Abstract
We have identified a cellular factor that interacts with the virus genome-linked proteins (VPgs) of a diverse range of potyviruses. The factor, called Potyvirus VPg-interacting protein (PVIP), is a plant-specific protein with homologues in all the species examined, i.e., pea, Arabidopsis thaliana, and Nicotiana benthamiana. The sequence of PVIP does not identify a specific function, although the existence of a “PHD finger” domain may implicate the protein in transcriptional control through chromatin remodeling. Deletion analysis using the yeast two-hybrid system showed that the determinants of the interaction lay close to the N terminus of VPg; indeed, the N-terminal 16 amino acids were shown to be both necessary and sufficient for the interaction with at least one PVIP protein. From a sequence comparison of different potyvirus VPg proteins, a specific amino acid at position 12 was directly implicated in the interaction. This part of VPg is distinct from regions associated with other functional roles of VPg. Through mutation of Turnip mosaic virus (TuMV) at VPg position 12, we showed that the interaction with PVIP affected systemic symptoms in infected plants. This resulted from reduced cell-to-cell and systemic movement more than reduced virus replication, as visualized by comparing green fluorescent protein-tagged wild-type and mutant viruses. Furthermore, by using RNA interference of PVIP in Arabidopsis, we showed that reduced expression of PVIP genes reduced susceptibility to TuMV infection. We conclude that PVIP functions as an ancillary factor to support potyvirus movement in plants.
The Potyvirus group is the major genus of the family Potyviridae. Potyviruses infect a broad range of monocot and dicot plants and can be responsible for severe damage to crops. Despite the economic importance of potyviruses, relatively little is understood about the detailed molecular interactions that occur in infected tissues that lead to the exploitation of cellular machinery and the accumulation of high virus titers. Notably, only the translation factors eIF4E (28, 30) and eIF(iso)4E (14, 15) have been identified as susceptibility factors supporting virus replication in potyvirus hosts. These proteins have been identified variously through mutational analysis of model host plants, such as Arabidopsis thaliana (hereafter referred to as Arabidopsis) (14), and through protein-protein interaction studies, initially in Saccharomyces cerevisiae (15, 30). We have also applied the latter approach to identify host proteins that interact with the potyvirus genome-linked protein, VPg.
VPg forms a covalent linkage to the 5′ end of the viral RNA and is one of the diagnostic features of the picornavirus supergroup, which includes the family Potyviridae (13). For potyviruses, it is one of 10 mature proteins that are proteolytic products of the primary translated protein product encoded by the viral RNA (6, 27). VPg is a multifunctional protein with several suggested roles in the viral infection cycle. It may act as a primer for RNA replicase during virus multiplication, possibly through direct interaction with the viral RNA polymerase (7, 11, 16). It has also been implicated indirectly in cell-to-cell movement of the virus through plasmodesmata and, more directly, through mutagenesis in the long-distance translocation of the virus (18, 22, 23, 31, 33). Additionally, the potyvirus VPg protein has been shown to be the avirulence factor for recessive resistance genes in various plants (12, 18).
Previous reports have identified eIF(iso)4E (37) and eIF4E (30) as host proteins that interact with VPg from Turnip mosaic virus (TuMV) and Tobacco etch virus (TEV), respectively. The location of VPg at the 5′ end of the viral RNA, in place of the mRNA cap structure, points to a role for VPg in the initiation of translation of the viral RNA. Competition for these translation factors has been proposed as a mechanism for reducing host gene expression during potyvirus infection (2). In addition to its covalent association with viral RNA, VPg or its immediate precursor protein, NIa, shows intracellular targeting to the nucleus (3, 25, 32). Whether or how this is related to the functional roles of VPg is unclear.
In this paper, we describe the identification, using the yeast two-hybrid system (YTHS), of a cellular factor that interacts with the VPg proteins of a diverse range of potyviruses. Initially identified from pea, the protein named PVIP (for Potyvirus VPg-interacting protein) has homologs in Nicotiana benthamiana and in Arabidopsis and interacts with the VPg proteins from potyviruses that infect one or more of these hosts. We have assessed the structural limitations of the interaction and used the information to obtain evidence to support a role for PVIP in vivo. Finally, using RNA interference (RNAi)-based silencing of PVIP in transgenic Arabidopsis, we confirmed that the protein acts as an ancillary factor to support potyvirus infection and movement.
MATERIALS AND METHODS
Virus and plant material.
The source clones for the VPg proteins of Pea seed borne mosaic virus (PSbMV) (isolate DPD1), TEV (HAT strain), Grapevine fan leaf virus (GFLV) (F13 strain), Tomato black ring virus (TBRV) (L strain), and Cowpea mosaic virus (CPMV) were gifts from E. Johansen, J. Carrington, C. Ritzenthaler, C. Fritsch, and G. Lomonossoff, respectively. Infectious clones, p35Tunos and pCB-TuMV-GFP, of TuMV UK1 (29) were gifts from F. Ponz. The latter plasmids give rise to infectious viral RNA in planta after expression from the upstream transcriptional promoters. The p35Tunos expression cassette was inserted into the binary vector pGreenII (10) as follows. The SacI site was deleted from pGreenII by digestion with SacI, polishing of the ends with Klenow fragment, and religation. After excision using flanking SmaI and ApaI sites, the complete expression cassette was inserted into similarly digested modified pGreenII to obtain pGreen-TuMVwt. This was transformed into Agrobacterium tumefaciens strain GV3101. The plasmid pCB-TuMV-GFP is a binary plasmid and was transformed directly into A. tumefaciens strain GV3101. Virus infections on N. benthamiana and Arabidopsis were established by stabbing the leaf tissue to introduce the bacteria. Alternatively, Agrobacterium was pressure infiltrated into the leaf lamina using a syringe, following established protocols (36), except when discrete foci of infection were required, in which case bacterial cultures (optical density at 600 nm, 1) were diluted 1 in 5,000 before infiltration. Arabidopsis ecotype Columbia (Col-0) plants were grown at 20 ± 3°C with a 10-h photoperiod. After inoculation, Arabidopsis plants and plants of N. benthamiana were grown at 25 ± 3°C with a 16-h photoperiod. Green fluorescent protein (GFP) expression was monitored with a handheld UV light (UV Products, Upland, Calif.) or under a Nikon SMZ1500 dissecting microscope coupled to a 100-W epifluorescence module (Nikon). A band pass filter allowed the removal of chlorophyll fluorescence.
Yeast two-hybrid analysis. (i) Yeast two-hybrid library.
A custom library from Pisum sativum L. cv. ‘Scout’ was made in the vector HybriZap 2.1 (Stratagene, La Jolla, Calif.). Excision of the library resulted in a yeast two-hybrid library in pAD-GAL4-2.1. This library was screened, using PSbMV P1 VPg as the bait, in pBD-GAL4 Cam in the yeast strain YRG-2 following Stratagene protocols. The strengths of the protein-protein interactions were measured using a chemiluminescent detection assay (Galacto-light kit; Applied Biosystems).
(ii) Yeast two-hybrid mating assay.
A yeast two-hybrid screen was performed using either the Matchmaker GAL4 or Matchmaker LexA two-hybrid system (Clontech, Palo Alto, Calif.) for detecting protein-protein interactions in yeast (4, 8). Bait constructs were cloned into pGBT9 and transformed into the yeast strain CG1945 or cloned into pLexA and transformed into the yeast strain EGY48[p8opLacZ], resulting in expression as a DNA-binding domain fusion protein. Prey constructs were cloned into pGAD424 and transformed into the yeast strain Y187 or cloned into pB42AD and transformed into the yeast strain YM4271, resulting in expression as an activation domain fusion protein. Protein-protein interactions were identified by yeast mating experiments. A positive interaction was indicated by the ability of cotransformed yeast to grow on synthetic medium lacking leucine, tryptophan, and histidine and containing 5 mM 3-aminotriazole in the case of the GAL4 YTHS or lacking leucine, tryptophan, histidine, and uracil and containing 5 mM 3-aminotriazole and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) in the case of the LexA YTHS. A positive control was provided by the interaction between a murine p53 protein and simian virus 40 large T antigen. A human lamin cDNA-binding domain fusion provided a negative control for the extraneous interaction of the binding domain with the prey. Methods were carried out as described in the Clontech Matchmaker and Matchmaker LexA manuals.
(iii) Protein extraction and Western blot analysis.
Yeasts containing pGBT-VPg constructs or pGAD424-VIP1 constructs were grown overnight in synthetic medium lacking tryptophan or synthetic medium lacking leucine, respectively. Yeast culture and urea-sodium dodecyl sulfate protein extraction were performed as described in the Clontech Yeast Protocol Handbook. Equalized loads of protein extracts (optical density at 600 nm, ∼0.8) were electrophoresed on a sodium dodecyl sulfate-12.5% polyacrylamide gel and blotted to Hybond-C (Amersham Pharmacia Biotech). The immunoblot was incubated with antibodies raised in rabbits against Potato virus A (PVA) VPg (21) at a dilution of 1/5,000 or antibodies raised in rabbits against a PVIP-specific synthetic peptide, SDQEPRRESPAESASS (Eurogentec), at a dilution of 1/500 and washed, and the specific reactivity was visualized using alkaline phosphatase-conjugated goat anti-rabbit serum and nitroblue tetrazolium as a colorimetric substrate, following standard techniques.
Deletion, site-specific mutagenesis, and mutant virus construction.
Site-directed mutagenesis was carried out by overlap extension PCR with specific mutagenic primers (sequences available upon request) and high-fidelity Pfu polymerase (Stratagene). The mutated sequences were cloned into the relevant vectors, and the mutations were confirmed by sequence analysis. The F12M point mutation was inserted into the TuMV cDNA expression plasmids pGreen-TuMVwt and pCB-TuMV-GFP as follows. pGreen-TuMVwt was digested with SacI, and the SacI-SacI fragment covering the VPg cistron was replaced with the corresponding mutant fragment obtained by overlap extension PCR to give pGreen-TuF12M. For pCB-TuMV-GFP, an NcoI-MluI fragment covering cistrons P3 to NIb was inserted into similarly digested pGemT to give pGemT-TuGFP. In turn, this was digested with ClaI to replace the ClaI-ClaI fragment covering the VPg region with the corresponding mutant fragment to give pGemT-TuF12MGFP. An NcoI-MluI fragment from pGemT-TuF12MGFP containing the F12M VPg was mobilized into similarly digested pCB-TuMVwt-GFP to give pCB-TuMVF12M-GFP.
RNAi transgene constructs and Arabidopsis transformation.
To generate PVIP1 and PVIP2 RNAi transgenic lines (pvip1 and pvip2), gene-specific 0.7-kb cDNA fragments were amplified using high-fidelity Pfu polymerase and the following primer pairs: PVIP1 forward primer (5′-TATACCCGGGGGCGCGCCACCTGCTGATGATTGCTACTG-3′) (XmaI and AscI sites underlined) and reverse primer (5′-TATATCTAGAATTTAAATCTGAAACTTCCTTGGCTTTGTC-3′) (XbaI and SwaI sites underlined) and PVIP2 forward primer (5′-TATATCTAGAGGCGCGCCGCTTGCCAAAACGAACTCCC-3′) (XbaI and AscI sites underlined) and reverse primer (5′-TATAGGATCCATTTAAATACCTCTCTCTCGCATTCCTC-3′) (BamHI and SwaI sites underlined). PVIP cDNA fragments were first cloned in the sense orientation between the AscI and SwaI sites of pFGC5941 (http://www.chromdb.org/fgc5941.html), after which the antisense PVIP cDNA fragment was inserted between XmaI and XbaI sites for PVIP1 and between XbaI and BamHI for PVIP2. A. tumefaciens strain GV3101 carrying these constructs was used to transform Arabidopsis (ecotype Col-0) plants by floral dipping (5). Seeds from these plants were sown, and transgenic plants were screened for resistance to the herbicide BASTA. Semiquantitative duplex reverse transcription (RT)-PCR was used to determine the efficiency of RNAi on PVIP1 and PVIP2 mRNAs. cDNA was synthesized using Expand reverse transcriptase (Roche Biodiagnostics GmbH, Mannheim, Germany) and used in a duplex PCR containing oligonucleotides specific for ubiquitin and PVIP1 or PVIP2 mRNA. Semiquantitative PCR of cDNA derived from the equivalent of 1 μg of total RNA was performed under the following conditions: 94°C for 2 min for 1 cycle; 94°C for 30 s, 52°C for 40 s, and 72°C for 1 min for 21, 26, and 31 cycles; and 72°C for 10 min for 1 cycle for each sample. The linear phase of DNA amplification (26 cycles) was determined by electrophoresing and ethidium bromide staining of the PCR products separated in a 1.5% agarose gel. Oligonucleotides to PVIP1 and PVIP2 were designed to detect PVIP1 or PVIP2 mRNA and not the pvip-1 or pvip-2 RNA sequences produced by the RNAi transgene.
RNA extraction and dot hybridization.
Total RNA was extracted from frozen plants using TriReagent (Sigma) according to the manufacturer's instructions. For infected plants, leaf tissue was pooled from at least three independent plants. For quantifying virus replication in infiltrated N. benthamiana leaves, at each time point, RNA was extracted from four replicate leaves from different plants and duplicate aliquots of 2.5 μg of total RNA were spotted to Hybond N (Amersham Pharmacia Biotech) before hybridization with virus-specific probes. Radiolabeled probes for detection of TuMV RNA were made in the presence of [α-32P]dCTP using either random hexanucleotides as primers for cDNA synthesis in the Prime-a-gene labeling system (Promega) to measure total viral RNA or TuMV strand-specific primers (sequences corresponding to nucleotides [nt] 5681 to 5700, 5903 to 5921, and 6469 to 6502) to synthesize a minus-strand viral RNA-specific probe using the Klenow fragment of DNA polymerase I. Dot blot quantification was carried out using AIDA software associated with the Fuji FLA500 phosphorimager, using a probe for actin RNA as an invariant standard for normalizing the data.
Nucleotide sequence accession numbers.
The nucleotide sequences of PVIP from P. sativum (pea; PVIPp) and its homolog from N. benthamiana (PVIPnb) have been deposited in GenBank under accession no. AY271743 (PVIPp) and AY271742 (PVIPnb).
RESULTS
A novel host factor interacts with potyvirus VPg.
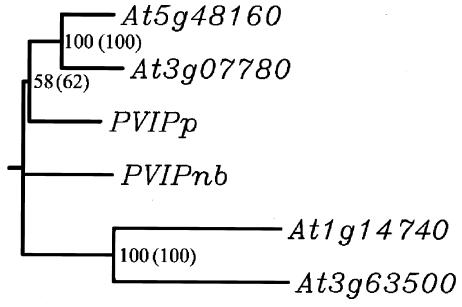
Viruses infect and multiply in their hosts through the deployment of a relatively small number of multifunctional gene products. To explore the diverse roles of potyvirus VPg (26), we screened plant cDNA libraries in yeast using VPg as the bait. Since our initial interest lay in the characterization of resistance genes from pea, we first used the VPg protein from PSbMV and a cDNA library constructed from pea leaf mRNA. PSbMV VPg was fused to the GAL4 DNA-binding domain, whereas the pea cDNA library was fused to the GAL4 activation domain. From ∼7 × 106 independent yeast transformants, 10 classes of potential interacting proteins were identified. Among them, one protein interacted more than twice as strongly as the positive control. This protein was studied further. The protein was named PVIPp (for Potyvirus VPg-interacting protein from pea). Based on nucleotide sequence and BLAST searches, PVIPp appeared to be related to the products of a small gene family in Arabidopsis (the genes At5g48160, At3g07780, At1g14740, and At3g63500). None of these proteins showed significant homology with proteins of known function. PVIPp showed the highest homology with At5g48160 and At3g07780 (Fig. 1).
FIG. 1.
Cluster analysis of the PVIP-related genes from pea, N. benthamiana, and Arabidopsis. Arabidopsis proteins translated from At5g48160 (PVIP1; 573 aa), At3g07780 (PVIP2; 566 aa), At1g14740 (protein 760 aa), At3g63500 (protein 1162 aa), and PVIPp (protein 503 aa) from P. sativum and PVIPnb (protein 549 aa) from N. benthamiana were aligned and clustered using the Clustal X program. Bootstrap values within and outside brackets correspond to calculations without and with inserted gaps, respectively, to optimize the alignment. The absence of a value indicates that the branch link is not supported by the analysis.
To determine whether the interaction with PVIP was specific for pea and PSbMV VPg, other plant virus VPgs and other plant PVIPs were tested using the YTHS. The VPg coding sequences for the potyviruses Lettuce mosaic virus (LMV) (strain 0) (24), TuMV, and TEV or related members of the family Comoviridae, CPMV, TBRV, and GFLV, were cloned into pGBT9. They were assayed for interaction with the products of At5g48160, At3g07780, At1g14740, and At3g63500; PVIPp; and a homologue of PVIPp isolated from N. benthamiana (PVIPnb), all cloned into pGAD424. The results (Table 1) showed that VPg proteins of PSbMV, LMV, and TuMV, but not TEV, interacted with products of At5g48160 and At3g07780 (named PVIP1 and PVIP2) and of PVIPp and PVIPnb. In contrast, interactions with PVIP were not observed for the VPg protein from CPMV, GFLV, or TBRV or for potyvirus VPg with products of At1g14740 and At3g63500. We also found that while the interaction between potyvirus VPg and PVIP was readily detected when transferred to the LexA YTHS, reversing the fusion partners so that VPg and PVIP were fused to the activation and DNA-binding domains, respectively, abolished the interaction (data not shown).
TABLE 1.
Analysis of the breadth of the interaction between VPgs and PVIP in yeast
| Bait | Growthd of yeast transformed with pGAD424 containing preyc:
|
||||||
|---|---|---|---|---|---|---|---|
| ADb | At1 | At2 | At3 | At4 | PVIPp | PVIPnb | |
| BDa | − | − | − | − | − | − | − |
| PSbMV | − | + | + | − | − | + | + |
| TuMV | − | + | + | − | − | + | + |
| LMV | − | + | + | − | − | + | + |
| TEV | − | − | − | − | − | − | − |
| CPMV | − | − | − | − | − | − | − |
| TBRV | − | − | − | − | − | − | − |
| GFLV | − | − | − | − | − | − | − |
Empty bait vector (confirming that PVIP was unable to autoactivate transcription).
Empty prey vector (confirming that VPg was unable to interact with the activation domain).
At1, At2, At3, At4, At5g48160, At3g07780, At1g14740, and At3g63500, respectively.
Growth of the yeast on selective media. Extent of the growth was equivalent for all positive interactions. −, no growth.
PVIP-VPg interaction in yeast depends on sequences within the N terminus of VPg.
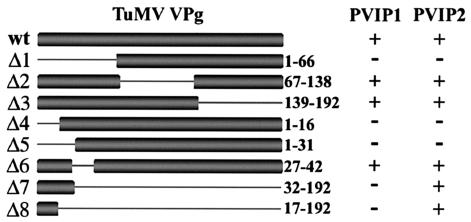
Deletion analysis was used to determine the sequence limitations of the interaction between VPg and PVIP. Two series of deletions for TuMV VPg were constructed and tested as fusions to the GAL4 DNA-binding domain in the YTHS, using pGAD-PVIP1 and -PVIP2 as the interaction partners. The first series contained deletions collectively spanning the entire VPg sequence (Fig. 2). The N-terminal 66 amino acids (aa), the central region (aa 67 to 138), or the C-terminal 54 aa, respectively, were deleted from the proteins encoded by pGBT-TuΔ1, pGBT-TuΔ2, and pGBT-TuΔ3. The second deletion series (Fig. 2), pGBT-TuΔ4, pGBT-TuΔ5, pGBT-TuΔ6, pGBT-TuΔ7, and pGBT-TuΔ8, removed aa 1 to 16, 1 to 31, 27 to 42, 32 to 192, and 17 to 192, respectively. Growth of the yeast transformants on selective media showed that the interaction with PVIP1 and -2 was absolutely dependent upon the first 66 aa of VPg (Fig. 2, compare TuΔ1 with TuΔ2 or TuΔ3). From the second deletion series, this region could be subdivided to identify aa 27 to 42 (TuΔ6) as not being required for the interaction while the N-terminal 16 aa (TuΔ4 and TuΔ5) were essential for the interaction with either PVIP1 or -2. Interestingly, the deletions TuΔ7 and TuΔ8, encoding only the first 31 and 16 aa, respectively, exhibited interaction with PVIP2 but not PVIP1. This showed that in the case of PVIP2, the first 16 aa of TuMV VPg were not only necessary but also sufficient for the interaction. A similar positive interaction was seen with PVIPp (data not shown). Comparison of the interaction assay for PVIP1 with those for TuΔ1 (aa 1 to 66) and TuΔ6 (aa 27 to 42) suggests that an additional component between aa 42 and 66 is required for the VPg-PVIP1 interaction to succeed.
FIG. 2.
Deletion analysis of the interaction of TuMV VPg with PVIP1 and PVIP2. TuMV wt VPg and two series of deletion mutants for TuMV VPg (Δ1 to Δ3 and Δ4 to Δ8) were tested for their interactions with PVIP1 and PVIP2 by yeast two-hybrid analysis. The positive (+) interactions were all equivalent, and the data were scored qualitatively.
Similarly, we tried to identify regions within PVIP that were necessary for the interaction with VPg. Using PVIP1, a series of four deletion mutants, together covering the entire PVIP1 sequence, were constructed in the pGAD-PVIP1 vector. Mutants pGAD-PVIP1Δ1, -PVIP1Δ2, -PVIP1Δ3, and -PVIP1Δ4 had aa 3 to 157, 157 to 283, 283 to 403, and 403 to 574 deleted, respectively. No growth was detected with these yeast strains in the presence of pGBT-TuMV VPg. Hence, we were unable to identify a functionally discrete PVIP domain for the interaction between VPg and PVIP1.
It is possible that the lack of detectable interaction reflected the stability and accumulation of the fusion proteins rather than a structural defect in either partner. Using antibodies to PVIP and VPg, we attempted to detect the respective fusion proteins in yeast expressing the Gal4 fusion proteins. These proteins accumulate at very low levels (Clontech instruction manual) and may not always be detectable. Nevertheless, both PVIP and VPg fusions were detected for most of the expressed constructs, but there was no correlation between detection of the fusion protein and a positive interaction (data not shown).
A single amino acid differentiates the abilities of two VPgs to interact with PVIP2.
Sequence comparisons of TuMV and three other potyviruses (PSbMV, TEV, and LMV) were made for the two domains (VPg aa 1 to 16 and 42 to 66) that controlled the interactions with the PVIP1 and PVIP2 proteins (Fig. 3A). The comparison shows that both regions are highly conserved among these potyviruses. To identify a residue(s) critical for the interaction with PVIP, it was useful to compare the sequence of TEV VPg, which showed no interaction, with the other sequences. Within the N-terminal 16 aa, which was sufficient for the interaction of TuMV VPg with PVIP2, 5 residues are identical and 2 are closely related for all four viruses (Fig. 3A). Among the nine remaining residues, the only consistent differences between the VPg proteins of PSbMV, TuMV, and LMV and that of TEV were a deleted glycine at aa position 3 in TEV; position 12, where an aromatic amino acid (F/Y) in TuMV, PSbMV, and LMV was replaced with methionine in TEV; and position 14, where TEV contained an acidic (E) rather than an uncharged (Q/N) residue. Site-directed mutagenesis was used to individually insert each of these changes into the sequence of TuMV VPg, and the mutants (G3-, F12 M, and N14E, respectively) were assessed for their interaction with PVIP1, PVIP2, PVIPnb, and PVIPp in yeast. A reciprocal mutant, TEV VPg M12F, was also constructed and tested. Mutations at positions 3 and 14 were benign with respect to the interaction with all PVIP proteins (Fig. 3B). In contrast, the F12M mutation in TuMV VPg abolished interactions with all PVIPs, whereas the reciprocal M12F mutation in TEV VPg conferred the interaction with PVIP2 and PVIPp. These results showed that F12 plays a crucial role in the physical interaction between VPg and PVIPs and that for TEV, M12 is the only lesion that prevents interaction with PVIP2 and PVIPp.
FIG. 3.
Analysis of amino acid determinants of VPg-PVIP interactions. (A) VPg sequences from TuMV, LMV, PSbMV, and TEV were aligned using the Clustal W program. Only regions 1 to 16 and 42 to 66 are shown. Identical (*) and related (:) amino acids for the four sequences are indicated. Amino acid positions where TEV differed from the other three with an unrelated residue are also identified (▴). (B) Mutational analysis of TuMV and TEV VPg proteins targeting aa 3, 12, or 14. The TuMV point mutants Tu G3-, F12M, and N14E and the TEV point mutant Te M12F (shaded) were tested for interaction with PVIP1 (column 1), PVIP2 (column 2), PVIPp (column p), and PVIPnb (column Nb) using yeast two-hybrid analysis. Interactions in yeast were scored qualitatively; all the positive (+) interactions were seen as equivalent yeast growth on selective media.
Mutations in VPg that block the interaction in yeast correlate with reduced virus movement in planta.
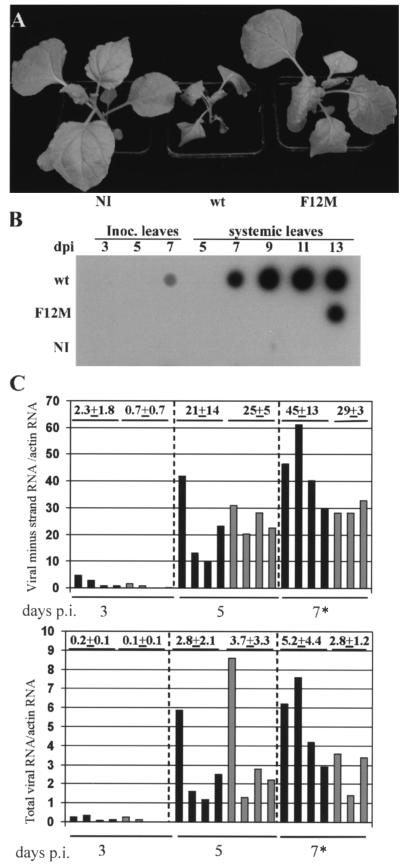
Interaction of two proteins in the YTHS is not proof of a biological role in vivo. We have also obtained evidence that the interaction is important for virus infection. Based upon the results obtained from the YTHS experiments, we tested the VPg F12M mutation in the complete TuMV virus genome for its effect upon virus multiplication. After stab inoculation to introduce Agrobacterium and the potential infection into N. benthamiana plants, the plants were observed for 15 days. At 7 days postinoculation (p.i.), all eight of the plants infected with the wild-type (wt) TuMV showed systemic chlorosis, some necrotic flecking, leaf distortion, and stunting (Fig. 4A). By 15 days p.i., all the plants showed extensive necrosis or had died. In contrast, all the plants inoculated with pGreen-TuF12M remained symptomless for 12 days p.i., developed weak chlorosis after 15 days p.i., and showed little growth reduction (Fig. 4A). The accumulation of viral RNA in these plants was assessed using dot hybridization of RNA samples extracted from leaves harvested over 13 days (Fig. 4B). Viral RNA was detected readily in inoculated and systemically infected leaves of plants inoculated with pGreen-TuMV as early as 7 days p.i. (Fig. 4B). In contrast, in plants inoculated with pGreen-TuF12M, viral RNA could not be detected in inoculated leaves and was first detected in systemically infected leaves of plants only at 13 days p.i. Since viruses show high efficiency in the selection of sequence variants, we checked that the virus appearing in pGreen-TuF12M-inoculated plants had retained the inserted mutation. Progeny viral RNA was analyzed by direct sequencing of RT-PCR products that spanned the VPg coding region between nt 5718 and 6538 of the TuMV genome. In all eight plants, the mutation had been retained, and no other mutations were present in the VPg. These results showed that the VPg F12M mutation directly or indirectly restricted TuMV multiplication and/or spread.
FIG. 4.
Phenotypic assessment of the VPg F12M mutation for TuMV multiplication in N. benthamiana. (A) Noninoculated (NI) N. benthamiana plants and plants stab inoculated with TuMV wt and TuMV F12M were compared. At 12 days p.i., plants infected with TuMV F12M showed significantly fewer symptoms than those infected with the wt TuMV. (B) Hybridization analysis of total viral RNA accumulation in plants illustrated in panel A. Stab-inoculated (Inoc.) and systemically infected (systemic) leaves were harvested 3, 5, and 7 and 5, 7, 9, 11, and 13 days p.i., respectively. Viral RNA accumulation was substantially less for the TuMV F12M mutant than for wt TuMV. (C) Hybridization analysis of minus-strand (top) and total (bottom) viral RNAs in patches of N. benthamiana leaves infiltrated to give infections with wt TuMV (solid bars) or TuMV F12M (shaded bars). Individual data points are expressed as arbitrary units relative to hybridization with a probe for actin RNA as an invariant standard. The mean data points for all the leaf samples are presented. The overall mean value ± standard deviation is given above each set. Because of the different specific activities of the probes for different viral RNAs, the upper and lower panels are not quantitatively comparable. *, only three samples were available for mutant infections at 7 days p.i.
To assess quantitatively the impact of the F12M mutation on virus replication, infections with pGreen-TuMV and pGreen-TuF12M were established after infiltrating the cultures of Agrobacterium into patches on the leaf lamina of N. benthamiana. This strategy provides high-efficiency transient expression of the transferred agrobacterium T-DNA in a large proportion of the leaf cells (36). Leaf samples were harvested 3, 5, and 7 days p.i., and the extracted RNA was analyzed for total or minus-strand viral RNA using dot hybridization (Fig. 4C). Minus-strand viral RNA is an essential and unique component of the replication mechanism for single-stranded viral RNAs. Total viral RNA measurements could feasibly include progeny viral RNA and RNA transcribed directly from the input cDNA expression cassette. For the wt and mutant virus infections, total and minus-strand viral RNAs were detected 3 days p.i. and increased substantially by 5 days p.i., but no further at 7 days p.i. Although there was variation among the individual samples at different times, there was no significant difference between wt and mutant virus infection for any of the times analyzed.
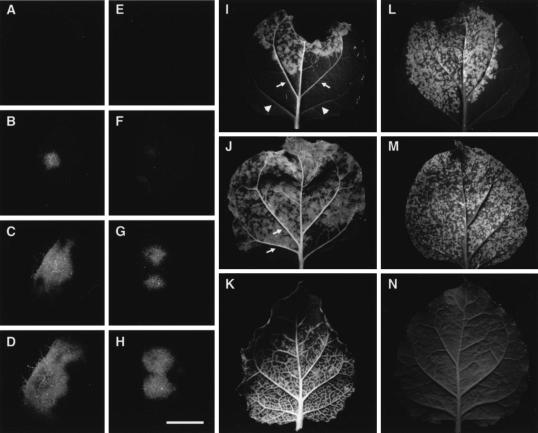
To visualize directly the impact of the F12M mutation on virus movement, the wt and mutant virus constructs were tagged with GFP. Leaf laminal tissues of N. benthamiana were infiltrated with suspensions of Agrobacterium at high dilution so that isolated foci of infection developed. The spread of the foci was monitored 2, 3, 4, 5, and 6 days p.i. as GFP fluorescence (Fig. 5). No fluorescence was visible 2 days p.i. Over days 3 to 5, GFP lesions were visible for both the wt and mutant virus, but the size and rate of expansion of the mutants were significantly less (Fig. 5A to H illustrate typical infection foci). At 6 days p.i., the wt infection had spread through virtually all of the infiltrated leaf area and showed significant fluorescence in the veinal tissues, including the petiole (Fig. 5I and J). At the same time, foci of infection for the F12M mutant were visible as abundant small patches of fluorescence, subjectively as bright as for the wt infection. Some of these patches overlapped the leaf veins, but there was no evidence of spread along the veins (Fig. 5L and M). Systemically infected tissue at 6 days p.i. showed extensive tissue invasion of the veins and lamina for the wt virus but no detectable spread of the mutant virus (compare Fig. 5K and N).
FIG. 5.
Differential local and systemic spread associated with the F12M mutation in TuMV VPg. Virus inocula pCB-TuMV-GFP (A to D and I to K) and pCB-TuMVF12M-GFP (E to H and L to N) were introduced into infiltrated leaf patches with Agrobacterium at a dilution appropriate to give isolated infection foci. The leaves were photographed at 2 (A and E), 3 (B and F), 4 (C and G), 5 (D and H), and 6 (I, J, L, and M) days p.i. Additionally, systemic leaves from infected plants were examined 6 days p.i. (K and N). Passage of the virus in vascular tissues (arrows) is seen for the wt virus only (compare panels I and J with L and M) as fluorescence along the veinal tissues within and beyond the infiltrated area (I and J); compare these with veins from the same leaf (I) without fluorescence (arrowheads). Photographs taken under UV light and printed as greyscale show bright (green) fluorescence against dark tissue background. Bar for panels A to H = 3 mm.
Reduced PVIP expression in planta reduces symptom production and virus accumulation.
The results presented above strongly suggest that VPg-PVIP interaction is important for potyvirus multiplication. However, as potyviral VPgs are multifunctional proteins, it remained possible that the F12M mutation could also have pleiotropic effects on one or more VPg functions. To assess directly the importance of PVIP for potyvirus infection, we generated Arabidopsis PVIP1 and PVIP2 mutant lines (designated pvip1 and pvip2, respectively) using RNAi (35) and challenged them by virus inoculation. Arabidopsis plants were transformed with an inverted-repeat construct specific for either PVIP1 or PVIP2 RNA and capable of forming a hairpin RNA after expression from the Cauliflower mosaic virus 35S promoter. Activation of RNAi by the double-stranded RNA leads to the targeted degradation of homologous plant RNAs. Since RNAi is functionally dominant, we expected all lines containing the transgene to show a depletion of the target transcript.
The expression of PVIP1 or PVIP2 transcripts was examined in 20 randomly chosen independent pvip1 or pvip2 RNAi lines by duplex RT-PCR, using ubiquitin RNA as an invariant control. PVIP1 or PVIP2 RNA was depleted to various extents in the RNAi lines tested, whereas our internal control, the ubiquitin transcript, showed no variation. We chose for further analysis two independent pvip1 and pvip2 lines that were most efficiently silenced for the corresponding PVIP, i.e., lines in which there was the least transcript accumulation (Fig. 6A). These lines showed no major morphological differences from wt plants except that they exhibited slightly slower growth (data not shown). Three plants of each of these RNAi lines were infected with pGreen-TuMV by stab inoculation. As for infection of N. benthamiana, TuMV-infected, untransformed Arabidopsis showed leaf necrosis and stunting. In contrast, no symptoms were visible on either the pvip1 or pvip2 RNAi lines (Fig. 6B). The level of progeny viral RNA in these plants was assessed by dot hybridization. Viral RNA was readily detected in systemic leaves of wt Arabidopsis at 23 days p.i. In contrast, at the same time, viral RNA accumulation in pvip1 and pvip2 RNAi lines was not detectable (Fig. 6C) unless the autoradiograph was exposed for at least 20 times as long, when a faint hybridization signal above background could be detected (data not shown). In inoculated leaves, we were able to detect progeny viral RNA in infected pvip1 and pvip2 plants 5 days p.i., although at a significantly lower level than in wt plants (Fig. 6C). Since all of the leaves received equal bacterial inocula, it is likely that most of the RNA detected for the wt inoculum resulted from virus replication rather than transcription of the input cDNA expression cassette.
FIG. 6.
TuMV infection phenotypes on plants with reduced PVIP1 or PVIP2 expression. Arabidopsis Col-0 plants were transformed with RNAi constructs to target PVIP1 or PVIP2 expression. (A) Duplicate samples from pvip1 and pvip2 transgenic plants and wt plants were assessed for the expression of PVIP1 and PVIP2 using semiquantitative RT-PCR. Products from duplex RT-PCRs with primers for PVIP RNAs and ubiquitin RNA (Ubi; as a control) were analyzed by agarose gel electrophoresis. Plant lines selected for inoculation (illustrated in lanes 1 and 2 and lanes 5 and 6) showed no detectable transcript accumulation compared with their wt parent plants (lanes 3 and 4 and lanes 7 and 8). Lane M shows DNA size markers. (B) wt uninfected (NI) plants and wt, pvip1, and pvip2 plants stab inoculated with wt TuMV. In contrast to the severe stunting seen with TuMV infection of wt plants, no disease symptoms were seen on the pvip1 and pvip2 plants. (C) Hybridization analysis of viral RNA for the plants illustrated in panel B. Leaves from noninfected (−) and infected (+) plants were analyzed for the presence of TuMV RNA. TuMV RNA was readily detected in inoculated (Inoc; 5 days p.i.) and systemically infected (Syst; 23 days p.i.) leaves of wt Arabidopsis plants. Analysis of triplicate samples from pvip1 and pvip2 plants showed reduced accumulation in inoculated leaves and no detectable accumulation in systemically infected leaves. However, exposure of the autoradiograph for at least 20 times as long showed a low level of accumulation in systemically infected leaves (data not shown).
DISCUSSION
We have identified an interaction in the YTHS between the VPg proteins of a range of potyviruses and a plant protein, PVIP. This interaction has also been confirmed in vitro (our unpublished data). PVIP appears not to be essential for virus replication but functions more as an ancillary factor for potyvirus movement and the development of disease. PVIP is part of a small gene family in Arabidopsis and has counterparts in pea and N. benthamiana. Alignment of all the PVIP-related sequences (Fig. 1) shows that At1g14740 and At3g63500 form a group discrete from PVIPs, data that reflect our YTHS interaction analysis, in which no interaction between VPg and these proteins could be demonstrated. Unfortunately, database searches using the PVIP sequences failed to identify any associated biological function. Furthermore, the phenotype of the uninfected pvip1 and pvip2 plants did not indicate an interruption in any particular stage of growth and development that might indicate a candidate function. Analysis of amino acid sequence motifs did, however, identify a “PHD finger-like” domain (34) present in all PVIPs analyzed. This domain is characterized as a series of histidine and cysteine residues in the order C4HC3 and is frequently associated with proteins that have roles in transcriptional regulation through chromosome remodeling (1). If this was true for PVIP, it would point to a role for VPg in the nucleus. VPg has a nuclear targeting sequence and has been shown to accumulate in the nucleus, at least in the form of its immediate proteolytic precursor molecule, nuclear inclusion a, or NIa, protein (3). Since potyvirus infection can be associated with altered host gene expression (17), the potential for the PVIP-VPg interaction to modulate host transcription following infection is intriguing.
The impact of breaking the PVIP-VPg interaction using a virus mutant or RNAi plants, pvip1 and pvip2, was to reduce disease symptoms significantly and to almost prevent virus accumulation in systemic uninoculated leaves. However, virus multiplication was not abolished, making PVIP an unlikely candidate as a complete resistance factor (e.g., in genetic knockout plants). Following the initial identification of PVIPp, we mapped the chromosomal position of PVIPp relative to that for the resistance genes sbm-1 and sbm-3 in pea. The map positions for all three loci were completely different (unpublished data).
VPg is attached to the 5′ end of the viral RNA through a covalent linkage to the tyrosine residues at position 64. Either as VPg or as part of NIa, it has also been functionally implicated in viral RNA replication and local or systemic virus movement and as an avirulence determinant. During viral RNA replication, VPg may act as a primer, but there is evidence (16) that NIa also interacts with the NIb polymerase. This interaction is dependent upon the C-terminal protease domain of NIa rather than the N-terminal VPg region. The structural determinants of virus movement have been determined for Tobacco vein mottling virus and PVA and have been shown to be associated with aa 110 to 115 and 116 or 118, respectively (18, 22, 23). In the latter case, the specific amino acids appeared to be a determinant of the host range (22, 23). Although our deletion analysis identified a different (N-terminal) region of VPg as the location of determinants of the interaction with PVIP, there was little effect of the TuMV F12M mutation on virus replication in infiltrated leaf patches and a stronger effect on the infection resulting from virus movement. Hence, the local and systemic infection of N. benthamiana with TuMV F12M was substantially delayed. Systemic infection of pvip1 and pvip2 plants by wt TuMV was also much delayed. Although replication in infiltrated leaf patches was unaffected, there was a marked reduction in the accumulation of the mutant virus (or wt TuMV on mutant plants) following stab inoculation. On balance, we conclude that the VPg-PVIP interaction has a predominant effect on virus cell-to-cell movement, with longer-term consequences for systemic movement. Using GFP-tagged wt and mutant virus, this effect was visualized as a slower expansion of isolated infection foci and reduced phloem loading, leading to systemic infection.
PVIP constitutes a small gene family in Arabidopsis. In addition to generating pvip1 and pvip2 RNAi plants, we also made pvip1-pvip2 double mutants (our unpublished data). These plants were unsuitable for virus inoculation due to their extremely small size, a phenotype not seen in either single-mutant line. While this suggests that PVIP1 and PVIP2 have redundant functions, it raises the question of why the single-mutant lines did not support virus multiplication and movement through complementation. We interpret this as indicating that in Arabidopsis, VPg interacts with a PVIP1/2 heterodimer. PVIP1 and PVIP2 self- and cross-interact in the YTHS (our unpublished data), and VPg can interact with itself in yeast (9, 19, 38).
A putative three-dimensional structure for the potyvirus VPg has been published (20). The VPg amino acid sequence showed the greatest structural homology to the folded structure for the enzyme malate dehydrogenase. This placed the N-terminal 16 aa on the surface of the folded VPg (20). In this location, F12 would be available for a surface interaction with PVIPs. From a comparison of VPg sequences for several potyviruses with TEV, we showed that F12 was crucial for interaction with PVIPs. Comparison of the VPg sequences for all four potyviruses for the region aa 42 to 66 revealed that only position 49 showed a consistent difference between PSbMV, TuMV, and LMV and TEV. Here, a basic amino acid (K or R) in TuMV, PSbMV, and LMV is replaced with a polar uncharged amino acid (T) in TEV. We speculate that a basic amino acid at position 49 might also be needed to achieve an interaction with PVIP1, although this has not been tested. This raises an important biological question about TEV VPg, which did not interact with any PVIP proteins. If PVIP makes a significant contribution to virus infection, how and why has TEV avoided a requirement for this interaction? From an alignment of the VPg sequences from 39 potyviruses (data not shown), it appears that the F/Y-M transition at position 12 in TEV is uncommon, and may be unique, among the potyviruses. Whether TEV has found an alternative means to generate the supportive potential of VPg-PVIP for infection or whether the selection pressure to force the M12F change is too weak in this virus remains to be determined.
Acknowledgments
We thank Margaret Boulton and Sue Angell for critical reading of the manuscript. We also thank F. Ponz, E. Johansen, J. Carrington, C. Ritzenthaler, C. Fritsch, and G. Lomonossoff for providing cDNA clones; J. Valkonen for providing PVA VPg antibodies; and Marc for assistance with the PVIP phylogenetic analysis.
During this work, P.D. was supported by an EMBO Fellowship, S.H. was supported as part of the European Union Framework IV project no. B104-CT97-2356, and F.R. was supported by INRA, Paris, France. The John Innes Centre is grant aided by the Biotechnology and Biological Sciences Research Council.
This work was carried out under the United Kingdom Department of the Environment, Food and Rural Affairs license PHL 185/4192.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Aranda, M., and A. Maule. 1998. Virus-induced host gene shutoff in animals and plants. Virology 243:261-267. [DOI] [PubMed] [Google Scholar]
- 3.Carrington, J. C., D. D. Freed, and A. J. Leinicke. 1991. Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell 3:953-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chien, C.-T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clough, S. J., and A. F. Bent. 1998. Technical advance. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735-743. [DOI] [PubMed] [Google Scholar]
- 6.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellers, J., J. Wan, Y. Hong, G. B. Collins, and A. G. Hunt. 1998. In vitro interactions between a potyvirus-encoded, genome-linked protein and RNA-dependent RNA polymerase. J. Gen. Virol. 79:2043-2049. [DOI] [PubMed] [Google Scholar]
- 8.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 9.Guo, D., M. L. Rajamaki, M. Saarma, and J. P. Valkonen. 2001. Towards a protein interaction map of potyviruses: protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J. Gen. Virol. 82:935-939. [DOI] [PubMed] [Google Scholar]
- 10.Hellens, R. P., E. A. Edwards, N. R. Leyland, S. Bean, and P. M. Mullineaux. 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42:819-832. [DOI] [PubMed] [Google Scholar]
- 11.Hong, Y., K. Levay, J. F. Murphy, P. G. Klein, J. G. Shaw, and A. G. Hunt. 1995. A potyvirus polymerase interacts with the viral coat protein and VPg in yeast cells. Virology 214:159-166. [DOI] [PubMed] [Google Scholar]
- 12.Keller, K. E., I. E. Johansen, R. R. Martin, and R. O. Hampton. 1998. Potyvirus genome-linked protein (VPg) determines pea seed-borne mosaic virus pathotype-specific virulence in Pisum sativum. Mol. Plant-Microbe Interact. 11:124-130. [DOI] [PubMed] [Google Scholar]
- 13.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 14.Lellis, A. D., K. D. Kasschau, S. A. Whitham, and J. C. Carrington. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12:1046-1051. [DOI] [PubMed] [Google Scholar]
- 15.Léonard, S., D. Plante, S. Wittmann, N. Daigneault, M. G. Fortin, and J.-F. Laliberté. 2000. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74:7730-7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, X. H., P. Valdez, R. E. Olvera, and J. C. Carrington. 1997. Functions of the tobacco etch virus RNA polymerase (NIb): subcellular transport and protein-protein interaction with VPg/proteinase (NIa). J. Virol. 71:1598-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maule, A. J., V. Leh, and C. Lederer. 2002. The dialogue between viruses and hosts in compatible interactions. Curr. Opin. Plant Biol. 5:279-284. [DOI] [PubMed] [Google Scholar]
- 18.Nicolas, O., S. W. Dunnington, L. F. Gotow, T. P. Pirone, and G. M. Hellmann. 1997. Variations in the VPg protein allow a potyvirus to overcome va gene resistance in tobacco. Virology 237:452-459. [DOI] [PubMed] [Google Scholar]
- 19.Oruetxebarria, I., D. Guo, A. Merits, K. Mäkinen, M. Saarma, and J. P. Valkonen. 2002. Identification of the genome-linked protein in virions of Potato virus A, with comparison to other members in genus Potyvirus. Virus Res. 73:103-112. [DOI] [PubMed] [Google Scholar]
- 20.Plochocka, D., M. Welnicki, P. Zielenkiewicz, and W. Ostoja-Zagorski. 1996. Three-dimensional model of the potyviral genome-linked protein. Proc. Natl. Acad. Sci. USA 93:12150-12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puustinen, P., M.-L. Rajamäki, K. I. Ivanov, J. P. T. Valkonen, and K. Mäkinen. 2002. Detection of the potyviral genome-linked protein VPg in virions and its phosphorylation by host kinases. J. Virol. 76:12703-12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajamäki, M. L., and J. P. Valkonen. 1999. The 6K2 protein and the VPg of potato virus A are determinants of systemic infection in Nicandra physaloides. Mol. Plant-Microbe Interact. 12:1074-1081. [DOI] [PubMed] [Google Scholar]
- 23.Rajamäki, M. L., and J. P. Valkonen. 2002. Viral genome-linked protein (VPg) controls accumulation and phloem-loading of a potyvirus in inoculated potato leaves. Mol. Plant-Microbe Interact. 15:138-149. [DOI] [PubMed] [Google Scholar]
- 24.Redondo, E., R. Krause-Sakate, S.-J. Yang, H. Lot, O. Le Gall, and T. Candresse. 2001. Lettuce mosaic virus pathogenicity determinants in susceptible and tolerant lettuce cultivars map to different regions of the viral genome. Mol. Plant-Microbe Interact. 14:804-810. [DOI] [PubMed] [Google Scholar]
- 25.Restrepo, M. A., D. D. Freed, and J. C. Carrington. 1990. Nuclear transport of plant potyviral proteins. Plant Cell. 2:987-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revers, F., O. Le Gall, T. Candresse, and A. J. Maule. 1999. New advances in understanding the molecular biology of plant/potyvirus interactions. Mol. Plant-Microbe Interact. 12:367-376. [Google Scholar]
- 27.Riechmann, J. L., S. Lain, and J. A. Garcia. 1992. Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 73:1-16. [DOI] [PubMed] [Google Scholar]
- 28.Ruffel, S., M. H. Dussault, A. Palloix, B. Moury, A. Bendahmane, C. Robaglia, and C. Caranta. 2002. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32:1067-1075. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez, F., D. Martinez-Herrera, I. Aguilar, and F. Ponz. 1998. Infectivity of turnip mosaic potyvirus cDNA clones and transcripts on the systemic host Arabidopsis thaliana and local lesion hosts. Virus Res. 55:207-219. [DOI] [PubMed] [Google Scholar]
- 30.Schaad, M. C., R. J. Anderberg, and J. C. Carrington. 2000. Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 273:300-306. [DOI] [PubMed] [Google Scholar]
- 31.Schaad, M. C., and J. C. Carrington. 1996. Suppression of long-distance movement of tobacco etch virus in a nonsusceptible host. J. Virol. 70:2556-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaad, M. C., R. Haldeman-Cahill, S. Cronin, and J. C. Carrington. 1996. Analysis of the VPg-proteinase (NIa) encoded by tobacco etch potyvirus: effects of mutations on subcellular transport, proteolytic processing, and genome amplification. J. Virol. 70:7039-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaad, M. C., A. D. Lellis, and J. C. Carrington. 1997. VPg of tobacco etch potyvirus is a host genotype-specific determinant for long-distance movement. J. Virol. 71:8624-8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindler, U., H. Beckmann, and A. R. Cashmore. 1993. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4:137-150. [DOI] [PubMed] [Google Scholar]
- 35.Smith, N. A., S. P. Singh, M. B. Wang, P. A. Stoutjesdijk, A. G. Green, and P. M. Waterhouse. 2000. Gene expression: total silencing by intron-spliced hairpin RNAs. Nature 407:319-320. [DOI] [PubMed] [Google Scholar]
- 36.Voinnet, O., P. Vain, S. Angell, and D. C. Baulcombe. 1998. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95:177-187. [DOI] [PubMed] [Google Scholar]
- 37.Wittmann, S., H. Chatel, M. G. Fortin, and J. F. Laliberté. 1997. Interaction of the viral protein genome link of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 234:84-92. [DOI] [PubMed] [Google Scholar]
- 38.Yambao, M. L. M., C. Masuta, K. Nakahara, and I. Uyeda. 2003. The central and C-terminal domains of VPg of Clover yellow vein virus are important for VPg-HCPro and VPg-VPg interactions. J. Gen. Virol. 84:2861-2869. [DOI] [PubMed] [Google Scholar]