Abstract
The human positive transcription elongation factor P-TEFb is composed of two subunits, cyclin T1 (hCycT1) and CDK9, and is involved in transcriptional regulation of cellular genes as well as human immunodeficiency virus type 1 (HIV-1) mRNA. Replication of HIV-1 requires the Tat protein, which activates elongation of RNA polymerase II at the HIV-1 promoter by interacting with hCycT1. To understand the cellular functions of P-TEFb and to test whether suppression of host proteins such as P-TEFb can modulate HIV infectivity without causing cellular toxicity or lethality, we used RNA interference (RNAi) to specifically knock down P-TEFb expression by degrading hCycT1 or CDK9 mRNA. RNAi-mediated gene silencing of P-TEFb in HeLa cells was not lethal and inhibited Tat transactivation and HIV-1 replication in host cells. We also found that CDK9 protein stability depended on hCycT1 protein levels, suggesting that the formation of P-TEFb CDK-cyclin complexes is required for CDK9 stability. Strikingly, P-TEFb knockdown cells showed normal P-TEFb kinase activity. Our studies suggest the existence of a dynamic equilibrium between active and inactive pools of P-TEFb in the cell and indicate that this equilibrium shifts towards the active kinase form to sustain cell viability when P-TEFb protein levels are reduced. The finding that a P-TEFb knockdown was not lethal and still showed normal P-TEFb kinase activity suggested that there is a critical threshold concentration of activated P-TEFb required for cell viability and HIV replication. These results provide new insights into the regulation of P-TEFb function and suggest the possibility that similar mechanisms for monitoring protein levels to modulate the activity of proteins may exist for the regulation of a variety of other enzymatic pathways.
The regulation of mRNA transcription is vital for mammalian cell growth and development. While transcription initiation is highly regulated and best understood, many eukaryotic and viral genes are specifically regulated at the level of transcription elongation. Among these genes are several proto-oncogenes (c-myc, c-myb, and c-fos), the gene for adenosine deaminase, stress response genes, and human immunodeficiency virus type 1 (HIV-1) and HIV-2 genes. The process of transcription elongation entails RNA polymerase II (RNA Pol II) pausing, arresting, and passing through or terminating transcription at terminator sequences. During these discrete steps of elongation that occur shortly after transcription initiation, RNA Pol II faces a barrier of negative transcription elongation factors and begins abortive elongation (42). Positive transcription elongation factors (P-TEF) lower the barrier of negative transcription elongation factors and help RNA Pol II to escape from this transition phase, which otherwise could lead to premature termination of transcription (42). P-TEFb, composed of subunits CDK9 and cyclin T1 (13), is one such P-TEF that mediates the transition to productive elongation, leading to the production of longer mRNAs (42).
One elegant example of transcription elongation control is the mechanism of HIV-1 gene expression (reviewed in references 8, 10, 23-25, and 52). HIV-1 encodes a small regulatory protein, Tat, which is required for the efficient transcription of viral genes. Tat enhances the processivity of RNA Pol II elongation complexes that initiate transcription in the HIV long terminal repeat (LTR) region. During elongation, Tat binds to a highly structured RNA element, transactivation-responsive (TAR) RNA, which is located at the 5′ end of nascent viral transcripts (43). Through its interactions with TAR RNA, Tat controls an early transcription elongation step that is sensitive to protein kinase inhibitors and requires the carboxyl-terminal domain (CTD) of the large subunit of RNA Pol II (24). The HIV-1 transcriptional activation mechanism requires Tat interactions with the human cyclin T1 (hCycT1) subunit of P-TEFb, which recruits the kinase complex to the Pol II elongation machinery (2, 16, 17, 19, 24, 30, 52, 55, 58, 62).
The recruitment of P-TEFb to TAR RNA is proposed to be both necessary and sufficient to activate transcription elongation from the HIV-1 LTR promoter (3). Neither hCycT1 nor the P-TEFb complex binds TAR RNA in the absence of Tat, signifying that binding to RNA is highly cooperative for both Tat and P-TEFb (13, 45, 55). Mutagenesis studies have shown that the hCycT1 sequence containing amino acids 1 to 272 is sufficient to form complexes with Tat-TAR (2, 12, 13, 21, 56, 61). hCycT1 residues 250 to 262 represent the Tat-TAR RNA recognition motif (13), which is required but not sufficient to form the hCycT1-Tat-TAR RNA ternary complex, and N-terminal residues in the cyclin box are also necessary for complex formation. Specifically, residues 252 to 260 of hCycT1 interact with one side of the TAR RNA loop, enhancing the interaction between Tat residue K50 and the other side of the loop (46). Thus, it has become clear that TAR RNA provides a scaffold for two protein partners to bind and assemble a regulatory switch for HIV replication.
The Pol II CTD and Spt5 are also intimately connected to this regulation of HIV gene expression by Tat and P-TEFb. Human Spt5 and its binding partner hSpt4 comprise the transcription elongation regulatory factor DRB sensitivity-inducing factor (DSIF) (53). DSIF binds to Pol II and, in concert with negative elongation factor (NELF), represses elongation at promoter-proximal positions in the transcription unit (44, 57). Escape from the repressive effect of DSIF-NELF requires the action of P-TEFb, which phosphorylates both the Pol II CTD and the Spt5 subunit of DSIF (20, 26, 40, 54). HIV usurps this mechanism to transcribe its own genome (Fig. 1A). During HIV transcription, P-TEFb, which is initially found as a component of the Pol II preinitiation complex (PIC), travels with the transcription elongation complex (TEC) as it moves along the HIV transcription unit (41). In contrast, DSIF and NELF are not present in the PIC but associate with the TEC at promoter-proximal positions and then travel with the TEC down the template (40).
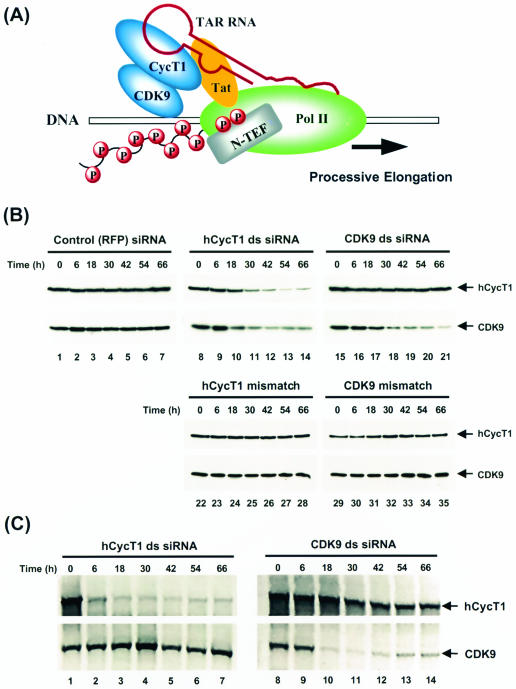
FIG. 1.
Specific silencing of P-TEFb expression by RNAi. (A) Model for HIV Tat transactivation involving the human P-TEFb (CycT1-CDK9) complex. See the text for details. (B) Analysis of specific hCycT1 and CDK9 RNAi activities by Western blotting. HeLa cells were transfected with double-stranded (ds) siRNAs targeting RFP (control; lanes 1 to 7), hCycT1 (lanes 8 to 14), or CDK9 (lanes 15 to 21). Cells were also transfected with mutant siRNAs (hCycT1 mismatch [lanes 22 to 28] or CDK9 mismatch [lanes 29 to 35]) with 2-nt mismatches between the target mRNA and the antisense strand of siRNA at the hypothetical cleavage site of the mRNA. Cells were harvested at various times posttransfection. Protein contents were resolved by SDS-10% PAGE, transferred onto PVDF membranes, and immunoblotted with antibodies against hCycT1 and CDK9. (C) Analysis of specific hCycT1 and CDK9 RNAi activities by RT-PCR. HeLa cells were transfected with hCycT1 double-stranded siRNA (lanes 1 to 7) and CDK9 double-stranded siRNA (lanes 8 to 14) and were harvested at various times after transfection, and mRNA was extracted. One-step RT-PCRs were performed, with specific primers for hCycT1 and CDK9 amplification (see Materials and Methods for details). RT-PCR products were resolved in 1% agarose gels and viewed by ethidium bromide staining.
To better understand the cellular functions of P-TEFb and to test whether the suppression of host proteins such as P-TEFb can modulate HIV infectivity without causing cell toxicity or lethality, we used RNA interference (RNAi) to specifically degrade hCycT1 or CDK9 mRNA. RNAi is a remarkably efficient process whereby double-stranded RNA (dsRNA) induces sequence-specific degradation of homologous mRNAs in animal and plant cells (18, 36). In mammalian cells, RNAi can be triggered by 21-nucleotide (nt) small interfering RNA (siRNA) duplexes, and a few dsRNA molecules are sufficient to inactivate a continuously transcribed target mRNA for an observable period of time (5, 9). RNAi has recently been used to successfully knock down the expression of a number of HIV genes, including p24, the reverse transcriptase (RT) gene, vif, nef, tat, and rev, and has led to pre- and postintegrative HIV-1 RNA degradation and reduced HIV infectivity (7, 22, 29, 38, 51). These results suggest that targeting of viral factors required for the HIV life cycle with siRNAs, including those required for HIV replication, is a viable method for treating HIV infections. Other groups have targeted cellular factors implicated in supporting the HIV life cycle, including T-cell coreceptors CD4, CXCR4, CCR5, and CD8 (31, 32, 35, 38) and transcription factor NF-κB (51), which has a role in HIV transcription initiation. Knockdown of the coreceptors reduced HIV infectivity, effectively blocking HIV entry into cells (31). Our results, presented here, show that siRNAs effectively and specifically target CDK9 and hCycT1 mRNA for RNAi-mediated degradation to silence P-TEFb expression. This RNAi-mediated gene silencing of P-TEFb in HeLa cells was not lethal but did inhibit Tat transactivation and HIV replication in host cells. Furthermore, despite the reduced levels of mRNA and protein resulting from RNAi, the P-TEFb kinase activity remained equivalent to that found in cells that expressed P-TEFb normally. These results suggest that there is a dynamic equilibrium between active and inactive P-TEFb pools that is regulated in response to protein concentrations and other stimuli, such as stress or HIV infection. These results provide new insights into P-TEFb function and offer a strong precedent for exploring knockdown of P-TEFb and other cellular proteins as a potential new strategy for the development of AIDS therapeutics.
MATERIALS AND METHODS
siRNA preparation.
Twenty-one-nucleotide dsRNAs were chemically synthesized as 2′ bis(acetoxyethoxy)-methyl ether-protected oligonucleotides by Dharmacon (Lafayette, Colo.). Synthetic oligonucleotides were deprotected, annealed, and purified according to the manufacturer's recommendations. Successful duplex formation was confirmed by 20% nondenaturing polyacrylamide gel electrophoresis (PAGE). All siRNAs were stored in 0.1% diethyl pyrocarbonate-treated water at −80°C. Sequences of siRNA duplexes were designed according to the manufacturer's recommendations and were subjected to a BLAST search of the National Center for Biotechnology Information expressed sequence tag library to ensure that they targeted only the desired genes. The siRNA sequences used for our experiments were as follows: hCycT1 ds, 5′-UCCCUUCCUGAUACUAGAAdTdT-3′; hCycT1 mm, 5′-UCCCUUCCGUAUACUAGAAdTdT-3′; CDK9 ds, 5′-CCAAAGCUUCCCCCUAUAAdTdT-3′; CDK9 mm, 5′-CCAAAGCUCUCCCC UAUAAdTdT-3′; CDK7 ds, 5′-UUGGUCUCCUUGAUGCUUUdTdT-3′; Tat ds, 5′-GAAACGUAGACAGCGCAGAdTdT-3′; GFP ds, 5′-GCAGCACGACUUCUUCAAGdTdT-3′; and RFP ds, 5′-GUGGGAGCGCGUGAUGAACdTdT-3′. Underlined residues represent sequences with mismatches with their targets.
Culturing and transfection of cells.
HeLa cells were maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 U of penicillin/ml, and 100 μg of streptomycin (Invitrogen)/ml. Magi (multinucleate activation of galactosidase indicator) cells harboring the endogenous HIV LTR β-galactosidase (β-Gal) gene were maintained at 37°C in DMEM supplemented with 10% FBS, 0.2 mg of Geneticin (G418)/ml, and 0.1 mg of hygromycin B (Roche Molecular Biochemicals)/ml. Cells were regularly passaged at subconfluence and plated at 70% confluence 16 h before transfection. Lipofectamine (Invitrogen)-mediated transient cotransfections of reporter plasmids and siRNAs were performed in duplicate 6-well plates (Falcon) as described by the manufacturer for adherent cell lines. A standard transfection mixture containing 100 nM siRNA and 10 μl of Lipofectamine in 1 ml of serum-reduced OPTI-MEM (Invitrogen) was added to each well. Cells were incubated in the transfection mixture for 6 h and further cultured in antibiotic-free DMEM. For Western blot analysis at various time intervals, the transfected cells were washed twice with phosphate-buffered saline (PBS; Invitrogen), flash frozen in liquid nitrogen, and stored at −80°C for analysis. For in vivo assays of Tat-mediated transactivation at 48 h posttransfection, Magi cells were subjected to β-Gal staining directly or were flash frozen in liquid nitrogen and stored at −80°C for the β-Gal assays described below.
Western blotting.
Cells treated with siRNA were harvested as described above and lysed in ice-cold reporter lysis buffer (Promega) containing protease inhibitors (complete, EDTA-free protease inhibitors; 1 tablet/10 ml of buffer) (Roche Molecular Biochemicals). After clearing of the resulting lysates by centrifugation, proteins in clear lysates were quantified by use of a Dc protein assay kit (Bio-Rad). Proteins in 60 μg of total cell lysate were resolved by SDS-10% PAGE, transferred onto a polyvinylidene difluoride membrane (PVDF; Bio-Rad), and immunoblotted with antibodies against hCycT1 and CDK9 (Santa Cruz). Protein contents were visualized with a BM chemiluminescence blotting kit (Roche Molecular Biochemicals). The blots were exposed to X-ray film (Kodak MR-1) for various times (30 s to 5 min).
RT-PCR for amplification of hCycT1 and CDK9 mRNA.
The total cellular mRNA was prepared from HeLa cells, with or without hCycT1 or CDK9 siRNA treatment, by using a Qiagen RNA mini kit followed by an Oligotex mRNA mini kit (Qiagen). RT-PCR was performed by using a SuperScript One-Step RT-PCR kit with platinum Taq (Invitrogen) and 40 cycles of amplification. Each RT-PCR mixture included 100 ng of total cellular mRNA, gene-specific primer sets for hCycT1 and CDK9 amplification (0.5 μM concentration of each primer), a 200 μM concentration of each deoxynucleoside triphosphate, 1.2 mM MgSO4, and 1 U of RT-platinum Taq mix. Primer sets for hCycT1 produced 2,178-bp products, while CDK9 primer sets produced 1,116-bp products. RT-PCR products were resolved in 1% agarose gels and viewed by ethidium bromide staining.
Plasmid harboring HIV-1 Tat sequence.
The pTat-RFP plasmid was constructed by fusing the DNA sequence of HIV-1 Tat with DNA sequences of DsRed1-N1, harboring coral (Discosoma sp.)-derived red fluorescent protein (RFP), per the manufacturer's recommendations (Clontech). The expression of the Tat-RFP fusion protein was driven by the cytomegalovirus promoter and was easily visualized in living cells by fluorescence microscopy (Zeiss). Tat-RFP fusion protein expression was quantified by directly exciting the RFP fluorophore in clear cell lysates and measuring the fluorescence, as described below.
β-Gal staining of cells.
Magi cells were transfected with Tat-containing plasmids in the absence or presence of siRNAs. At 48 h posttransfection, cells were washed twice with PBS and fixed for 5 min in fixative (1% formaldehyde and 0.2% glutaraldehyde in PBS) at room temperature. After two washes with PBS, cells were covered with staining solution (PBS containing 4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2, and 0.4 mg of X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] [Promega]/ml) and incubated at 37°C for exactly 50 min. Plates were washed twice with PBS. Cell counts represent numbers of β-Gal-positive (blue) cells per 100-power field.
β-Gal enzyme assay.
Magi cells were transfected with Tat-containing plasmids in the absence or presence of siRNAs. At 48 h posttransfection, cells were harvested and clear cell lysates were prepared and quantified as described above. The total cell lysate (120 μg) in reporter lysis buffer (150 μl) was subjected to a standard β-Gal assay by the addition of 150 μl of 2× β-Gal assay buffer (Promega) and incubation at 37°C for 30 min. The reactions were stopped by the addition of 500 μl of 1 M sodium carbonate and brief mixing on a vortex machine. The absorbance was read immediately at 420 nm. The same amount of cell lysate was subjected to fluorescence measurements in a Photon Technology International fluorescence spectrophotometer, with slit widths set at 4 nm for both excitation and emission wavelengths. All experiments were performed at room temperature. The fluorescence of Tat-RFP in the cell lysate was detected by excitation at 568 nm and recording of the emission spectrum from 588 to 650 nm; the spectrum peak at 583 nm represents the maximum fluorescence intensity of Tat-RFP. Tat transactivation was determined by calculating the ratio of β-Gal activity (absorbance at 420 nm) of pTat-RFP-transfected cells to that of cells without pTat-RFP plasmid treatment. The inhibitory effect of siRNA treatment was determined by normalizing the Tat transactivation activity to the amount of Tat-RFP protein (represented by RFP fluorescence intensity) in the presence and absence of siRNA.
In vivo fluorescence analysis.
A pEGFP-C1 reporter plasmid (1 μg) and siRNA (100 nM) were cotransfected into HeLa cells by use of Lipofectamine, as described above, except that cells were cultured in 35-mm-diameter plates with glass bottoms (MatTek Corporation, Ashland, Mass.) instead of standard 6-well plates. The fluorescence in living cells was visualized at 50 h posttransfection by conventional fluorescence microscopy (Zeiss). For green fluorescent protein (GFP) fluorescence detection, a fluorescein isothiocyanate filter was used.
HIV-1 replication assay.
HeLa CD4-LTR-β-Gal indicator (Magi) cells (27) were plated in 24-well plates (7.5 × 105 cells per well) and transfected with siRNAs as previously described (22). siRNAs (60 pmol) were transfected into cells by use of Oligofectamine (2 μl) (Invitrogen) for 3 h in serum-free DMEM (Gibco). Cells were rinsed twice, and 500 μl of DMEM-10% FBS was layered onto the cells. Sixteen hours after transfection, cells were trypsinized and seeded in 96-well microtiter plates (4 × 104 cells per well), incubated for 3 h, and infected. HIV-1 virions (normalized to the RT activity [in counts per minute]) were added in doubling dilutions to duplicate wells. At 36 h postinfection, cells were harvested for quantification of β-Gal activity.
Immunoprecipitation.
Protein extracts were prepared by sonicating cells in RIPA buffer (20 mM Tris-HCl [pH 8.0], 0.5% Nonidet P-40, 1% Triton X-100, 150 mM KCl, 5 mM dithiothreitol) containing protease inhibitors (1 tablet/10 ml) (Roche) and recombinant RNase inhibitor (1 U/μl) (Promega). Cell lysates (300 μg of protein) were immunoprecipitated with 3 μg of anti-CDK9 antibody (Santa Cruz) which had been adsorbed to protein G-Sepharose beads (Amersham Pharmacia Biotech) in RIPA buffer during an overnight incubation at 4°C. The beads were then washed three times with 300 μl of RIPA buffer containing 0.5% NP-40 and 1 M KCl and once with 300 μl of RIPA buffer. The beads were resuspended in 200 μl of RIPA buffer and split into two equal aliquots. One of the aliquots was treated with 10 U of RNase A (Amersham Pharmacia Biotech) for 15 min at 30°C. RNase A treatment was stopped by washing the beads with 300 μl of RIPA buffer three times. The second aliquot was not treated with RNase A. The beads, treated or not treated with RNase A, were then split into three equal aliquots, one for silver stain analysis, one for Western blot analysis, and one for kinase activity analysis.
Kinase activity analysis of immunoprecipitates.
Kinase assays were performed with anti-CDK9 immunoprecipitates (with or without RNase A treatment as described above) at 37°C for 1 h in a mixture of 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 60 mM NaCl, and 10 μM ATP and [γ-32P]ATP in a total volume of 45 μl. The reaction was terminated by the addition of 15 μl of 4× Laemmli sample buffer. The phosphorylated protein was visualized by autoradiography after electrophoresis in an SDS-10% polyacrylamide gel. The incorporation of 32P was quantified by phosphorimager analysis.
RESULTS
Specific silencing of P-TEFb expression by siRNA in HeLa cells.
For inhibition of hCycT1 and CDK9 expression in cultured human (HeLa) cell lines by RNAi, siRNAs targeting hCycT1 from positions 347 to 367 relative to the start codon and CDK9 from positions 258 to 278 relative to the start codon were constructed. HeLa cells were transfected by use of Lipofectamine with hCycT1 or CDK9 siRNA duplexes targeting either hCycT1 or CDK9. For analysis of the RNAi effects, lysates from siRNA duplex-treated cells were prepared at various times after transfection, and hCycT1 and CDK9 protein levels were examined by use of anti-hCycT1 and anti-CDK9 antibodies (Fig. 1B). An immunoblot analysis revealed that siRNAs targeting hCycT1 inhibited hCycT1 protein expression (Fig. 1B, lanes 8 to 14, upper panel) and siRNAs targeting CDK9 were similarly specific for inhibiting CDK9 expression (Fig. 1B, lanes 15 to 21, lower panel). This RNAi effect depended on the presence of a 21-nt duplex siRNA harboring a sequence complementary to the target mRNA, but not on the presence of single-stranded antisense siRNAs (data not shown) or an unrelated control siRNA which targeted coral (Discosoma sp.)-derived RFP (Fig. 1B, lanes 1 to 7). As a specificity control, cells were also transfected with mutant siRNAs (mismatched siRNAs) for hCycT1 or CDK9 which had 2-nt mismatches between the target mRNA and the antisense strand of the siRNA at the putative cleavage site of the mRNA. These mutant siRNAs showed no RNAi activity (Fig. 1B, lanes 22 to 35), indicating the specificity of the RNAi effect.
Immunoblot experiments (Fig. 1B) using anti-hCycT1 and anti-CDK9 antibodies revealed the kinetics of gene suppression and the persistence of RNAi in HeLa cells. Although RNAi suppressed the expression of hCycT1 and CDK9 proteins up to 66 h posttransfection, maximum activities were observed at 42 to 54 h, and inhibition by siRNAs did not persist. After reaching a maximal activity at 42 to 54 h posttransfection, RNAi started to decrease at 54 h, with protein levels gradually recovering to normal between 66 and 90 h (3 to 4 days) posttransfection (data not shown). Similar phenomena were demonstrated in 293T cells, a kidney epithelial cell line (data not shown). The recovery of target gene expression in both cell lines indicated that RNAi of CDK9 or hCycT1 by exogenous siRNA duplexes lasted comparably to that of other genes silenced in mammalian cells (51) and did not last indefinitely.
CDK9 protein stability depends on the presence of hCycT1.
An intriguing finding revealed by the above analysis of hCycT1 knockdown was that cells transfected with hCycT1 siRNA had significantly down-regulated CDK9 protein levels. In fact, the kinetics of CDK9 knockdown by hCycT1 siRNA showed a pattern similar to the kinetics of hCycT1 knockdown (Fig. 1B, lanes 8 to 14, lower panel). CDK9 knockdown by hCycT1 siRNA was most likely not due to cleavage of CDK9 mRNA through the RNAi pathway because homologous sequences have not been found between the CDK9 mRNA and hCycT1 siRNA used in this assay. However, two other possibilities could explain this unexpected down-regulation. First, hCycT1 knockdown may have affected CDK9 protein stability. If protein-protein contacts between CDK9 and hCycT1 are involved in forming a stable P-TEFb complex, these hCycT1-CDK9 interactions may be required to stabilize CDK9 in the cell. Second, since P-TEFb is a positive transcription factor, it is possible that the P-TEFb complex is required for transcription of the CDK9 gene. If this were the case, down-regulation of hCycT1 by hCycT1 siRNA would decrease the amount of P-TEFb complexes in the cell available for transcription and in turn down-regulate intracellular CDK9 transcription.
To distinguish between these two proposed hypotheses and determine the specificity of P-TEFb knockdown by siRNAs at the mRNA level, we performed RT-PCR to evaluate the effect of siRNAs on P-TEFb mRNA expression. As shown in Fig. 1C, cells transfected with siRNA duplexes targeting hCycT1 (hCycT1 ds siRNA) had significantly reduced hCycT1 mRNA expression (Fig. 1C, lanes 1 to 7, upper panel), but there was no effect on CDK9 mRNA (Fig. 1C, lanes 1 to 7, lower panel). On the other hand, the transfection of cells with siRNA duplexes targeted to CDK9 (CDK9 ds siRNA) significantly interfered with the expression of CDK9 mRNA, but not hCycT1 (Fig. 1C, lanes 8 to 14). These results suggest that hCycT1 knockdown did not result in decreased transcription of CDK9 mRNA. The siRNA duplex started to cause an RNAi effect as early as 6 to 18 h posttransfection and gradually increased with time, peaking at 30 h and then decreasing between 54 and 66 h. Protein levels were not down-regulated until 18 to 30 h post-siRNA transfection (Fig. 1B), indicating that there was a lag time between the degradation of target mRNA (starting at 6 h post-siRNA transfection, as shown by semiquantitative RT-PCR [Fig. 1C]) and the half-life of the existing protein expressed by the target gene. These results demonstrated that hCycT1 knockdown specifically affected the protein stability of CDK9. Collectively, these studies confirmed the specific knockdown of P-TEFb by siRNA at the mRNA level and suggested that the formation of a complex with hCycT1 was a prerequisite for CDK9 protein stability in living cells.
Knockdown of hCycT1 and CDK9 is not lethal to HeLa cells.
For analysis of the viability of cells subjected to P-TEFb gene silencing, a pEGFP-C1 reporter plasmid, harboring enhanced GFP under the control of the cytomegalovirus immediate-early promoter, plus hCycT1 and CDK9 siRNAs were cotransfected into HeLa cells by use of Lipofectamine. GFP reporter gene expression and cellular morphology and density were then monitored by fluorescence and phase-contrast microscopy, respectively. GFP expression was not affected by hCycT1 or CDK9 knockdown (Fig. 2A, panels a to c), indicating that normal levels of transcription occurred during P-TEFb gene silencing. Cells with P-TEFb knockdown were also normal in both shape and growth rate, reaching a cell density of ∼90 to 100% confluence at 50 h posttransfection (Fig. 2A, panels e to g).
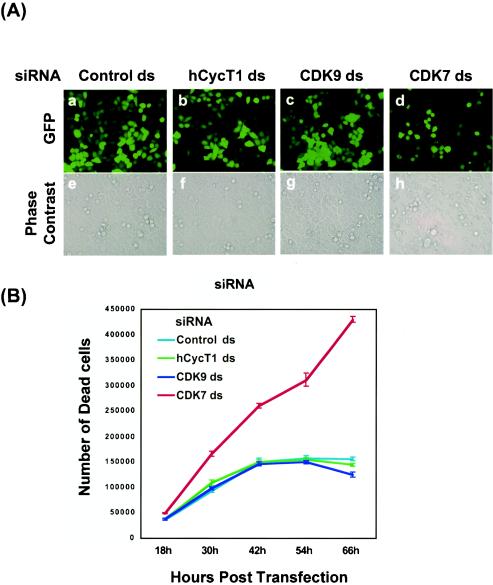
FIG. 2.
P-TEFb silencing is not lethal to HeLa cells. (A) Analysis of cell viability by in vivo fluorescence analysis. HeLa cells were cotransfected by use of Lipofectamine with a pEGFP-C1 reporter (GFP) plasmid and siRNAs (see Materials and Methods). Four siRNA duplexes, including a control duplex targeting RFP (panels a and e) and three duplexes targeting hCycT1 (panels b and f), CDK9 (panels c and g), and CDK7 (panels d and h), were used in these experiments. Reporter gene expression was monitored at 50 h posttransfection by fluorescence imaging of living cells (upper panels). Cellular shapes and densities were recorded by phase-contrast microscopy (lower panels). (B) Analysis of cell viability by counting of trypan blue-stained cells. HeLa cells were cotransfected by use of Lipofectamine with a pEGFP-C1 reporter (GFP) plasmid and siRNAs (see Materials and Methods). Four siRNA duplexes, including a control unrelated duplex (light blue) and three duplexes targeting hCycT1 (green), CDK9 (dark blue), and CDK7 (red), were used in these experiments. At various times after transfection, cells floating in the medium were collected and counted in the presence of 0.2% trypan blue. Cells that took up dye (stained blue) were not viable.
For a comparison of the seemingly normal appearance of P-TEFb knockdown cells to that of cells transfected with siRNA targeting an mRNA likely to be required for cell viability, a siRNA targeting CDK7 was designed. The CDK7 kinase is required for stimulation of the transcription factor TFIIH to phosphorylate the RNA Pol II CTD at the step of promoter clearance during transcription initiation. The Saccharomyces cerevisiae CDK7 homolog Kin28 is an essential gene product that phosphorylates Ser5 of the CTD YSPTSPS repeat region (28, 47, 49) and is required for recruitment of the mRNA capping enzyme to the transcription machinery (6, 33, 34, 60). Similarly, in larger eukaryotes, CDK7 is a bifunctional enzyme that promotes both CDK activation and transcription (15), suggesting that, like Kin28 expression in yeast, CDK7 expression is essential in mammalian cells. As predicted, the reduction of CDK7 levels by RNAi led to lower reporter (GFP) expression and an arrest in cellular growth (Fig. 2A, panel d). Moreover, CDK7 knockdown cells were smaller than control cells and showed blebbing (Fig. 2A, panel h), indicating that unlike RNAi of P-TEFb, CDK7 gene silencing had an adverse affect on transcription, cell morphology, and cell growth.
The cell viability of P-TEFb and CDK7 knockdown cells was also analyzed. At various times after siRNA transfection of the various siRNAs, cell viability was assessed by trypan blue exclusion. Over a 66-h time course experiment, the rate of cell death in P-TEFb (hCycT1 or CDK9) knockdown cells was comparable to that in control cells with an unrelated siRNA treatment (Fig. 2B). In contrast, CDK7 knockdown cells showed a significant increase in cell death (Fig. 2B). These results indicated that P-TEFb knockdown was not lethal to HeLa cells, while a much more stringent threshold for CDK7 expression was required to maintain cell viability and growth.
hCycT1 and CDK9 RNAi inhibit HIV-1 Tat transactivation in HeLa cells.
A dominant paradigm for Tat up-regulation of HIV gene expression revolves around the ability of the Tat-TAR RNA complex to bind P-TEFb and stimulate phosphorylation of the CTD and Spt5 to override the elongation arrest caused by DSIF and NELF (40, 42). To test whether gene silencing of P-TEFb would specifically block Tat transactivation, we evaluated P-TEFb knockdown in a Magi cell line. Magi cells were derived from a HeLa cell line harboring a single copy of a persistently transfected HIV-1 LTR β-Gal gene and were programmed to express the CD4 receptor and the CCR5 coreceptor for HIV-1, making them a model cell line for measuring HIV replication (27). For analysis of Tat transactivation in the presence of various siRNAs, the Tat expression construct pTat-RFP, hCycT1 or CDK9 double-stranded siRNA, antisense hCycT1 or CDK9 siRNA, mutant hCycT1 or CDK9 siRNA, or a non-P-TEFb duplex siRNA control was transfected into Magi cells. An immunoblot analysis with anti-RFP antibody confirmed that the HIV-1 Tat-RFP fusion protein was expressed under the control of the cytomegalovirus early promoter in all transfected cells (data not shown). Tat transactivation was determined by calculating the ratio of β-Gal activity in pTat-RFP transfected cells to the β-Gal activity of cells without pTat-RFP treatment. Inhibitory activity was determined by normalizing the Tat transactivation activity to the amount of Tat-RFP protein (represented by RFP fluorescence intensity as described in Materials and Methods) in the presence and absence of siRNA.
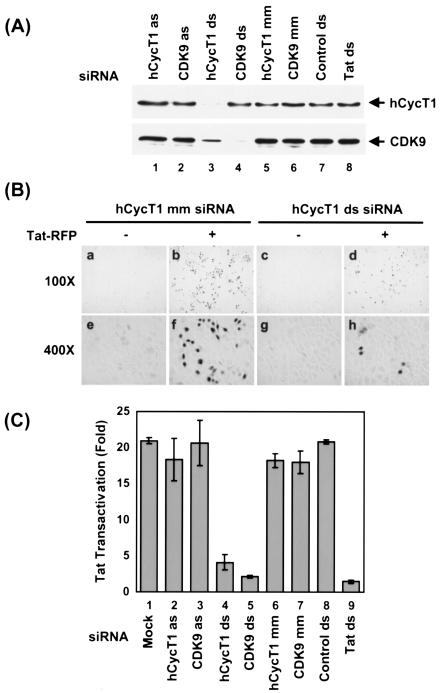
Under standard experimental conditions, Tat-RFP alone strongly enhanced β-Gal gene expression (20- to 25-fold) from the HIV-1 LTR promoter (Fig. 3C, bar 1). This activation was strongly inhibited by cotransfection of host Magi cells with duplex siRNAs specifically targeting hCycT1 and CDK9 (Fig. 3C, bars 4 and 5), but not with antisense (as) RNA strands (Fig. 3C, bars 2 and 3), mutant (mm) siRNAs (Fig. 3C, bars 6 and 7), or an unrelated control siRNA (Fig. 3C, bar 8). Specific RNAi of hCycT1 and CDK9 expression in Magi cells was demonstrated by immunoblot analysis (Fig. 3A, lanes 3 and 4), and the inhibition of Tat transactivation correlated well with the knockdown of hCycT1 and CDK9 protein levels by hCycT1 and CDK9 siRNAs (compare Fig. 3C, bars 4 and 5, to 3A, lanes 3 and 4). As shown in Fig. 3B, LTR activation (represented by cells with blue β-Gal staining) was reduced in hCycT1 double-stranded siRNA-treated cells (Fig. 3B, compare panels d and b and panels h and f). Altogether, these results indicated that siRNA targeting P-TEFb inhibited Tat transactivation in HeLa cells without affecting cellular viability.
FIG. 3.
hCycT1 and CDK9 duplex siRNAs inhibit HIV-1 Tat transactivation in Magi cells. (A) Analysis of hCycT1 and CDK9 RNAi activities in Magi cells by Western blotting. Magi cells were cotransfected with pTat-RFP plasmid and various siRNAs. Cells were harvested at 48 h posttransfection. Proteins were resolved by SDS-10% PAGE, transferred onto PVDF membranes, and immunoblotted with antibodies against hCycT1 (upper panel) and CDK9 (lower panel). Lanes 1 and 2, RNAi activities in Magi cells treated with antisense (as) strands of hCycT1 and CDK9 siRNAs; lanes 3 and 4, RNAi activities of cells treated with double-stranded siRNAs targeting hCycT1 and CDK9; lanes 5 and 6, RNAi activities in cells treated with mutant hCycT1 siRNA (hCycT1 mm) or mutant CDK9 siRNA (CDK9 mm). A double-stranded GFP siRNA was used as an unrelated control (lane 7), while double-stranded Tat siRNA was used to target mRNA encoding Tat (lane 8). (B) Photomicrographs of β-Gal-stained Magi cells. Magi cells were untransfected (panels a, c, e, and g) or transfected (panels b, d, f, and h) with pTat-RFP in the presence of mismatched hCycT1 siRNA (mm) (panels b and f) or hCycT1 double-stranded siRNA (panels d and h). LTR activation (represented by β-Gal-stained cells) was reduced in the hCycT1 double-stranded siRNA-treated cells (panels d and h). (C) Effect of P-TEFb silencing by RNAi on Tat transactivation in Magi cells. Twenty-four hours after pretreatment of Magi cells with siRNA, the cells were cotransfected with pTat-RFP plasmid and various siRNAs. Cells were harvested at 48 h post-pTat-RFP transfection, and the activity of β-Gal in clear cell lysates was measured (see Materials and Methods). Tat transactivation was determined by the ratio of β-Gal activity in pTat-RFP-transfected cells to that in cells without pTat-RFP treatment. Inhibitory activity was determined by normalizing the Tat transactivation activity to the amount of Tat-RFP protein (see Materials and Methods) in the presence or absence of siRNA treatment. Bar 1, Tat-RFP transfection (mock). Magi cells were cotransfected with double-stranded siRNAs targeting hCycT1 and CDK9 (bars 4 and 5), with antisense (as) RNA strands (bars 2 and 3), or with mutant (mm) siRNAs (bars 6 and 7). Double-stranded GFP siRNA was used as an unrelated control (bar 8), while a double-stranded Tat siRNA targeting the mRNA encoding the Tat sequence was used as a positive control (bar 9). Means ± standard deviations (SD) of two experiments are shown.
hCycT1 and CDK9 RNAi inhibit HIV-1 replication.
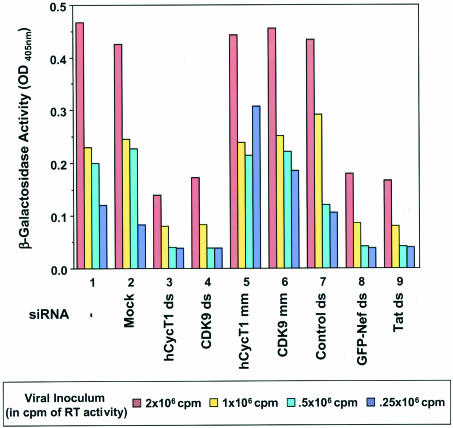
The next question addressed was whether targeting the human P-TEFb complex with RNAi inhibited HIV replication. For investigation of this question, Magi cells were transfected with homologous and mismatched siRNAs directed against hCycT1 or CDK9 and 16 h later were infected with various concentrations of HIVNL-GFP, an infectious molecular clone of HIV-1. HIV-1 Tat-mediated transactivation of the LTR led to β-Gal production and was quantified at 36 h postinfection. For this single-cycle replication assay, β-Gal activity reflected the activity of RT and viral replication of various amounts of viral inoculum so that changes in β-Gal activity could be directly correlated to changes in the efficacy of HIV replication. Positive controls were siRNAs targeting HIV Nef and Tat that previously showed decreased levels of β-Gal activity and viral infectivity (22). As expected, these control siRNAs exhibited a significant decrease in β-Gal activity in our assay compared to untreated or mock-treated cells and also showed a reduced level of viral replication even with the largest amount of viral inoculum (Fig. 4). Double-stranded siRNA directed against hCycT1 or CDK9 showed a similar decrease in β-Gal activity when compared with the Nef or Tat knockdown (Fig. 4). This observed decrease activity was equivalent to the β-Gal activity measured when eight times less viral inoculum was used with untransfected or mock-treated cells (Fig. 4), indicating that the P-TEFb knockdown had significantly reduced HIV replication. In addition, we monitored p24 levels during these experiments and observed a decrease in p24 in the context of P-TEFb knockdown (data not shown), further supporting the conclusion that the P-TEFb knockdown inhibited HIV replication. Control experiments using hCycT1 or CDK9 siRNA duplexes containing mismatched sequences or an unrelated double-stranded siRNA against the RFP sequence showed no antiviral activities. In addition, no significant toxicity or cell death was observed during these experiments, further suggesting that P-TEFb knockdown is not lethal. These results demonstrated that HIV replication is modulated by siRNAs targeting P-TEFb component CycT1 or CDK9.
FIG. 4.
siRNAs targeting CycT1 or CDK9 modulate HIV-1 replication. HeLa CD4-LTR-β-Gal (Magi) cells were transfected with homologous (ds; bars 3 and 4) and mismatched (mm; bars 5 and 6) siRNAs directed against CycT1 or CDK9. Cells were also mock transfected without siRNA (bar 2) or transfected with an unrelated double-stranded siRNA against the RFP sequence (bar 7). Sixteen hours later, cells were infected with HIVNL-GFP, an infectious molecular clone of HIV-1. Data for cells infected with virus and not treated with Oligofectamine are shown with bar 1. HIV-1 Tat-mediated transactivation of the LTR led to β-Gal production, which was quantified at 36 h postinfection. Cells treated with double-stranded siRNAs targeting Nef (bar 8; note that in this clone, Nef is fused to GFP, as previously reported [22]) and targeting the mRNA encoding the Tat sequence (lane 9) served as positive controls. Serial double dilutions of the viral inoculum (RT activity in counts per minute) are consistent with eightfold decreases in viral replication.
P-TEFb kinase activity remains unchanged in P-TEFb knockdown cells.
The observation that knockdown of P-TEFb did not lead to lethality was a surprising result, as P-TEFb plays an important role in transcription elongation and a P-TEFb knockdown was previously shown to inhibit the early embryonic development of Caenorhabditis elegans (50). There were a number of possible explanations for these findings. One possibility was that mRNA and protein levels were significantly decreased in HeLa cells transfected with siRNAs, but the low levels of P-TEFb that remained during RNAi may have been sufficient for sustaining cell viability and growth. Clarifying this further, cells may have compensated for lower P-TEFb levels by converting inactive forms of the kinase to an activated form. This would suggest that a shift in equilibrium between inactive and active forms of P-TEFb may have occurred during hCycT1 or CDK9 knockdown that ultimately allowed cells to survive with lower levels of P-TEFb.
Recent studies have shown that P-TEFb is inhibited when it is in a complex with 7SK RNA and this 7SK-P-TEFb complex is the predominant form of P-TEFb present in the cell (37, 59). If cells compensate for decreased P-TEFb levels by converting inactive 7SK-P-TEFb complexes to active P-TEFb complexes, then P-TEFb kinase activity in P-TEFb knockdown cells should be equal to the kinase activity in cells without P-TEFb knockdown. To determine if levels of P-TEFb kinase activity were knocked down concomitantly with protein levels during RNAi, we evaluated P-TEFb kinase activity over the course of an hCycT1 knockdown time course experiment (see experimental design in Fig. 5A).
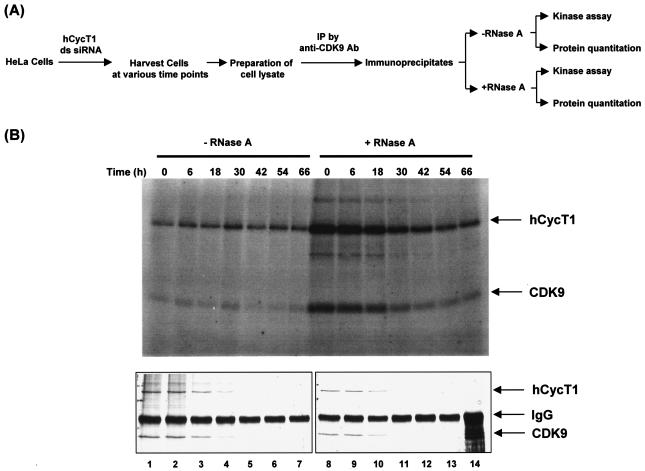
FIG. 5.
Evaluation of P-TEFb kinase activity in P-TEFb knockdown cells. (A) Experimental procedure for assaying P-TEFb kinase activity in cells with or without hCycT1 siRNA treatment. See Materials and Methods for details. IP, immunoprecipitation. (B) Kinase activity of P-TEFb. P-TEFb and its associated factors were affinity purified (anti-CDK9 immunoprecipitation) from HeLa cell extracts and were treated (lanes 8 to 14) or not treated (lanes 1 to 7) with RNase A as outlined in panel A. Kinase assays were performed with anti-CDK9 immunoprecipitates at 37°C for 1 h in a solution of 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 60 mM NaCl, and 10 μM ATP and [γ-32P]ATP in a total volume of 45 μl. The reaction was terminated by the addition of 15 μl of 4× Laemmli sample buffer. Phosphorylated proteins were visualized by autoradiography after electrophoresis in an SDS-10% polyacrylamide gel (upper panel). hCycT1 and CDK9 proteins in the immunoprecipitates were eluted with SDS, resolved by SDS-10% PAGE, and stained with a Bio-Rad silver stain plus kit (bottom panel). The specificity of the protein bands was confirmed by immunoblotting with anti-hCycT1 or anti-CDK9 antibody (data not shown).
We used immunoprecipitation with an anti-CDK9 antibody to isolate P-TEFb complexes to ensure the specificity of P-TEFb-associated kinase activity in our assays. Briefly, P-TEFb and its associated factors were affinity purified by anti-CDK9 immunoprecipitation from HeLa cell extracts at various time points after hCycT1 siRNA transfection. Immunoprecipitates were then treated (Fig. 5B, lanes 8 to 14, upper panel) or not treated (Fig. 5B, lanes 1 to 7, upper panel) with RNase A. Both the RNase A-treated and nontreated immunoprecipitates were split into three equal pools to evaluate the kinase activity and protein levels of P-TEFb isolated by immunoprecipitation. Kinase assays were performed on the anti-CDK9 immunoprecipitates at 37°C for 1 h. hCycT1 and CDK9 proteins in the immunoprecipitates were eluted with SDS and resolved by SDS-10% PAGE to evaluate P-TEFb autophosphorylation. The specificity of the protein bands was confirmed by immunoblotting with anti-hCycT1 or anti-CDK9 antibody (data not shown). For evaluation of P-TEFb knockdown and of protein levels of the immunoprecipitates used in the kinase assay, hCycT1, CDK9, and anti-CDK9 immunoglobulin G (IgG) were visualized by silver staining. As shown in Fig. 5B (bottom panels), the silver stain showed that both hCycT1 and CDK9 levels had been knocked down during the time course experiment (time = 30 to 66 h) and that the same amount of immunoprecipitate was loaded for each time point (IgG band).
The P-TEFb kinase activity of cells transfected with hCycT1 siRNA and showing hCycT1 and CDK9 knockdown between 30 and 66 h was compared to the P-TEFb kinase activity at the time of transfection (time = 0 h). Time zero was prior to any observable P-TEFb knockdown and should be representative of normal levels of P-TEFb kinase activity. The observed kinase activities of immunoprecipitates that were not treated with RNase A were quantitatively the same at all time points, despite the reduction in hCycT1 protein levels observed over time (Fig. 5B, lanes 1 to 7). These results indicated that although P-TEFb protein levels were knocked down in cells transfected with hCycT1 or CDK9 siRNA, the P-TEFb kinase activity was not significantly affected. These observations therefore support the hypothesis that cells are able to survive P-TEFb knockdown because the level of P-TEFb kinase activity normally present in cells was retained even after hCycT1 or CDK9 knockdown.
The kinase activities of immunoprecipitates that were treated with RNase A prior to the kinase assay were also compared at time zero and at various time points post-siRNA transfection. Immunoprecipitates from time zero that were treated with RNase A showed a significant increase in kinase activity compared to immunoprecipitates from time zero that were not treated with RNase A (Fig. 5B, compare lanes 1 and 8). This increase in activity indicated that 7SK RNA had been degraded by RNase A, relieving the inhibition of P-TEFb kinase activity, as was observed previously (37, 59). At later time points, the increase in P-TEFb kinase activity began to decrease as P-TEFb protein levels decreased. However, through the last time points of the experiment, when the highest levels of P-TEFb knockdown were sustained, P-TEFb kinase activity never decreased to levels below the kinase activities observed with immunoprecipitates that were not treated with RNase A (Fig. 5B, compare lanes 5 to 7 and 12 to 14). This result suggested that there was a critical threshold of kinase activity that was consistently maintained during P-TEFb knockdown in the presence or absence of 7SK. These results also indicated that in the presence of 7SK, a shift in the equilibrium between pools of kinase-inactive 7SK-P-TEFb and kinase-active P-TEFb complexes occurred to achieve the critical threshold required for viability. Altogether, these results strongly support the hypothesis that the lower level of P-TEFb protein expressed during RNAi was sufficient for sustaining a critical threshold of P-TEFb kinase activity required for the performance of essential cellular functions.
DISCUSSION
The results presented here provided new mechanistic data about the regulation of P-TEFb kinase activity in response to changes in protein expression during RNAi. First, this analysis demonstrated that P-TEFb mRNA and protein levels could be knocked down with siRNAs targeted to either CDK9 or hCycT1. Another important discovery was that CDK9 protein levels depended on hCycT1 protein levels, indicating that the formation of the P-TEFb complex is essential for the stabilization of CDK9 in cells. Interestingly, CDK9 levels, although showing a striking initial decline, seemed to stabilize instead of steadily declining further over time along with hCycT1 (Fig. 1B, lanes 8 to 14), suggesting that a separate pool of CDK9 was bound to other proteins that could stabilize the kinase subunit. CDK9 has been shown to form chaperone complexes with the heat shock protein Hsp70 and the kinase-specific chaperone Hsp90/Cdc37 (39), raising the possibility that the pool of CDK9 still present during hCycT1 knockdown may have been stabilized by these chaperone interactions.
The most surprising finding of this study was that cells survived knockdown of P-TEFb by maintaining normal kinase activity in the wake of reduced protein expression. The cell viability associated with RNAi of P-TEFb could have suggested that P-TEFb function was not essential for cell viability, a conclusion that was contradictory to its central role in transcription elongation and its essential roles in transcription during C. elegans embryonic development (50). At later stages in the life cycle, however, it was still not entirely clear whether the complete knockout of P-TEFb was lethal to cells. Insight into whether P-TEFb has some essential functions in adult cells was revealed by the studies herein. With P-TEFb kinase activity remaining virtually the same under both normal and P-TEFb knockdown conditions, significantly reducing P-TEFb protein levels did not appear to limit its ability to carry out essential functions. However, our kinase assays indicated that there is a critical threshold of P-TEFb kinase activity that is required to sustain cell viability.
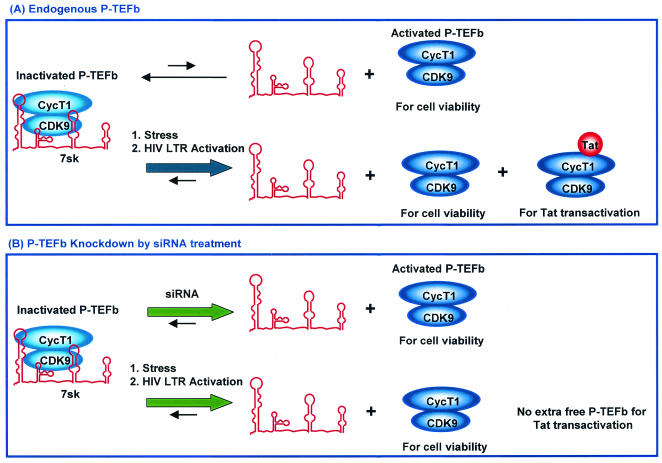
The mechanism by which cells appeared to compensate for lower P-TEFb protein levels, via converting kinase-inactive P-TEFb to active P-TEFb, suggested that at the very least P-TEFb kinase activity was essential for cell viability. With such a compensatory mechanism in place, however, cells must first have a system for monitoring P-TEFb proteins levels and second have a mechanism established for signaling the conversion of inactive P-TEFb to kinase-active P-TEFb. Recent studies that uncovered an inhibitory role for 7SK RNA in regulating P-TEFb activity (37, 59) may explain in part how the switch between inactive and active forms is signaled. Under normal conditions, 7SK interacts with the majority of P-TEFb available in the cell, rendering it inactive, an assertion supported by biochemical data documenting strong interactions between 7SK and P-TEFb and its negative effect on P-TEFb kinase activity (37, 48, 59). A shift in this standard equilibrium between inactive P-TEFb-7SK complexes and active P-TEFb may then be triggered in several ways, as depicted in Fig. 6 and described below.
FIG. 6.
Model for how siRNA-mediated P-TEFb silencing modulates HIV-1 transcription without causing a major lethal effect on host cells. See the text for details. (A) Endogenous P-TEFb. (B) P-TEFb knockdown by siRNA treatment.
One possible trigger may result from exposure to a stressor, such as UV irradiation or the transcriptional inhibitor actinomycin D, which induces the release of 7SK from P-TEFb and increases the levels of kinase-active P-TEFb in the cell (37, 59). Recently, cardiac hypertrophy signaling pathways have also been shown to cause the release of P-TEFb from the 7SK inhibitor (48). Similarly, HIV may elicit a shift in the equilibrium between P-TEFb and its interactions with 7SK to increase Tat transactivation. Our results showing that HIV replication is reduced despite normal levels of P-TEFb kinase activity suggested that Tat transactivation associated with normal rates of HIV replication requires a higher concentration of activated P-TEFb than is normally available in the cell. Therefore, HIV has likely developed a signaling mechanism for shifting the equilibrium between 7SK and P-TEFb. Although it is clear that there must be a signaling mechanism involved in regulating 7SK-P-TEFb interactions, what the signaling components are and how they associate with the 7SK-P-TEFb complexes to trigger release of P-TEFb remain unknown.
Although knockdown of P-TEFb was able to support cell viability and showed no significant growth defects, P-TEFb knockdown did have an effect on gene expression. Most significantly, RNAi of P-TEFb resulted in a decrease in both Tat transactivation and HIV replication, suggesting that the amount of P-TEFb in cells is important for the fidelity of Tat transactivation and HIV replication. Previous studies, including P-TEFb immunodepletion analyses and studies with small-molecule inhibitors of P-TEFb or anti-hCycT1 intrabodies, also supported an important role for P-TEFb in Tat transactivation and HIV replication (1, 4, 11, 14, 30). However, none of these studies specifically showed the effects of altering P-TEFb protein levels in vivo. The studies herein are the first to show that targeting of P-TEFb by RNAi is not lethal and also decreases HIV transcription and viral replication. These results suggest that the use of siRNAs to target P-TEFb is a feasible new approach for the development of therapeutic agents for AIDS.
In addition to the P-TEFb RNAi effects on HIV replication, changes in global gene expression observed in microarray analyses demonstrated that knockdown of P-TEFb also has an effect on the expression of a variety of other genes (Y. L. Chiu and T. M. Rana, unpublished results). A microarray analysis showed that P-TEFb is required for the regulation of expression of a broad range of genes involved in a host of different cellular processes, including cell division, development, and the stress response. These results provided additional evidence that P-TEFb knockdown occurred and indicated the cellular concentration of activated P-TEFb-regulated expression of a broad range of genes. These data also suggested that only a small fraction of P-TEFb regulatory functions have truly been defined, and more directed studies of the genes and related cell processes controlled by P-TEFb will be performed in the future.
The results presented here raise new and intriguing questions about P-TEFb function and how it is regulated in the cell. This analysis suggests that there is a critical threshold for P-TEFb kinase activity that is required for cell viability and Tat transactivation and that there are built-in intracellular mechanisms that allow cells to cope with changes in P-TEFb protein levels. These results also demonstrate that knockdown of P-TEFb by RNAi is a novel tool that can be used to understand P-TEFb cellular functions and the functions of genes regulated by P-TEFb.
Acknowledgments
We thank Tamara J. Richman for editorial assistance and Bryan Cullen, B. Matija Peterlin, and David Price for helpful discussions. We also acknowledge assay support provided by the University of Massachusetts Center for AIDS Research.
This work was supported by grants from the NIH to T.M.R. (AI43198 and AI41404) and M.S. (AI37475 and P30AI42845).
REFERENCES
- 1.Bai, J., J. Sui, R. Y. Zhu, A. S. Tallarico, F. Gennari, D. Zhang, and W. A. Marasco. 2003. Inhibition of Tat-mediated transactivation and HIV-1 replication by human anti-hCyclinT1 intrabodies. J. Biol. Chem. 278:1433-1442. [DOI] [PubMed] [Google Scholar]
- 2.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 96:7791-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, S. H., K. Fujinaga, J. E. Marion, R. Taube, E. A. Sausville, A. M. Senderowicz, B. M. Peterlin, and D. H. Price. 2000. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275:28345-28348. [DOI] [PubMed] [Google Scholar]
- 5.Chiu, Y. L., and T. M. Rana. 2002. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell 10:549-561. [DOI] [PubMed] [Google Scholar]
- 6.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 1998. HIV-1 auxiliary proteins: making connections in a dying cell. Cell 93:685-692. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 10.Emerman, M., and M. H. Malim. 1998. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science 280:1880-1884. [DOI] [PubMed] [Google Scholar]
- 11.Flores, O., G. Lee, J. Kessler, M. Miller, W. Schlief, J. Tomassini, and D. Hazuda. 1999. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc. Natl. Acad. Sci. USA 96:7208-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghose, R., L. Y. Liou, C. H. Herrmann, and A. P. Rice. 2001. Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4+ T lymphocytes by combination of cytokines. J. Virol. 75:11336-11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper, J. W., and S. J. Elledge. 1998. The role of Cdk7 in CAK function, a retro-retrospective. Genes Dev. 12:285-289. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrmann, C. H., and A. P. Rice. 1993. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology 197:601-608. [DOI] [PubMed] [Google Scholar]
- 18.Hutvagner, G., and P. D. Zamore. 2002. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12:225-232. [DOI] [PubMed] [Google Scholar]
- 19.Isel, C., and J. Karn. 1999. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J. Mol. Biol. 290:929-941. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanov, D., Y. T. Kwak, E. Nee, J. Guo, L. F. Garcia-Martinez, and R. B. Gaynor. 1999. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for Tat-activation. J. Mol. Biol. 288:41-56. [DOI] [PubMed] [Google Scholar]
- 22.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeang, K. T., H. Xiao, and E. A. Rich. 1999. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274:28837-28840. [DOI] [PubMed] [Google Scholar]
- 24.Jones, K. A. 1997. Taking a new TAK on Tat transactivation. Genes Dev. 11:2593-2599. [DOI] [PubMed] [Google Scholar]
- 25.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J. B., and P. A. Sharp. 2001. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276:12317-12323. [DOI] [PubMed] [Google Scholar]
- 27.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 30.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez, M. A., B. Clotet, and J. A. Este. 2002. RNA interference of HIV replication. Trends Immunol. 23:559-561. [DOI] [PubMed] [Google Scholar]
- 32.Martinez, M. A., A. Gutierrez, M. Armand-Ugon, J. Blanco, M. Parera, J. Gomez, B. Clotet, and J. A. Este. 2002. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS 16:2385-2390. [DOI] [PubMed] [Google Scholar]
- 33.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 35.McManus, M. T., B. B. Haines, C. P. Dillon, C. E. Whitehurst, L. van Parijs, J. Chen, and P. A. Sharp. 2002. Small interfering RNA-mediated gene silencing in T lymphocytes. J. Immunol. 169:5754-5760. [DOI] [PubMed] [Google Scholar]
- 36.McManus, M. T., and P. A. Sharp. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3:737-747. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 38.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 39.O'Keeffe, B., Y. Fong, D. Chen, S. Zhou, and Q. Zhou. 2000. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated Tat stimulation of HIV-1 transcription. J. Biol. Chem. 275:279-287. [DOI] [PubMed] [Google Scholar]
- 40.Ping, Y. H., and T. M. Rana. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276:12951-12958. [DOI] [PubMed] [Google Scholar]
- 41.Ping, Y. H., and T. M. Rana. 1999. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J. Biol. Chem. 274:7399-7404. [DOI] [PubMed] [Google Scholar]
- 42.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rana, T. M., and K. T. Jeang. 1999. Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA. Arch. Biochem. Biophys. 365:175-185. [DOI] [PubMed] [Google Scholar]
- 44.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 45.Richter, S., H. Cao, and T. M. Rana. 2002. Specific HIV-1 TAR RNA loop sequence and functional groups are required for human cyclin T1-Tat-TAR ternary complex formation. Biochemistry 41:6391-6397. [DOI] [PubMed] [Google Scholar]
- 46.Richter, S., Y. H. Ping, and T. M. Rana. 2002. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA 99:7928-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez, C. R., E. J. Cho, M. C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sano, M., M. Abdellatif, H. Oh, M. Xie, L. Bagella, A. Giordano, L. H. Michael, F. J. DeMayo, and M. D. Schneider. 2002. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 8:1310-1317. [DOI] [PubMed] [Google Scholar]
- 49.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surabhi, R. M., and R. B. Gaynor. 2002. RNA interference directed against viral and cellular targets inhibits human immunodeficiency virus type 1 replication. J. Virol. 76:12963-12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 53.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 56.Wimmer, J., K. Fujinaga, R. Taube, T. P. Cujec, Y. Zhu, J. Peng, D. H. Price, and B. M. Peterlin. 1999. Interactions between Tat and TAR and human immunodeficiency virus replication are facilitated by human cyclin T1 but not cyclins T2a or T2b. Virology 255:182-189. [DOI] [PubMed] [Google Scholar]
- 57.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 58.Yang, X., M. O. Gold, D. N. Tang, D. E. Lewis, C. E. Aguilar, A. P. Rice, and C. H. Herrmann. 1997. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc. Natl. Acad. Sci. USA 94:12331-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 60.Yue, Z., E. Maldonado, R. Pillutla, H. Cho, D. Reinberg, and A. J. Shatkin. 1997. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:12898-12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]