95:6733-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pardali, E., X. Q. Xie, P. Tsapogas, S. Itoh, K. Arvanitidis, C. H. Heldin, P. ten Dijke, T. Grundstrom, and P. Sideras. 2000. Smad and AML proteins synergistically confer transforming growth factor β1 responsiveness to human germ-line IgA genes. J. Biol. Chem. 275:3552-3560. [DOI] [PubMed] [Google Scholar]
- 56.Park, S. R., J. H. Lee, and P. H. Kim. 2001. Smad3 and Smad4 mediate transforming growth factor-β1-induced IgA expression in murine B lymphocytes. Eur. J. Immunol. 31:1706-1715. [DOI] [PubMed] [Google Scholar]
- 57.Penttila, I. A., A. B. van Spriel, M. F. Zhang, C. J. Xian, C. B. Steeb, A. G. Cummins, H. Zola, and L. C. Read. 1998. Transforming growth factor-beta levels in maternal milk and expression in postnatal rat duodenum and ileum. Pediatr. Res. 44:524-531. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Shi, Y., Y. F. Wang, L. Jayaraman, H. Yang, J. Massagué, and N. P. Pavletich. 1998. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell 94:585-594. [DOI] [PubMed] [Google Scholar]
- 60.Song, C. Z., T. E. Siok, and T. D. Gelehrter. 1998. Smad4/DPC4 and Smad3 mediate transforming growth factor-beta (TGF-β) signaling through direct binding to a novel TGF-β-responsive element in the human plasminogen activator inhibitor-1 promoter. J. Biol. Chem. 273:29287-29290. [DOI] [PubMed] [Google Scholar]
- 61.Song, C. Z., X. Tian, and T. D. Gelehrter. 1999. Glucocorticoid receptor inhibits transforming growth factor-beta signaling by directly targeting the transcriptional activation function of Smad3. Proc. Natl. Acad. Sci. USA 96:11776-11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramaniam, N., G. M. Leong, T. A. Cock, J. L. Flanagan, C. Fong, J. A. Eisman, and A. P. Kouzmenko. 2001. Cross-talk between 1,25-dihydroxyvitamin D3 and transforming growth factor-beta signaling requires binding of VDR and Smad3 proteins to their cognate DNA recognition elements. J. Biol. Chem. 276:15741-15746. [DOI] [PubMed] [Google Scholar]
- 63.ten Dijke, P., I. Miyazono, and I. Heldin. 2000. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem. Sci. 25:64-70. [DOI] [PubMed] [Google Scholar]
- 64.Tsai, R. Y., and R. R. Reed. 1997. Using a eukaryotic GST fusion vector for proteins difficult to express in Escherichia coli. BioTechniques 23:794-800. [DOI] [PubMed] [Google Scholar]
- 65.Tsubura, A., M. Inaba, S. Imai, A. Murakami, N. Oyaizu, R. Yasumizu, Y. Ohnishi, H. Tanaka, S. Morii, and S. Ikehara. 1988. Intervention of T cells in transportation of mouse mammary tumor virus (milk factor) to mammary gland cells in vivo. Cancer Res. 48:6555-6559. [PubMed] [Google Scholar]
- 66.van Leeuwen, F., and R. Nusse. 1995. Oncogene activation and oncogene cooperation in MMTV-induced mouse mammary cancer. Semin. Cancer Biol. 6:127-133. [DOI] [PubMed] [Google Scholar]
- 67.Wieland, S., M. D. Schatt, and S. Rusconi. 1990. Role of TATA-element in transcription from glucocorticoid receptor-responsive model promoters. Nucleic Acids Res. 18:5113-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu, R. Y., Y. Zhang, X. H. Feng, and R. Derynck. 1997. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol. Cell. Biol. 17:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao, Z., X. Liu, Y. I. Henis, and H. F. Lodish. 2000. A distinct nuclear localization signal in the N terminus of Smad 3 determines its ligand-induced nuclear translocation. Proc. Natl. Acad. Sci. USA 97:7853-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanagisawa, J., Y. Yanagi, Y. Masuhiro, M. Suzawa, M. Watanabe, K. Kashiwagi, T. Toriyabe, M. Kawabata, K. Miyazono, and S. Kato. 1999. Convergence of transforming growth factor-beta and vitamin D signaling pathways on SMAD transcriptional coactivators. Science 283:1317-1321. [DOI] [PubMed] [Google Scholar]
- 71.Zhu, Q., K. Gregg, M. Lozano, J. Liu, and J. P. Dudley. 2000. CDP is a repressor of mouse mammary tumor virus expression in the mammary gland. J. Virol. 74:6348-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]