Abstract
Background:
Periodontitis is a disease mainly caused by a chronic infection of tissues that support the teeth. Several factors, such as diabetes, smoking and oral care, as well as genetic susceptibility can influence both the risk to develop periodontitis and its progression. The aim of the investigation was to test whether alleles of candidate genes were associated with periodontitis.
Materials and Methods:
A case control study was performed with a cohort of 184 patients with chronic periodontitis and 231 healthy controls from the Italian population. A total of six single nucleotide polymorphisms from five candidate genes, i.e., IL1A, IL1B, IL6, IL10 and vitamin D receptor, were investigated.
Results:
Evidence of association were obtained for rs1800795 mapping in IL6 (P value = 0.01) as well as for the rs1800872 mapping in IL10 (P = 0.04). The rarer variant allele lowered the risk to develop periodontitis at IL6 (Odds Ratio [OR] = 0.69 [95% confidence interval {CI} 0.51-0.93]) and increased the risk at IL10 (OR = 1.38 [95% CI 1.01-1.86]).
Conclusions:
The present investigation indicated that polymorphisms of IL6 and IL10 constitute risk factors for chronic periodontitis, while there was no evidence implicating a specific IL1A or IL1B genotype.
Keywords: Bone resorption, genes, inflammation, periodontal disease, polymorphism
INTRODUCTION
Periodontitis is a multifactorial disease in which both environmental and genetic factors play a role. A major role in the progression of periodontitis is played by the periodontal pocket microbiota, where both the amount and the presence of specific bacteria species represent risk factors. Additional risk factors are smoking and diabetes. However, a range of host genetic factors can influence individual susceptibility to periodontitis, and are able to influence the clinical aspects and rate of progression of the disease.[1,2]
Genetic susceptibility to multifactorial diseases, as well as to periodontitis, is usually due to several gene polymorphisms instead of a single, or few, gene mutations. Accordingly, common variation in the genetic code may result in either altered expression or in functional changes of the encoded molecules; therefore, making individuals with genotypes more susceptible to a given disease or resulting in an increase of disease severity.[3,4]
In recent years, investigations on the susceptibility factors of periodontitis have mainly focused on genes that modulate immunoregulation, such as cytokines, cell-surface receptors, chemokines, enzymes and proteins related to antigen recognition. Cytokines, such as IL1A, IL1B, IL10 and IL6, are key factors that mediate the inflammatory process during periodontal disease. They have a role in B-cell activation, proliferation and differentiation, and are the majority of infiltrating cells in advanced periodontitis lesions.[5] Thus, these variations can interfere with the progression of disease[6] because they may be responsible for the repeated cycles of tissue inflammation observed in these disorders.[7]
In periodontal disease, periodonto–pathogenic bacteria accumulated in the subgingival region are the environmental factors that influence the inflammatory response in periodontal tissues.[8] However, cytokines are also considered to indirectly contribute to connective tissue destruction and bone resorption.[9] Because alveolar bone resorption is a key factor in periodontal disease, vitamin D receptor (VDR) has been considered as a periodontitis susceptibility factor. Recent articles reported a revision of the scientific literature regarding genetic association analysis between common polymorphisms of candidate genes and periodontitis.[10,11]
It is noteworthy that the vast majority of the genetic studies on periodontitis have employed small-sized cohorts, resulting in a large potential for false-positive and false-negative results; thus, having a low statistical power to properly detect an association. Additionally, the number and types of disease-modifying genes in periodontal disease may be different in different ethnic populations or disease subgroups.
We analyzed six specific polymorphisms of the IL1A, IL1B, IL6, IL10 and VDR genes in order to test whether they act as susceptibility factors of chronic periodontitis in the Italian population.
MATERIALS AND METHODS
Patients
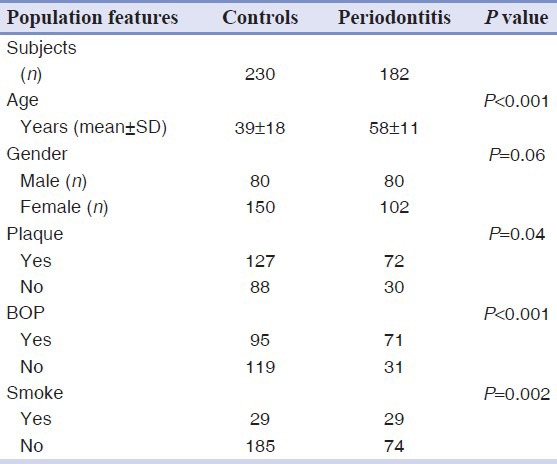
A total of 412 individuals participated in the study; 183 were affected by chronic periodontitis while 230 constituted the control group, being healthy or affected by gingivitis. Table 1 summarizes the principal characteristics and clinical data of the two groups.
Table 1.
Baseline characteristics of the study population
Genotyping
The following six polymorphisms were investigated: At IL1A the NM_000575.3:c.-949C >T (rs1800587); at IL1B the NM_000576.2:c.315C >T (rs1143634); at IL6 the XR_108749.1:n.50-321G >C (rs1800795); at IL10 the NG_012088.1:g.3943A >G (rs1800896) and NG_012088.1:g.4433A >C (rs1800872); and at VDR the NM_000376.2:c.1056T >C (rs731236). Single nucleotide polymorphism (SNP) assays were selected using the Applied Biosystems SNPbrowser Software (Applied Biosystems, Foster City, CA, USA). Genotyping were performed using an ABI PRISM 7500 Sequence Detection System and TaqMan chemistry according to the manufacturers’ protocols (Applied Biosystems). Genotypes were scored by researchers in a blind system (biologists had no information about the clinical disease classification of the samples).
Statistical analysis
The distribution of genotypes in the patient and control groups was tested for deviations from the Hardy–Weinberg equilibrium using Pearson's χ2 test. Genetic association was investigated by both allelic and genotypic tests with a likelihood ratio approach using Unphased software v3.1.5 within a Windows Vista operative system.[12] Odds Ratios (ORs) were calculated in order to evaluate the level of association of the rare allele carriers as well as of the heterozygote and homozygote individuals. Haplotype association was evaluated for the IL1 cluster (IL1A and IL1B) and IL10 cluster because two linked polymorphisms were available. Specifically, a global test of association was performed, which tested whether any haplotype was associated with the disease. A specific association test for each haplotype was performed as well.
RESULTS
Genetic analysis of patient genomic DNA purified from crevicular fluid was highly effective. Indeed, a success rate between 96% and 99% was obtained with the six different assays in the genotyping step. Only 20 out of 412 DNA samples produced three or fewer genotypes.
Genotype frequencies were in agreement with the Hardy–Weinberg law in the whole sample and among the cases and controls groups.
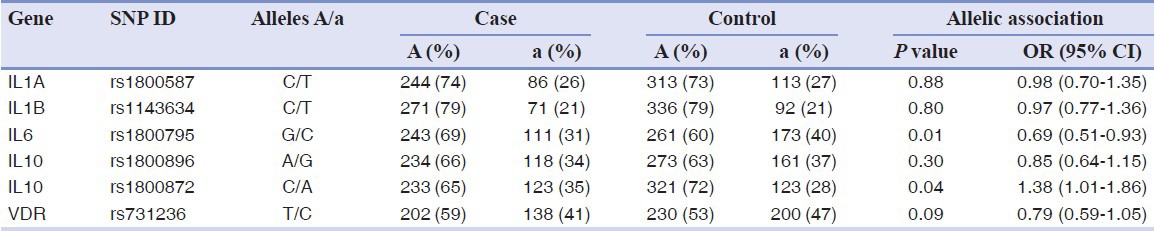
A comparison of the observed allelic frequencies observed in the case and controls is reported in Table 2. A significant difference between the cases and the controls was observed at rs1800795 in IL6 (P = 0.01) and rs1800872 mapping in IL10 (P = 0.04). Marginal values of association were obtained for rs731236 of VDR (P = 0.09). Indeed, individuals having the rarer allele at IL6 and VDR polymorphisms appeared less-susceptible to periodontitis OR = 0.69 (95% confidence interval [CI] 0.51-0.93) and OR = 0.79 (95% CI 0.59-1.05), respectively. The rarer allele at IL10 increased the risk OR = 1.38 (95% CI 1.01-1.86).
Table 2.
Allelic association analysis of periodontitis
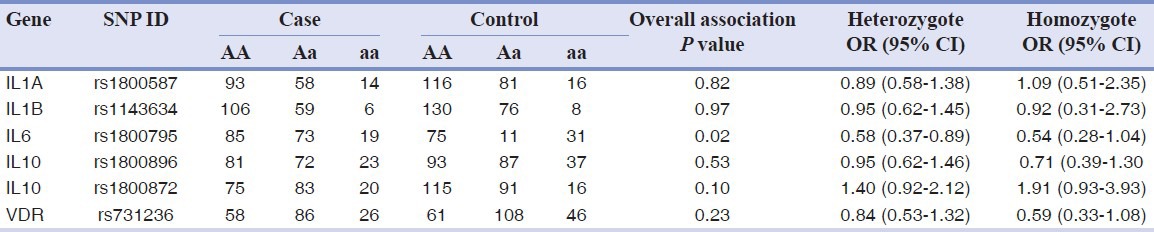
Genotypic association testing provided similar results [Table 3]. The rs1800795 polymorphism at IL6 showed a significant overall genotypic association (P value = 0.02) and a lower risk for both heterozygote and homozygote carrier of the rare allele OR = 0.58 (95% CI 0.37-0.89) and OR = 0.54 (95% CI 0.28-1.04), respectively, which was close to the nominal levels of significance.
Table 3.
Genotype distribution among groups and genotype association analysis data
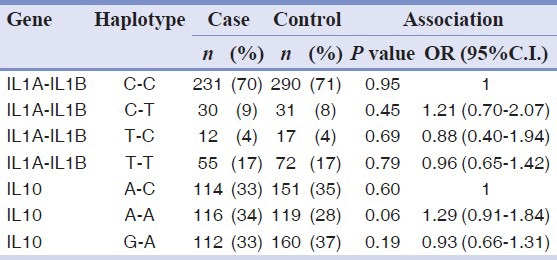
High level of linkage disequilibrium between SNPs of the IL1 gene cluster (D’ =0.75; r2= 0.44) and between SNPs of the IL10 gene (D’ =0.93; r2= 0.22) were detected. This allowed us to combine data from two adjacent markers and to perform a haplotype association analysis. The overall statistics, which tested whether any haplotypes were associated, provided no evidence of association with the IL1 gene cluster (P = 0.87), also not for the IL10 locus (P = 0.16). Specific association values and estimated ORs for each of the haplotypes are reported in Table 4. The haplotype rs1800896 (A)-rs1800872 (A) showed a borderline level of association with periodontitis OR = 1.29 (95% CI 0.91-1.84), P value = 0.06.
Table 4.
Haplotype analysis of two candidate loci and periodontitis
DISCUSSION
Many studies have assessed a genetic association between genetic variation and periodontitis; however, there is still no agreement regarding the genetic bases of this disease. Indeed, conflicting results have been reported. These are mainly related to the different ethnicities of the studied populations and also to the small sample size. Indeed, small studies–with less than 100 patients and 100 controls–cannot provide adequate statistical power to detect a moderate genetic effect, i.e., ORs of 1.5 or less. Another source of variability could be related to the study design, being different studies focused on specific phenotypes, such as aggressive periodontitis, chronic periodontitis or response to treatments. Herein, we report the results of a genetic study performed with 182 patients affected by chronic periodontitis and 182 controls of Italian ancestry. Crevicular fluids collected with a paper probe were used as a source of genomic DNA. The efficiency of a genetic test on this DNA was high; indeed, it provided about 98% of the genotypes. The results of the genetic association analyses of the different loci are discussed, below by locus.
IL1A and IL1B
IL1A and IL1B genes, and seven other interleukin 1 family genes, form a cytokine gene cluster on chromosome 2. IL1A and IL1B polymorphisms have been among the first and most frequently investigated molecules for the association with periodontitis. We found no evidence of an association with rs1800587 (also known as IL1A -889C >T) or rs1143634 (also known as IL1B + 3954C >T). Noteworthy, the ORs were close to 1 in both allele and genotype association analyses, indicating that there was no trend of association. Similar results were obtained when both polymorphisms were analyzed together in the haplotype analysis [Table 4]. In a recent literature review of the investigation with at least 100 individuals in either the case or the control group, the authors concluded that the IL1 gene cluster polymorphisms cannot be considered as risk factors for aggressive periodontitis or chronic periodontitis susceptibility.[11] Our results, obtained with a sample from the Italian population, further supports this statement. On the other hand, a recent meta-analysis of Caucasian chronic periodontitis that pooled data from 13 investigations found significant effects for the two individual gene variations (IL1A OR = 1.48 and IL1B OR = 1.54). Considering that the sample size of the present investigation is more than adequate to detect such a level of association (power calculation not shown), we may conclude that genetic heterogeneity is probably present among Caucasians, being IL1 polymorphisms not associated with periodontitis in the Italian population. Other authors reported no association with aggressive periodontitis in Italy.[13]
IL6
Polymorphism mapping in IL6 has been recurrently reported as risk factors for both aggressive and chronic periodontitis in different populations, although several investigations have shown negative association results.[11] Thus, the authors state that no clear conclusion can be drawn about this locus. In this study, we found the strongest level of association; indeed, the variant allele rs1800795-C (also known as IL6 -174G >C) was significantly less represented among periodontitis patients. This indicated that carriers of rs1800795-C allele had a lower risk of developing periodontitis, as previously reported by other researchers.[11]
IL10
It has been proposed that IL10 could attenuate periodontal tissue destruction through the induction of tissue inhibitors of metalloproteinases and the inhibitor of osteoclastogenesis osteoprotegerin.[14] Here, we tested two polymorphisms of the IL10 promoter region and showed that the variant allele rs1800872-A increased the risk of developing periodontitis, with an observed OR of 1.38 (95% CI 1.01-1.86). Interestingly, Reichert et al. demonstrated that this allele is functional in chronic periodontitis, being associated with a lower expression level of IL10.[15]
VDR
The VDR regulates a variety of biological processes, including bone metabolism and immune response to microbial infections.[16] As alveolar bone resorption and bacterial accumulation are major characteristics of periodontal disease, it is possible that the VDR) and its genetic polymorphisms play a role in periodontitis susceptibility. Here, we found a suggestive association between VDR allele and periodontitis, although at a borderline statistical significance. Additional investigations with independent data or different markers could provide more information to elucidate the role of VDR in periodontitis.
CONCLUSION
Here, we demonstrated a statistically significant association between common variant alleles, IL6 rs1800795-G and IL 10 rs1800872-A, and periodontal disease. These data suggest a possible use of these polymorphisms in a DNA-based diagnostic test of periodontitis.
ACKNOWLEDGEMENTS
This work was supported by FAR from the University of Ferrara (FC), Ferrara, Italy and LAB® s.r.l., Ferrara, Italy.
Footnotes
Source of Support: This work was supported by FAR from the University of Ferrara (FC), Ferrara, Italy and LAB® s.r.l., Ferrara, Italy
Conflict of Interest: None declared
REFERENCES
- 1.Heitz-Mayfield LJ. Disease progression: Identification of high-risk groups and individuals for periodontitis. J Clin Periodontol. 2005;32:196–209. doi: 10.1111/j.1600-051X.2005.00803.x. [DOI] [PubMed] [Google Scholar]
- 2.Shah Abouei M, Abrishami MR, Nasr A, Fateh A. Association between chronic periodontitis and acute myocardial infarction: A case-control study in Isfahan. Dent Res J (Isfahan) 2006;3:1–7. [Google Scholar]
- 3.Craandijk J, van Krugten MV, Verweij CL, van der Velden U, Loos BG. Tumor necrosis factor-alpha gene polymorphisms in relation to periodontitis. J Clin Periodontol. 2002;29:28–34. doi: 10.1034/j.1600-051x.2002.290105.x. [DOI] [PubMed] [Google Scholar]
- 4.Behfarnia P, Birang R, Andalib AR, Asadi S. Comparative Evaluation of IFNγ, IL4 and IL17 Cytokines in Healthy Gingiva and Moderate to Advanced Chronic Periodontitis. Dent Res J (Isfahan) 2010;7:45–50. [PMC free article] [PubMed] [Google Scholar]
- 5.Yamazaki K, Nakajima T, Gemmell E, Polak B, Seymour GJ, Hara K. IL-4- and IL-6-producing cells in human periodontal disease tissue. J Oral Pathol Med. 1994;23:347–53. doi: 10.1111/j.1600-0714.1994.tb00074.x. [DOI] [PubMed] [Google Scholar]
- 6.Scarel-Caminaga RM, Trevilatto PC, Souza AP, Brito RB, Jr, Line SR. Investigation of IL4 gene polymorphism in individuals with different levels of chronic periodontitis in a Brazilian population. J Clin Periodontol. 2003;30:341–5. doi: 10.1034/j.1600-051x.2003.00317.x. [DOI] [PubMed] [Google Scholar]
- 7.Duff GW. Molecular genetics of cytokines: Cytokines in chronic inflammatory disease. In: Thompson, editor. The cytokine handbook. London: Academic Press; 1998. pp. 21–33. [Google Scholar]
- 8.Wilson M, Reddi K, Henderson B. Cytokine-inducing components of periodontopathogenic bacteria. J Periodontal Res. 1996;31:393–407. doi: 10.1111/j.1600-0765.1996.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 9.Okada H, Murakami S. Cytokine expression in periodontal health and disease. Crit Rev Oral Biol Med. 1998;9:248–66. doi: 10.1177/10454411980090030101. [DOI] [PubMed] [Google Scholar]
- 10.Karimbux NY, Saraiya VM, Elangovan S, Allareddy V, Kinnunen T, Kornman KS, et al. Interleukin-1 gene polymorphisms and chronic periodontitis in adult caucasians: A systematic review and meta-analysis. J Periodontol. 2012;83:1407–19. doi: 10.1902/jop.2012.110655. [DOI] [PubMed] [Google Scholar]
- 11.Laine ML, Loos BG, Crielaard W. Gene polymorphisms in chronic periodontitis. Int J Dent 2010. 2010 doi: 10.1155/2010/324719. 324719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scapoli C, Borzani I, Guarnelli ME, Mamolini E, Annunziata M, Guida L, et al. IL-1 gene cluster is not linked to aggressive periodontitis. J Dent Res. 2010;89:457–61. doi: 10.1177/0022034510363232. [DOI] [PubMed] [Google Scholar]
- 14.Claudino M, Trombone AP, Cardoso CR, Ferreira SB, Jr, Martins W, Jr, Assis GF, et al. The broad effects of the functional IL-10 promoter-592 polymorphism: Modulation of IL-10, TIMP-3, and OPG expression and their association with periodontal disease outcome. J Leukoc Biol. 2008;84:1565–73. doi: 10.1189/jlb.0308184. [DOI] [PubMed] [Google Scholar]
- 15.Reichert S, Machulla HK, Klapproth J, Zimmermann U, Reichert Y, Glaser CH, et al. The interleukin-10 promoter haplotype ATA is a putative risk factor for aggressive periodontitis. J Periodontal Res. 2008;43:40–7. doi: 10.1111/j.1600-0765.2007.00992.x. [DOI] [PubMed] [Google Scholar]
- 16.Amano Y, Cho Y, Matsunawa M, Komiyama K, Makishima M. Increased nuclear expression and transactivation of vitamin D receptor by the cardiotonic steroid bufalin in human myeloid leukemia cells. J Steroid Biochem Mol Biol. 2009;114:144–51. doi: 10.1016/j.jsbmb.2009.01.022. [DOI] [PubMed] [Google Scholar]


