Abstract
Previous work demonstrated that intracellular enveloped vaccinia virus virions use microtubules to move from the site of membrane wrapping to the cell periphery. The mechanism and direction of intracellular virion movement predicted that viral proteins directly or indirectly interact with the microtubule motor protein kinesin. The yeast two-hybrid assay was used to test for interactions between the light chain of kinesin and the cytoplasmic tails from five viral envelope proteins. We found that the N-terminal tetratricopeptide repeat region of the kinesin light chain (KLC-TPR) interacted with the cytoplasmic tail of the viral A36R protein. A series of C- and N-terminal truncations of A36R further defined a region from residues 81 to 111 that was sufficient for interaction with KLC-TPR. Interactions were confirmed by using pull-down assays with purified glutathione S-transferase (GST)-A36R and 35S-labeled KLC-TPR. The defined region on A36R for interaction with kinesin overlaps the recently defined region (residues 91 to 111) for interaction with the A33R envelope protein. The yeast three-hybrid system was used to demonstrate that expression of A33R interrupted the interaction between A36R and KLC-TPR, indicating that the binding of A36R is mutually exclusive to either A33R or kinesin. Pull-down assays with purified GST-A36R and 35S-labeled KLC-TPR in the presence of competing A33R corroborated these findings. Collectively, these results demonstrated that the viral A36R protein interacts directly with the microtubule motor protein kinesin and that the viral protein A33R may regulate this interaction.
Vaccinia virus, the prototype member of the poxvirus family, replicates entirely in the cytoplasm and produces both intracellular and extracellular forms of infectious virions (24). Intracellular mature virions (IMVs) contain a lipoprotein membrane and are the first infectious form produced (9, 16, 28, 38). A subset of IMVs is wrapped with an extra double membrane derived from the trans-Golgi network (TGN) or endosomal cisternae (34, 42). These wrapped forms are called intracellular enveloped virions (IEVs). IEVs are transported via microtubules from the site of wrapping to the cell periphery, in which the outermost IEV membrane fuses with the plasma membrane, depositing cell-associated enveloped virions (CEVs) on the cell surface (15, 20, 27, 48, 49). Actin polymerization occurs on the cytosolic side of the plasma membrane, directly beneath the CEV. The thick actin structures that result, called actin tails, propel CEVs away from the surface of the cell (3, 8, 17, 39). Eventually, some enveloped virions are released from the cell surface and are called extracellular enveloped virions (EEVs) (1, 26). While IMVs make up the majority of progeny virions, the enveloped forms of the virus, CEVs and EEVs, are critical for cell-to-cell and long-range spread (1, 3, 6, 26).
Seven proteins exclusive to enveloped forms of the virus are encoded by the following open reading frames (ORFs): A33R (29), A34R (11), A36R (46), A56R (36), B5R (12, 21), F12L (45), and F13L (19). In this article, vaccinia virus ORFs are italicized and designated by a capital letter indicating a HindIII restriction endonuclease fragment, a number indicating the position in the HindIII fragment, and a letter (L or R) indicating the direction of transcription (e.g., A36R). Proteins encoded by the ORFs are in plain type (e.g., A36R). Except for A56R, deletion of any one of these ORFs results in a mutant that produces small plaques on cell monolayers (11, 22, 25, 30, 31, 52, 53, 55). Of the seven proteins encoded by the ORFs described above, A36R and F12L are uniquely restricted to the IEVs. F13L (2) and B5R (13, 51) are required for IMV envelopment, because deletion of either protein reduces the numbers of IEVs and decreases EEV production. Although deletion of the A33R, A34R, or A36R ORF results in the loss of actin tails (30, 31, 52, 53), only A36R has been shown to be directly involved in actin tail formation. Phosphorylation of Tyr 112 and 132 of A36R activates the Arp2-Arp3 complex and leads to actin nucleation (14, 23, 33). While the role of A34R in actin tail formation still needs to be determined, recent studies showed that the cytoplasmic domain of A33R interacts with residues 91 to 111 of A36R (50). The interaction of A36R with A33R is required for the incorporation of A36R into the viral envelope and subsequent actin tail formation (54).
Recent work dispelled a previously accepted belief that actin tails propel IEVs through the cytoplasm and demonstrated that IEV movement is dependent on microtubules (15, 20, 27, 48, 49). The cellular protein kinesin is a molecular motor that moves cargo from the Golgi network to the plasma membrane (18, 35), making it a likely candidate for movement of nascent enveloped virions to the cell surface. Kinesin is found in the cell as a heterotetramer consisting of two copies of the light chain (KLC) and two copies of the heavy chain (KHC). The KHC contains the microtubule-binding domain along with ATP-dependent motor activity. The KLCs interact with the central coiled stalk of the KHCs through their N-terminal α-helical domain. The C-terminal end of KLC contains a degenerate 34-amino-acid repeat motif termed the tetratricopeptide repeat (TPR) (10, 44). TPR motifs from several diverse proteins are involved in protein-protein interactions (4). Recently KLC-TPR was shown to interact with a class of scaffolding proteins for the c-Jun NH2-terminal kinase-signaling pathway (7, 47). These scaffolding proteins mediate the binding of cellular cargo to kinesin through attachment at the KLC-TPR.
The binding of kinesin-specific antibody to enveloped virions and evidence that overexpression of TPR results in a decrease in the accumulation of enveloped virus at the cell periphery were recently reported (27). Residues 71 to 100 of A36R were required for dispersion of IEV to the periphery of the cell, although binding to kinesin of either the whole A36R protein or segments of it was not demonstrated (27). Because residues 91 to 111 of A36R mediate an interaction with the A33R envelope protein (50), the role of A36R in movement could be indirect. Here, we used the yeast two-hybrid assay to screen for interactions between the KLC-TPR and the cytoplasmic domains of five viral envelope proteins. A strong interaction was detected between A36R and the KLC-TPR. The site of binding to the KLC was further mapped to residues 81 to 111 of A36R, which overlaps with the A33R interaction site (50). Using the yeast three-hybrid assay, we demonstrated that the binding of A36R is mutually exclusive to either the A33R protein or kinesin, suggesting that A33R regulates the intracellular transport of enveloped virions.
MATERIALS AND METHODS
Yeast two-hybrid constructs and assays.
Cloning of coding sequences for the cytoplasmic domains of A33R, A34R, A36R, B5R, and F12L and the series of A36R truncations into pGADT7 and pGBKT7 (BD Biosciences Clontech) has been described previously (50), with the exception of A36R24-101. Primers CCTCGAGTCACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCTTCGTTATCCCATATTAAACT and GCATATGATTTGTAGGAAAAAGATACGTACT were used to amplify the coding sequence of residues 24 to 101 of A36R and to add a 5′ NdeI restriction endonuclease site (underlined) and a 3′ V5 tag (double underlined) followed by an XhoI (underlined) site. Amplified fragments were cloned into pGEM-T (Promega), sequenced, and subsequently excised from pGEM-T by digestion with NdeI-XhoI, and ligated into similarly cleaved pGBKT7 and pGADT7. Plasmids containing the coding sequence for either the six TPR motifs of rat KLC or the TPR motifs from protein phosphatase 5 (47) were a gift from K. J. Verhey. The coding sequence of the TPR motifs was excised from pGBDU-C1 with EcoRI-SalI and ligated into pGADT7 that had been cleaved with EcoRI-XhoI.
To test for interactions, yeast strain AH109 was cotransformed with purified plasmids by using the Yeastmaker yeast transformation system 2 (BD Biosciences Clontech) according to manufacturer's instructions. Transformed yeast cells were plated onto standard dropout medium minus leucine and tryptophan. Resulting yeast colonies were tested for interaction by streaking on standard quadruple-dropout (QDO) medium minus leucine, tryptophan, histidine, and adenine.
Yeast three-hybrid constructs.
The coding sequence of KLC-TPR was excised from pGBDU-KLC-TPR6 by using EcoRI-SalI and ligated into similarly cleaved pBridge (BD Biosciences Clontech) to create pBridgeBD-KLC-TPR. Primer AGCGGCCGCGGAGGACCTGCATATG was used in conjunction with the 3′ DNA binding domain (BD) sequencing primer (BD Biosciences Clontech) to amplify the coding sequence of either residues 24 to 111 of A36R from pGAD-A36R24-111 or residues 1 to 40 of A33R from pGAD-A33R1-40 and to add a 5′ NotI (underlined) restriction endonuclease site. Amplified fragments were cloned into pGEM-T, sequenced and subsequently excised from pGEM-T by digestion with NotI-BamHI, and ligated into pBridgeBD-KLC-TPR that had been cleaved with NotI-BglII.
Western blots.
Doubly transformed yeast cells were grown overnight in standard dropout medium lacking leucine and tryptophan that either did or did not contain methionine. The next day, cultures were diluted to equal optical densities, and an equal volume was extracted with Y-PER (Pierce). Proteins from the extracts were mixed with protein loading buffer, resolved by electrophoresis on a sodium dodecyl sulfate (SDS)-polyacrylamide (10 to 20%) gel (Invitrogen), and transferred to a nitrocellulose membrane. Membranes were incubated with antihemagglutinin (HA) monoclonal antibody (MAb) (Roche) followed by horseradish peroxidase-conjugated sheep anti-mouse antibody (Amersham). Bound antibodies were detected with chemiluminescence reagents (Pierce).
GST fusion constructs and protein interaction assays.
Plasmids pGAD-A36R24-111 and pGAD-A33R1-40 were digested with NheI and made blunt by filling in the overhang. Subsequently, the coding sequence for residues 24 to 111 of A36R and 1 to 40 of A33R were removed from pGAD-A36R24-111 and pGAD-A33R1-40, respectively, by digestion with XhoI and ligated into pGEX-5X-1 (Amersham Biosciences) that had been digested with SmaI-XhoI. The resulting plasmids were transformed into Escherichia coli strain BL21, and the glutathione S-transferase (GST) fusions were induced and purified.
For pull-down assays, KLC-TPR was labeled in vitro with [35S]methionine by using the TNT coupled reticulocyte lysate system (Promega). Dilutions of the labeled protein were resolved on SDS-polyacrylamide (10 to 20%) gels and transferred to nitrocellulose. Full-length products were excised and quantified by scintillation counting. Five micrograms of purified GST or GST-A36R24-111 was incubated with 20 μl of glutathione Sepharose 4B (Amersham Pharmacia) in phosphate-buffered saline (PBS) for 1 h at 4°C followed by three washes with PBS. Washed beads were used with 0.5 fmol of labeled KLC-TPR for pull-down assays as described previously (40).
For inhibition assays, 0.13 nmol of purified GST-A36R24-111 was incubated with 0.5 fmol of [35S]methionine-labeled KLC-TPR in 200 μl of PBS containing 200 μg of bacterial protein extract. For competition, either 1.3 or 13 nmol of purified GST-A33R1-40 or 13 nmol of purified GST was also included. After 1 h at 4°C, 50 μl of anti-V5 antibody-conjugated agarose (Sigma) was added. After another 1-h incubation at 4°C, the beads were washed three times with PBS. Proteins were eluted in SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer, resolved on SDS-polyacrylamide (10 to 20%) gels, transferred to nitrocellulose, and quantified by scintillation counting.
For preparation of cytosolic extracts, 108 HeLa cells were placed on ice, washed twice with ice-cold PBS, and scraped in ice-cold homogenization buffer (100 mM PIPES [pH 6.9], 2 mM MgSO4, 1 mM EGTA, complete protease inhibitor tablet [Roche], 0.2 mM phenylmethylsulfonyl fluoride). Cells were chilled in a 7-ml Dounce homogenizer on ice for 5 min and disrupted by 20 strokes of the pestle. The resulting extract was clarified by centrifugation at 50,000 × g for 1 h. Two milliliters of extract was incubated with 10 μg of purified GST or GST-A36R24-111 bound to glutathione Sepharose 4B (Amersham Pharmacia) for 3 h at 4°C. After incubation, the beads were washed three times with homogenization buffer. Proteins were eluted in SDS-PAGE loading buffer, resolved on SDS-polyacrylamide (10%) gels, and transferred to nitrocellulose. Membranes were incubated with either anti-KHC or anti-KLC antibody (Santa Cruz Biotechnology) followed by horseradish peroxidase-conjugated donkey anti-goat antibody (Jackson ImmunoResearch). Bound antibodies were detected with chemiluminescence reagents (Pierce).
RESULTS
Interaction of the A36R protein with kinesin in a yeast two-hybrid system.
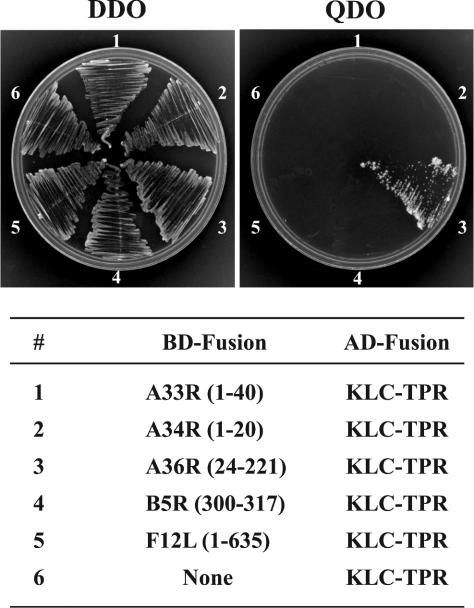
After envelopment, IEVs are transported from the site of wrapping at the TGN or late endosomal cisternae to the cell surface. We considered that the microtubule motor protein kinesin would be involved in virion transport because of its known role in positive-end-directed movement on microtubules (18, 35) and data of Rietdorf and coworkers (27). Previously, we demonstrated that the yeast two-hybrid system was an effective tool for determining the interactions between the cytoplasmic domains of various IEV proteins (50) and thought that it would also be useful for determining interactions between the same domains and the microtubule motor kinesin. Cellular cargo interacts with kinesin through a series of six TPRs in the C terminus of KLC (7, 47). Based on this information, we decided to test the TPR domain of KLC (KLC-TPR) for interaction with the cytoplasmic tails of five virally encoded envelope proteins: A33R, A34R, A36R, B5R, and F12L. DNA encoding the cytoplasmic domain of each of these proteins was fused to DNA encoding the GAL4 BD. Correspondingly, the DNA encoding residues 199 to 489 of rat KLC, which encode the TPR domain, was fused to DNA encoding the GAL4 activation domain (AD). The AD KLC-TPR plasmid was cotransfected into yeast with one of the BD plasmids. In each case, the doubly transfected yeast grew on double-dropout (DDO) medium lacking leucine and tryptophan, indicating that they contained both the AD KLC-TPR plasmid and a BD construct plasmid (Fig. 1). However, only the yeast transfected with the KLC-TPR plasmid and the BD construct with the cytoplasmic tail of A36R grew on the selective QDO medium (Fig. 1). We did not detect growth on selective QDO medium when the BD plasmid contained the GAL4 DNA BD unfused or fused to the cytoplasmic domain of any of the other viral proteins (Fig. 1). Furthermore, we did not detect growth on selective QDO medium when the AD plasmid contained an unrelated TPR motif from protein phosphatase 5 and the BD plasmid contained the cytoplasmic domain of A36R (data not shown). Taken together, these data indicated a specific interaction between the KLC-TPR and the cytoplasmic domain of A36R.
FIG. 1.
Yeast two-hybrid assay. The cytoplasmic domains of various IEV proteins were fused to the GAL4 BD (BD-Fusion) and tested for interaction with KLC-TPR that had been fused to the GAL4 AD (AD-Fusion) by cotransfection of the two plasmids into yeast. DDO medium was streaked with cotransformed yeast cells to confirm the presence of both BD and AD fusions. Positive interactions were determined by growth on QDO medium.
We tested our series of previously constructed BD-A36R truncations (50) for interactions with AD-KLC-TPR to further define the region of A36R responsible for the interaction with KLC-TPR. The genes for the truncated proteins encoding amino acids 24 to 221, 24 to 123, and 24 to 111 interacted with KLC-TPR, while those encoding amino acids 24 to 101, 24 to 93, and 24 to 80 failed to interact as determined by growth on selective QDO medium (Table 1), indicating that the kinesin interaction region was contained within the first 111 residues of A36R. Next we tested for interaction between AD KLC-TPR and four truncations of A36R in which the first 60, 70, 80, or 90 residues of the cytoplasmic domain were removed in conjunction with the deletion of the last 108 residues to give constructs expressing amino acids 61 to 111, 71 to 111, 81 to 111, or 91 to 111. Positive interactions were detected with the first three truncations, but we were unable to detect an interaction with residues 91 to 111 (Table 1), indicating that residues 81 to 111 of A36R were sufficient for interaction with the TPR domain of KLC in the yeast two-hybrid system.
TABLE 1.
Test for interaction of AD-KLC-TPR with various BD-A36R constructs by the yeast two-hybrid assay
| BD fusion | Growth on QDOa |
|---|---|
| A36R (24-221) | + |
| A36R (24-123) | + |
| A36R (24-111) | + |
| A36R (24-101) | − |
| A36R (24-93) | − |
| A36R (24-80) | − |
| A36R (61-111) | + |
| A36R (71-111) | + |
| A36R (81-111) | + |
| A36R (91-111) | − |
Growth with the KLC-TPR AD fusion.
Interaction of A36R with KLC-TPR in vitro.
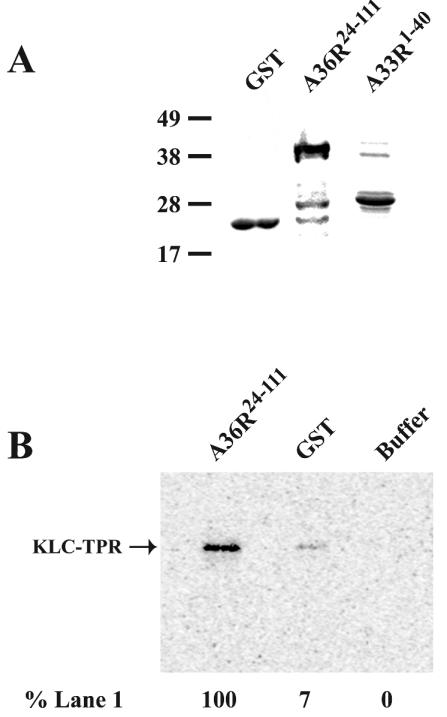
We next used an in vitro GST pull-down assay to confirm the A36R-KLC-TPR interaction determined in the yeast two-hybrid system. DNA sequences corresponding to residues 24 to 111 of A36R were genetically fused to GST (GST-A36R24-111) sequences. Subsequent expression in E. coli and one-step purification of GST-A36R24-111 yielded a prominent, Coomassie-stained band of ∼38 kDa that agreed with the predicted size of GST-A36R24-111 and showed a slower mobility than GST alone (Fig. 2A). Equivalent molar amounts of either GST or GST-A36R24-111 were bound to glutathione beads and incubated with 0.5 fmol of in vitro 35S-labeled KLC-TPR. Following extensive washing, complexes were eluted from glutathione beads by boiling in SDS sample buffer and resolved by SDS-PAGE. After transfer to nitrocellulose, the amount of 35S-labeled KLC-TPR was imaged and quantified with a PhosphorImager. GST-A36R24-111 was able to bind over 10 times more KLC-TPR than GST alone (Fig. 2B), indicating a specific interaction between KLC-TPR and A36R and confirming our yeast two-hybrid results.
FIG. 2.
In vitro interaction assay. Fusion proteins were overexpressed in E. coli and affinity purified with glutathione resin. (A) Purified proteins were analyzed by SDS-PAGE followed by Coomassie blue staining. (B) GST pull-down assays were carried out with either GST-A36R24-111, GST, or buffer alone bound to glutathione resin and incubated with in vitro 35S-labeled KLC-TPR. The masses (in kilodaltons) and positions of migration of markers are indicated on the left. Complexes were analyzed by SDS-PAGE. A PhosphorImager was used to detect and quantify the amount of radioactivity in each band with the relative amounts shown at the bottom.
Interaction of A36R with cytosolic kinesin.
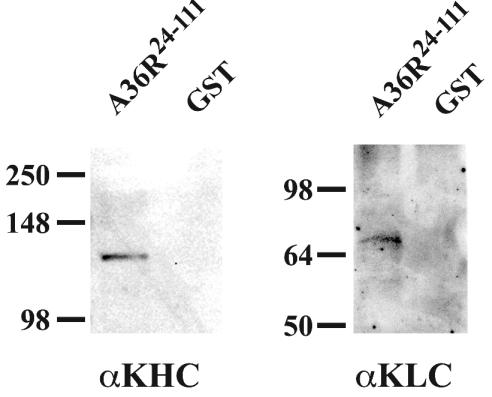
Until this point, we had only showed an interaction of A36R with the isolated TPR domain of kinesin. We wanted to determine if A36R could interact with the TPR domain of KLC from the complete kinesin complex. To do this, a cytosolic extract containing endogenous kinesin from HeLa cells was prepared and incubated with either GST or GST-A36R24-111 that had been bound to glutathione beads. Following extensive washing, complexes were eluted from glutathione beads by boiling in SDS sample buffer, and the presence of both KLC and KHC was determined by SDS-PAGE followed by Western blotting with either KLC- or KHC-specific MAbs. GST- A36R24-111 was able to bring down both KLC and KHC, while GST alone was not (Fig. 3).
FIG. 3.
In vivo interaction assay. GST pull-down assays were carried out with either GST-A36R24-111 or GST bound to glutathione resin and incubated with a HeLa cell cytoplasmic extract. Complexes were analyzed by SDS-PAGE followed by Western blotting with either anti-KHC MAb (αKHC) or anti-KLC MAb (αKLC). The masses (in kilodaltons) and migration positions of markers are indicated on the left.
The A33R protein competes with KLC-TPR for binding to the A36R protein.
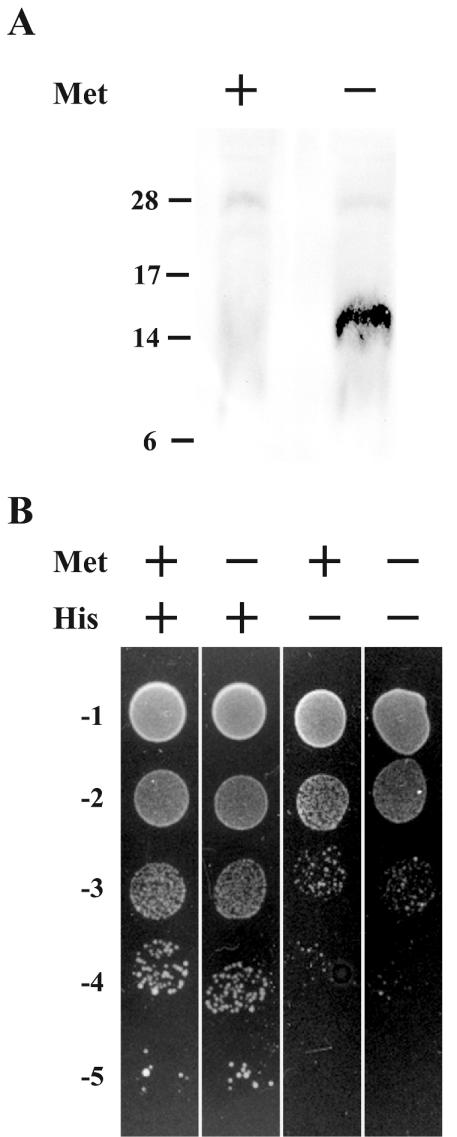
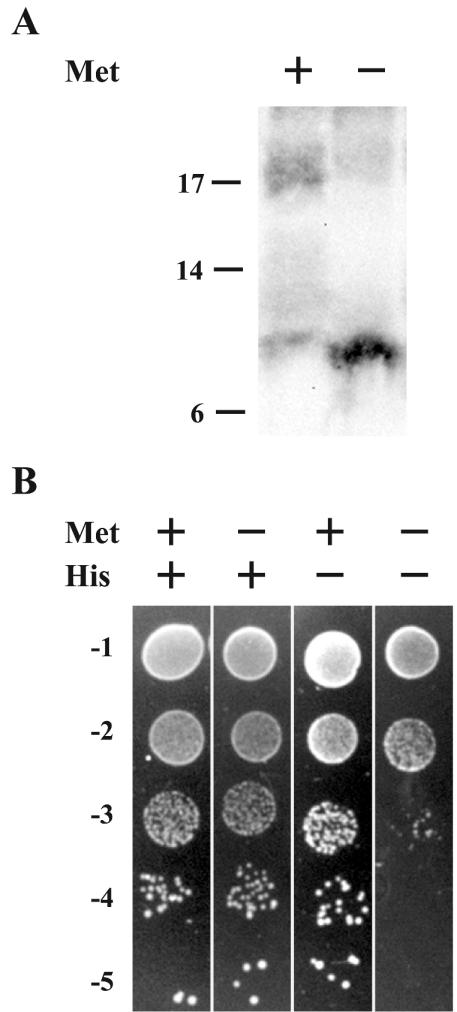
We previously reported that residues 91 to 111 of A36R were sufficient for interaction with the cytoplasmic tail of the A33R envelope protein (50). We wanted to determine if A36R could bind A33R and KLC-TPR simultaneously, because the sites on A36R for binding these two proteins overlap. To test for the ability of A36R to simultaneously interact with A33R and KLC-TPR, we used the yeast three-hybrid system (32). The KLC-TPR domain was inserted into the BD of pBridge (Clontech). Subsequently, DNA containing codons 24 to 111 of A36R was inserted into MCS II of the pBridge vector, placing its expression under control of the M25 promoter and creating pBridgeBD-KLC-TPR/A36R24-111. Proteins controlled by the M25 promoter are only expressed when yeast are grown on medium deficient in methionine, since growth in the presence of methionine inactivates the promoter. The conditional expression of A36R24-111 in this system allowed us to check for a specific simultaneous interaction with BD-KLC-TPR and AD-A33R1-40 by scoring for growth on medium deficient in both histidine and methionine. Before testing for simultaneous interactions, we checked the induction and stable expression of A36R24-111 from the M25 promoter. Yeast cells were cotransformed with pBridgeBD-KLC-TPR/A36R24-111 and pAD-A33R1-40 and plated on standard DDO medium lacking leucine and tryptophan in order to select for the presence of both plasmids. Doubly transformed yeast was grown overnight in DDO medium that either did or did not contain methionine. Extracts from yeast cultures were separated by SDS-PAGE, transferred to nitrocellulose, and probed with an anti-HA MAb. The anti-HA MAb interacted with an ∼15-kDa band of the predicted size of A36R24-111 (Fig. 4A). Importantly this band was only seen in extracts from yeast grown in the absence of methionine, confirming both the regulation of the M25 promoter and the stable expression of A36R24-111.
FIG. 4.
Yeast three-hybrid interaction assay. Yeast strain CG1945 was cotransfected with pBridgeBD-KLC-TPR/A36R24-111 and pADA33R1-40. (A) Extracts from cotransformed yeast that had been grown in medium with (+) or without (−) methionine were analyzed by SDS-PAGE followed by Western blotting with anti-HA MAb. The masses (in kilodaltons) and positions of migration of markers are indicated on the left. (B) Tenfold dilutions of overnight cultures of cotransformed yeast were applied as spots to selective medium with (+) or without (−) methionine and histidine.
After confirming the regulation of the M25 promoter, we checked for simultaneous interaction of KLC-TPR and A33R with A36R by making 10-fold dilutions from overnight cultures of yeast that had been cotransformed with pBridgeBD-KLC-TPR/A36R24-111 and pAD-A33R1-40. Subsequently, equal volumes of each dilution were applied as spots to DDO, DDO-Met, DDO−His, and DDO−His/−Met media. Equal levels of growth were seen on nonselective DDO and DDO−Met media, indicating that the expression of A36R24-111 has no toxic effects on yeast growth (Fig. 4B). Yeast grown on DDO−His medium, which selects for positive interactions, showed almost a 2-log decrease in growth when compared to nonselective DDO medium (Fig. 4B). This was in accordance with our initial screen, indicating that BD-KLC-TPR and AD-A33R1-40 were unable to interact with each other. Likewise, yeast grown on DDO−His/−Met medium, which selects for positive interaction in the presence of A36R24-111, showed a similar 2-log decrease, as was seen for growth on DDO−His medium, indicating that the expression of A36R24-111 did not increase the ability of BD-KLC-TPR and AD-A33R1-40 to interact. These results suggested that A36R cannot simultaneously interact with KLC-TPR and A33R and that their binding may be mutually exclusive.
We considered that if the binding of A36R was mutually exclusive to either KLC-TPR or A33R, then A33R should be able to compete with KLC-TPR for binding to A36R. We decided to use the yeast three-hybrid assay (32, 41) to see if the expression of A33R could compete and disrupt the interaction between BD-KLC-TPR and AD-A36R24-111. To test this idea, pBridgeBD-KLC-TPR/A33R1-40 was generated by replacing A36R24-111 in pBridgeBD-KLC-TPR/A36R24-111 with residues 1 to 40 of A33R. Induction and stable expression of A33R1-40 under the M25 promoter were checked by growing yeast cotransformed with pBridgeBD-KLC-TPR/A33R1-40 and pADA36R24-111 in DDO medium with and without methionine. Extracts from yeast cultures were separated on SDS-PAGE, transferred to nitrocellulose, and probed with an anti-HA MAb. The anti-HA MAb interacted with a band of ∼12.5 kDa, which is the predicted size of A33R1-40 (Fig. 5A). As anticipated, this band was only detected in extracts from yeast grown in the absence of methionine, confirming the stable expression of A33R1-40 from the M25 promoter.
FIG. 5.
Yeast three-hybrid competition assay. Yeast strain CG1945 was cotransfected with pBridgeBD-KLC-TPR/A33R1-40 and pADA36R24-111. (A) Extracts from cotransformed yeast that had been grown in medium with (+) or without (−) methionine were analyzed by SDS-PAGE followed by Western blotting with anti-HA MAb. The masses (in kilodaltons) and migration positions of markers are indicated on the left. (B) Tenfold dilutions of overnight cultures of cotransformed yeast were applied as spots to selective medium with (+) or without (−) methionine and histidine.
The possible inhibition of the BD-KLC-TPR:AD-A36R24-111 interaction by A33R1-40 was determined with yeast cotransformed with pBridgeBD-KLC-TPR/A33R1-40 and pADA36R24-111 using the same dilution assay as before. Yeast cotransformed with pBridgeBD-KLC-TPR/A33R1-40 and pADA36R24-111 grew equally well on DDO and DDO−Met media, indicating that the expression of A33R1-40 had no detectable toxic effects on yeast (Fig. 5B). Equal amounts of growth were also seen for yeast plated on DDO and DDO−His media, indicating a positive interaction between BD-KLC-TPR and AD-A36R24-111 (Fig. 5B). In contrast, expression of A33R1-40 by growth on DDO−His/−Met resulted in almost a 3-log reduction in growth as compared to growth on DDO−Met medium (Fig. 5B). The most straightforward interpretation of this assay result is that A33R effectively competed with KLC-TPR for interaction with A36R and disrupted the interaction between BD-KLC-TPR and AD-A36R24-111.
A33R competes with TPR for A36R binding in vitro.
In order to confirm the yeast three-hybrid assay results, we performed pull-down assays in the presence of competing A33R. For this purpose, a fusion between GST and residues 1 to 40 of A33R was constructed (GST-A33R1-40). The fusion protein was overexpressed in E. coli and purified with glutathione resin in a one-step purification process. SDS-PAGE analysis of the purified protein resulted in a prominent ∼21-kDa band that agreed with the predicted size of GST-A33R1-40 (Fig. 2A). Complexes between GST-A36R24-111 and in vitro 35S-labeled KLC-TPR were allowed to form in the presence of either a 10- or 100-fold molar excess of GST-A33R1-40 or a 100-fold molar excess of GST alone. GST-A36R24-111 complexes were purified with anti-V5 MAb-conjugated agarose by virtue of a V5 epitope tag that is present in GST-A36R24-111 but absent in GST-A33R1-40. Following extensive washing, complexes were eluted from V5 MAb agarose by boiling in SDS sample buffer and resolved by SDS-PAGE. After transfer to nitrocellulose, the amount of 35S-labeled KLC-TPR was imaged and quantified with a PhosphorImager. In the presence of 10-fold-more A33R, the amount of KLC-TPR bound to A36R was markedly decreased when compared to the amount of complexes that were formed in its absence (Table 2). Furthermore, in the presence of 100-fold-more A33R, the interaction between A36R and KLC-TPR was reduced almost to the background level of KLC-TPR binding in the absence of A36R. Importantly, in the presence of a 100-fold molar excess of GST, there was no significant reduction in the interaction between A36R and KLC-TPR. These data confirm our results from the yeast three-hybrid assay.
TABLE 2.
Test for the ability of GST-A33R1-40 to compete with in vitro 35S-labeled TPR for interaction with GST-A36R24-111
| Competitor | Expt 1
|
Expt 2
|
||
|---|---|---|---|---|
| Amt of TPRa | % of no competitor | Amt of TPRa | % of no competitor | |
| None | 10,270 | 100 | 14,525 | 100 |
| 10× A33R | 5,884 | 57 | 10,636 | 73 |
| 100× A33R | 3,838 | 37 | 7,096 | 49 |
| 100× GST | 9,252 | 90 | 15,013 | 103 |
| Backgroundb | 4,094 | 40 | 4,408 | 30 |
Raw densitometry values.
GST-A36R24-111 was excluded from the assay.
DISCUSSION
Simple diffusion through the viscous cytoplasm would be prohibitively slow for progeny virions to escape the cell and continue the infectious process (37). For this reason, poxviruses employ the same microtubule network that the host cell uses for the transport of cargo to the cell surface. Previous reports established the dependency of enveloped vaccinia virus virions on the microtubule network for intracellular transport from the site of wrapping at the TGN to the cell surface. Movement of cellular cargo from the TGN and late endosomes to the cell surface is carried out by the molecular motor kinesin (18, 35). A recent report demonstrated that kinesin-specific antibodies labeled enveloped virions and that overexpression of TPR resulted in a decrease in the accumulation of enveloped virus at the cell periphery (27). However, that study did not demonstrate a direct interaction between kinesin and any viral protein. Furthermore, an inability to demonstrate an interaction between residues 71 to 100 of the A36R protein and the TPRs of KLC expressed in E. coli was stated (27). In the present work, we used the yeast two-hybrid assay to demonstrate a direct interaction between the TPR domain of the KLC and the virus-encoded A36R envelope protein. The finding that A36R provides this link fits well with our previous results. We reported that IEV of a recombinant virus with a deletion of the entire A36R protein or a region that contains residues 81 to 111 displayed a short sporadic motion that was difficult to track. IEV of a recombinant virus with most of the cytoplasmic tail of A33R deleted resulted in a similar defect (50). It is now evident why both of these mutations resulted in similar IEV intracellular movement. Removal of residues 81 to 111 from A36R not only disrupts the ability of A36R to interact with A33R but also disrupts the interaction of A36R with kinesin and severs the link between IEVs and microtubules. Likewise, the deletion of the cytoplasmic tail of A33R prohibited the incorporation of A36R into IEVs and also removes this link between IEVs and microtubules.
Recent reports have identified three cellular scaffolding proteins from the c-Jun NH2-terminal kinase signaling pathway (JIP-1, JIP-2, and JIP-3) (7, 47) that also interact with KLC through the TPR region. Interactions with the cytoplasmic domains of different membrane receptors have been established for both JIP-1 and JIP-2, providing a link between vesicle cargo and KLC. Multiple adapter proteins linking different vesicle attachments to kinesin raise the possibility that the adapter proteins regulate the binding of cargo to kinesin. Poxviruses appear to have circumvented this potential cellular regulation by encoding an envelope protein that interacts directly with the KLC. This would allow enveloped virions the fastest access to microtubule motor transport and exit from the cell.
If poxviruses bypass the normal cellular regulation of kinesin binding, then what regulates the IEV-kinesin interaction—or, more precisely, the A36R-KLC-TPR interaction? One possibility is A33R. In this report, we have demonstrated that the TPR interaction site on A36R overlaps with the previously determined A33R binding site. Furthermore, we have shown that interactions at this site on A36R are mutually exclusive to either A33R or KLC-TPR. The exclusivity of these interactions may act as a regulatory mechanism. The interaction between A33R and A36R is required for the incorporation of A36R into the IEV membrane, implying that all of the A36R in the viral envelope is interacting with A33R. Logically, the A33R-A36R interaction precedes the A36R-KLC-TPR interaction. At some point, A36R must disassociate from A33R to allow for interaction with kinesin. Dissociation of A36R from A33R may also relate to the presence of A36R only on the outer of the two IEV membranes. The envelopment of IMV could serve as a signal for the release of A33R by A36R and a subsequent interaction with KLC-TPR followed by trafficking of newly enveloped virions out to the cell periphery.
Interestingly, cells infected with recombinant vaccinia viruses containing deletions in the A36R gene still release enveloped virions, albeit at a reduced level (25, 53). We can think of three possibilities to explain this apparent discrepancy. The first is that the envelopment of some virions occurs close to or at the cell surface overcoming the requirement for kinesin-mediated transport. Alternatively, some enveloped virions may be produced by budding of IMV at the plasma membrane, once again forgoing the requirement for kinesin. Budding has been described for the IHD strain of vaccinia virus (43) as well as for the distantly related fowlpox virus (5). Third, it is possible that other proteins on the IEV envelope also interact with kinesin or that there are other modes of intracellular transport for IEV. Indeed, in the absence of A36R, we were able to observe occasional infrequent abnormal IEV movement that consisted of short sporadic motions (50).
Acknowledgments
We thank members of the Laboratory of Viral Diseases for their interest and suggestions, especially Tom Kristie and Jodi Vogel. Kristen J. Verhey kindly provided plasmids.
REFERENCES
- 1.Appleyard, G., A. J. Hapel, and E. A. Boulter. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9-17. [DOI] [PubMed] [Google Scholar]
- 2.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 65:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 5.Boulanger, D., T. Smith, and M. A. Skinner. 2000. Morphogenesis and release of fowlpox virus. J. Gen. Virol. 81:675-687. [DOI] [PubMed] [Google Scholar]
- 6.Boulter, E. A., and G. Appleyard. 1973. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog. Med. Virol. 16:86-108. [PubMed] [Google Scholar]
- 7.Bowman, A. B., A. Kamal, B. W. Ritchings, A. V. Philp, M. McGrail, J. G. Gindhart, and L. S. Goldstein. 2000. Kinesin-dependent axonal transport is mediated by the Sunday driver (SYD) protein. Cell 103:583-594. [DOI] [PubMed] [Google Scholar]
- 8.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 9.Dales, S., and L. Siminovitch. 1961. The development of vaccinia virus in Earle's L strain cells as examined by electron microscopy. J. Biophys. Biochem. Cytol. 10:475-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diefenbach, R. J., J. P. Mackay, P. J. Armati, and A. L. Cunningham. 1998. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry 37:16663-16670. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, S. A., and G. L. Smith. 1992. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol. 66:1610-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelstad, M., S. T. Howard, and G. L. Smith. 1992. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology 188:801-810. [DOI] [PubMed] [Google Scholar]
- 13.Engelstad, M., and G. L. Smith. 1993. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology 194:627-637. [DOI] [PubMed] [Google Scholar]
- 14.Frischknecht, F., V. Moreau, S. Rottger, S. Gonfloni, I. Reckmann, G. Superti-Furga, and M. Way. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401:926-929. [DOI] [PubMed] [Google Scholar]
- 15.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 16.Grimley, P. M., E. N. Rosenblum, S. J. Mims, and B. Moss. 1970. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J. Virol. 6:519-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiller, G., K. Weber, L. Schneider, C. Parajsz, and C. Jungwirth. 1979. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology 98:142-153. [DOI] [PubMed] [Google Scholar]
- 18.Hirokawa, N. 1998. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279:519-526. [DOI] [PubMed] [Google Scholar]
- 19.Hirt, P., G. Hiller, and R. Wittek. 1986. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J. Virol. 58:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaacs, S. N., E. J. Wolffe, L. G. Payne, and B. Moss. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 66:7217-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIntosh, A. A. G., and G. L. Smith. 1996. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J. Virol. 70:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreau, V., F. Frischknecht, I. Reckmann, R. Vincentelli, G. Rabut, D. Stewart, and M. Way. 2000. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol. 2:441-448. [DOI] [PubMed] [Google Scholar]
- 24.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 25.Parkinson, J. E., and G. L. Smith. 1994. Vaccinia virus gene A36R encodes a Mr 43-50 K protein on the surface of extracellular enveloped virus. Virology 204:376-390. [DOI] [PubMed] [Google Scholar]
- 26.Payne, L. G. 1980. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol. 50:89-100. [DOI] [PubMed] [Google Scholar]
- 27.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 28.Risco, C., J. R. Rodriguez, C. López-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roper, R. L., L. G. Payne, and B. Moss. 1996. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 70:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roper, R. L., E. J. Wolffe, A. Weisberg, and B. Moss. 1998. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 72:4192-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanderson, C. M., F. Frischknecht, M. Way, M. Hollinshead, and G. L. Smith. 1998. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J. Gen. Virol. 79:1415-1425. [DOI] [PubMed] [Google Scholar]
- 32.Sandrock, B., F. Tirode, and J. M. Egly. 2001. Three-hybrid screens. Inducible third-party systems. Methods Mol. Biol. 177:271-289. [DOI] [PubMed] [Google Scholar]
- 33.Scaplehorn, N., A. Holmstrom, V. Moreau, F. Frischknecht, I. Reckmann, and M. Way. 2002. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr. Biol. 12:740-745. [DOI] [PubMed] [Google Scholar]
- 34.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seiler, S., F. E. Nargang, G. Steinberg, and M. Schliwa. 1997. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16:3025-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shida, H. 1986. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology 150:451-462. [DOI] [PubMed] [Google Scholar]
- 37.Sodeik, B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 8:465-472. [DOI] [PubMed] [Google Scholar]
- 38.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stokes, G. V. 1976. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J. Virol. 18:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swaffield, J. C., and S. A. Johnston. 1998. Affinity purification of proteins binding to GST fusion proteins. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 3. Greene Publishing Associates and Wiley Interscience, New York, N.Y. [DOI] [PubMed]
- 41.Tirode, F., C. Malaguti, F. Romero, R. Attar, J. Camonis, and J. M. Egly. 1997. A conditionally expressed third partner stabilizes or prevents the formation of a transcriptional activator in a three-hybrid system. J. Biol. Chem. 272:22995-22999. [DOI] [PubMed] [Google Scholar]
- 42.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 43.Tsutsui, K. 1983. Release of vaccinia virus from FL cells infected with the IHD-W strain. J. Electron Microsc. 32:125-140. [PubMed] [Google Scholar]
- 44.Vale, R. D., and R. J. Fletterick. 1997. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13:745-777. [DOI] [PubMed] [Google Scholar]
- 45.van Eijl, H., M. Hollinshead, G. Rodger, W. H. Zhang, and G. L. Smith. 2002. The vaccinia virus F12L protein is associated with intracellular enveloped virus particles and is required for their egress to the cell surface. J. Gen. Virol. 83:195-207. [DOI] [PubMed] [Google Scholar]
- 46.van Eijl, H., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology 271:26-36. [DOI] [PubMed] [Google Scholar]
- 47.Verhey, K. J., D. Meyer, R. Deehan, J. Blenis, B. J. Schnapp, T. A. Rapoport, and B. Margolis. 2001. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152:959-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward, B. M., A. S. Weisberg, and B. Moss. 2003. Mapping and functional analysis of interaction sites within the cytoplasmic domains of the vaccinia virus A33R and A36R envelope proteins. J. Virol. 77:4113-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolffe, E. J., S. N. Isaacs, and B. Moss. 1993. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol. 67:4732-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolffe, E. J., E. Katz, A. Weisberg, and B. Moss. 1997. The A34R glyco-protein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J. Virol. 71:3904-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolffe, E. J., A. S. Weisberg, and B. Moss. 1998. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology 244:20-26. [DOI] [PubMed] [Google Scholar]
- 54.Wolffe, E. J., A. S. Weisberg, and B. Moss. 2001. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 75:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, W.-H., D. Wilcock, and G. L. Smith. 2000. Vaccinia virus F12L protein is required for actin tail formation, normal plaque size, and virulence. J. Virol. 74:11654-11662. [DOI] [PMC free article] [PubMed] [Google Scholar]