Abstract
Integrase interactor 1 (INI1)/hSNF5 is a host factor that directly interacts with human immunodeficiency virus type 1 (HIV-1) integrase and is incorporated into HIV-1 virions. Here, we show that while INI1/hSNF5 is completely absent from purified microvesicular fractions, it is specifically incorporated into HIV-1 virions with an integrase-to-INI1/hSNF5 stoichiometry of approximately 2:1 (molar ratio). In addition, we show that INI1/hSNF5 is not incorporated into related primate lentiviral and murine retroviral particles despite the abundance of the protein in producer cells. We have found that the specificity in incorporation of INI1/hSNF5 into HIV-1 virions is directly correlated with its ability to exclusively interact with HIV-1 integrase but not with other retroviral integrases. This specificity is also reflected in our finding that the transdominant mutant S6, harboring the minimal integrase interaction domain of INI1/hSNF5, blocks HIV-1 particle production but not that of the other retroviruses in 293T cells. Taken together, these results suggest that INI1/hNSF5 is a host factor restricted for HIV-1 and that S6 acts as a highly specific and potent inhibitor of HIV-1 replication.
Despite the effectiveness of highly active antiretroviral therapy in controlling human immunodeficiency virus type 1 (HIV-1) replication, the emergence of drug-resistant viruses in infected patients and the severe side effects caused by the currently used drug regimen necessitate continued search for new and improved therapeutic strategies for controlling AIDS (12, 42). HIV-1-encoded proteins such as integrase (IN) and cellular proteins that are implicated in the HIV-1 life cycle are attractive targets for the development of antivirals (22, 38).
IN catalyzes the integration of viral cDNA into the host genomic DNA, a process that is essential for the replication of all retroviruses and a step that results in the latent form of the virus (8). During the life cycle of a retrovirus, IN is produced as part of the Gag-Pol polyprotein, which is assembled into virions and subsequently cleaved into individual components during maturation (8). IN consists of three distinct structural domains (2), the N-terminal zinc-binding domain (HHCC), the central core domain with a highly conserved D,D(35)E motif required for the catalytic activity, and the less highly conserved C-terminal domain. Although crystal structure data exist for single and double domains of IN, no structural data exist yet for the entire IN protein (18). Various biochemical and genetic approaches have been used to demonstrate the multimerization of IN (1, 16, 25), but the exact nature of the IN multimer is still unknown (11, 24).
Studies of IN have demonstrated that it may affect steps in viral replication besides integration itself. For example, the presence of IN mutations affects viral morphology, particle formation, particle release, and infectivity (9, 17, 43). Deletions or mutations in the Zn finger region of IN (for example, H12A) have been shown to result in decreased particle production, defects before or at the initiation of reverse transcription, and decrease in the particle-associated reverse transcriptase (RT) activity (28, 29, 50). IN also plays a role in the nuclear localization of the preintegration complex (PIC), and although this is a controversial hypothesis, several putative nuclear localization signals in its protein sequence have been implicated in this process (5, 15, 32). Taken together, these studies indicate that mutations of IN are pleiotropic and may alter viral replication by blocking various steps other than integration itself. Although the mechanistic basis for the pleiotropic effects of IN mutants remains elusive, it is likely that therapeutic strategies that target IN could affect multiple steps in viral replication.
INI1/hSNF5 is one of the proteins that directly interacts with HIV-1 IN (26). It is a 385-amino-acid protein with three highly conserved regions including two direct imperfect repeats, repeat 1 (Rpt1) and repeat 2 (Rpt2), a C-terminal coiled-coiled domain, and homology region 3 (HR3) (33). Previously, we have demonstrated that the Rpt1 region of INI1/hSNF5 is necessary and sufficient to bind to HIV-1 IN (33). Subsequent studies from several laboratories, including ours, demonstrated that INI1/hSNF5 interacts with various viral and cellular proteins such as EBNA2 (49), c-MYC (10), ALL1 (41), HPV18-E1 (31), and p53 (30) and that many of these interactions involve the Rpt regions of INI1/hSNF5, suggesting that these regions include domains important for protein-protein interactions. Furthermore, studies from our laboratory indicated that INI1/hSNF5 has a masked nuclear export sequence located at the beginning of Rpt2 (13) and a nonspecific DNA-binding activity upstream of Rpt1 (33). INI1/hSNF5 is a homologue of yeast transcription factor SNF5 and is a component of the ATP-dependent chromatin-remodeling mammalian SWI/SNF complex (48). Reconstitution of SWI/SNF activity from purified proteins revealed that INI1/hSNF5, BAF170, BAF155, and the ATPase subunit, BRG1 or hBRM, form the critical core components of the complex (37). Recent studies have suggested that INI1/hSNF5 is also a tumor suppressor mutated in atypical teratoid and rhabdoid tumors, or malignant rhabdoid tumors, an aggressive pediatric cancer with a poor prognosis (46) and a mortality rate of nearly 100% in early childhood.
Targeting the interaction between IN and the host cellular factors may prove to be a fruitful area of investigation for the development of antivirals against HIV-1. In an attempt to develop IN inhibitors by using cellular proteins, we previously isolated and characterized a transdominant mutant of INI1/hSNF5 (51). We demonstrated that a fragment of INI1/hSNF5 (S6) spanning the minimal IN interaction domain profoundly inhibited HIV-1 particle production (10,000- to 100,000-fold). Stable expression of S6 resulted in protection of the T cells from infection by full- length clones of HIV-1. Mutations in S6 or IN that disrupt the IN-INI1 interaction abrogated this inhibitory effect, suggesting that S6 inhibits particle production by directly binding to IN. An IN-deficient HIV-1 strain containing Vpr-RT-IN in trans was not affected by S6 (51), suggesting that IN within the context of Gag-Pol is required for this inhibition. In addition, we found that the truncation fragment S6 is ectopically expressed in the cytoplasm while INI1 is nuclear. Taken together, the results of our studies suggest that ectopic overexpression of a minimal IN-binding domain of INI1 transdominantly inhibits HIV-1 particle production and replication (51). Furthermore, by analyzing the purified and subtilisin-treated virions, we found that INI1/hSNF5 is incorporated into the HIV-1 particles (51).
Gene therapy strategies to deliver the trandominant mutant, S6, into hematopoietic stem cells could be a useful approach to protect T cells from infection. However, since the transdominant mutant inhibits HIV-1 assembly and particle production, lentiviruses cannot be used as a delivery system for this purpose. Furthermore, it is not clear whether the transdominant mutant affects the assembly and particle production of other retroviruses and whether INI1/hSNF5 interacts with other retroviral integrases. Therefore, in this study we have examined (i) the specificity of the interaction of INI1/hSNF5 with retroviral integrases, (ii) the ability of INI1/hSNF5 to be incorporated into various primate lentiviral and murine retroviral particles, and (iii) the effect of transdominant mutants on particle production of these other retroviruses. We found that INI1/hSNF5 is specifically incorporated into HIV-1 but not the other retroviral particles. This HIV-1-specific incorporation was correlated with the ability of INI1/hSNF5 to interact with HIV-1 IN but not the other retroviral INs. In addition, INI1/hSNF5 transdominant mutant S6 inhibited particle production of HIV-1 but not that of the other retroviruses. Taken together, these results demonstrate a specific incorporation of INI1/hSNF5 into HIV-1 virions and a potent and specific inhibitory effect of S6 on HIV-1 replication.
MATERIALS AND METHODS
Yeast two-hybrid system.
DNA fragments containing the IN open reading frames of HIV-2ROD10 (23), simian immunodeficiency virus mac239 strain (SIVmac239) (27), and human T-cell leukemia virus type 1 (HTLV-1) (40) were cloned into yeast vectors as fusions to both the LexA DNA-binding domain (LexADB) and the GAL4 activation domain (GAL4AD). The DNA fragments were PCR amplified using Vent polymerase and the following primer pairs: GKSIV-A (5′CGCGGATCCTCTTCTTGGAAAAGATAGAGCCA3′) plus GKSIV-C (5′CGGAATTCCTATGCCACCTCTCTAGA3′) for SIV IN, GKHIV2-A (5′CGCGGATCCTGTTCCTGGAAAAAATAGAG3′) plus GKHIV2-C (5′CGGAATTCTATGCCATTTCTCCATCC3′) for HIV-2 IN, and HTLV-1F (5′CGGAATTCGTCCTGCAGCTC3′) plus HTLV-1R (5′GCGAATTCTTACCCATGGTG3′) for HTLV-1 IN.
The amplified HIV-2 IN (IN-2) and SIV IN (IN-S) fragments were digested with BamHI and EcoRI and were first cloned into pGEX3xPL at the same sites to generate glutathione S-transferase (GST) fusions. The amplified HTLV IN (IN-T) fragment was digested with EcoRI and cloned into the EcoRI site of pGEX1λT. The gene fusions were sequenced to ensure the absence of PCR-mediated errors. The BamHI and EcoRI fragments of IN-2 and IN-S were then excised from the recombinants and ligated with EcoRI-SalI adapters. These fragments were cloned into the BamHI and SalI sites of pSH2-1 (encoding LexADB) and pGADNot (encoding GAL4AC). The two EcoRI ends of the HTLV-1 IN (IN-T) fragment were modified to BamHI by the addition of EcoRI-XmnI-BamHI adapters (New England Biolabs no. S1106S) and cloned into the BamHI site of pSH2-1 and pGADNot.
In vitro binding assays.
In vitro assays were carried out by testing the ability of GST-IN proteins to pull down the hexahistidine (His6) fusion of INI1 proteins from bacterial lysates. For the purpose of expressing His6-INI1, a BamHI/SalI fragment of INI1 was isolated from pSP72-INI1 and cloned into pQE32 vector (Qiagen) to generate pQE32-INI1 (10). The stable expression of recombinant protein in bacteria was confirmed using both anti-INI1 antibodies (INI1-PB3, produced using the GST-N-INI1 fusion protein containing the N-terminal 130-amino-acid fragment of INI1/hSNF5) and anti-His6 antibodies. Various GST-IN proteins were induced and then immobilized onto G-beads as described previously for GST-INI1 (10, 33). In brief, the GST-IN proteins were induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to bacterial cultures harboring the appropriate GST-IN expression constructs. The cells were harvested, suspended in high-salt buffer Y (500 mM NaCl, 50 mM Tris-Cl [pH 8.0], 1 mM EDTA, 0.5% IGEPAL (Sigma; catalog no. I-3021), and protease inhibitors including 2 mg of aprotinin per ml, 2 mg of leupeptin per ml 2 mg of pepstatin A per ml and 18 mg of phenylmethylsulfonyl fluoride [PMSF] per ml) and lysed by six cycles of freezing and thawing followed by sonication. The lysed cells were pelleted to remove the cell debris, and the supernatant was collected by centrifugation. To the cleared lysate, preswollen and equilibrated glutathione-agarose beads (Sigma no. G-4510) were added, and the mixture was incubated for 1 h at 4°C with gentle rocking. The proteins bound to beads were washed with buffer Y containing 50 mM NaCl (low-salt buffer Y).
To carry out the in vitro binding reaction, the GST and GST-IN proteins that were immobilized onto G-beads were incubated with 200 μl of crude bacterial lysates containing His6-INI1 in a final volume of 1 ml of reaction buffer containing 20 mM HEPES (pH 7.0), 200 mM NaCl, 4 mM MgCl2, 5 mM dithiothreitol (DTT), 0.1% IGEPAL, 100 mg of bovine serum albumin (BSA) per ml, 2 mg of aprotinin per ml, 2 mg of leupeptin per ml, 2 mg of pepstatin A per ml and 18 mg of PMSF per ml (HND buffer) as described before (33). The mixture was incubated for 1 h with gentle rocking at 4°C. After the binding reaction, the beads were washed and the bound proteins were eluted, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblotted, and detected by the enhanced chemiluminescence detection method using anti-His6 antibodies.
Antibodies.
The antibodies used in the Western analysis are as follows. Monoclonal anti-IN antibodies were obtained by collecting culture supernatants of the 8E51F9 hybridoma cell line (a kind gift from Dag Helland, University of Bergen, and kindly provided by A. Engelman, Dana Farber Cancer Institute). Polyclonal anti-INI1 antibody, INI1-PB3, was raised against the GST-N-INI1 protein (a 1-to 130-amino-acid fragment of INI1 fused to GST) in rabbits. The antibody was affinity purified by depleting the serum of contaminating anti-GST antibody and then subjecting the depleted serum to affinity purification on a nitrocellulose strip containing GST-N-INI1 protein. The anti-p24 antibody is a goat polyclonal antibody (goat no. 81, bleed no. 001090), anti-gp41 is a mouse monoclonal antibody (New England Nuclear no. NEA-9303), and anti-CD45 is a polyclonal antibody (BD Biosciences no. 610266). The anti-tubulin antibody is a mouse monoclonal antibody from Sigma (no. T5168, clone B-5-1-2), and anti-His6 antibody was purchased from Clontech (no. 8904-1).
Purification of His6-IN and His6-INI1 proteins.
Bacterially expressed recombinant His6-HIV-1 IN was purified using a Ni-nitrilotriacetate (NTA) affinity column under nondenaturing conditions. Exponentially growing bacterial cells harboring pIN-C6H (HIV-1 IN coding sequence with the His6 tag at the carboxy terminus; a kind gift from S. LeGrice, HIV Drug Resistance Program, National Cancer Institute) were induced with 1 mM IPTG for 5 to 6 h. The cells were harvested and lysed in lysis buffer A (50 mM NaH2PO4 [pH 7.4], 0.1 mM EDTA, 5% glycerol, 10 mM β-mercaptoethanol [βME], 1 mM PMSF) containing 1 mg of lysozyme per ml. After the lysis, NaCl was added to a final concentration of 1 M. The cells were further lysed by three rounds of sonication, and the DNA was sheared by being passed through a 21G1.5-gauge needle. The debris was removed by centrifugation, and the supernatant was diluted 1:1 with the lysis buffer to bring the NaCl concentration to 0.5 M. A 50% slurry of equilibrated Ni-NTA beads was added to the above lysate, and the mixture was incubated for 30 min with gentle rocking at 4°C to bind the His6 fusion proteins. The bound beads were then packed into columns, extensively washed with washing buffer B (50 mM NaH2PO4 [pH 6.3], 0.1 mM EDTA, 5% glycerol, 10 mM β-ME, 0.5 mM NaCl, 1 mM PMSF), and given a high-stringency wash with 10 to 15 column volumes of wash buffer (50 mM NaPO4 [pH 7.2], 0.1 mM EDTA, 5% glycerol, 10 mM β-ME, 0.5 mM NaCl, 1 mM PMSF) containing 40 mM imidazole. The bound protein was then eluted with a continuous gradient of 40 to 800 mM imidazole in elution buffer (50 mM NaPO4 [pH 7.2], 0.1 mM EDTA, 5% glycerol, 10 mM β-ME, 0.5 mM NaCl, 1 mM PMSF), and 0.5-ml fractions were collected. Fractions were separated by SDS-PAGE and stained with Coomassie blue to detect fractions containing purified protein. The peak fractions were pooled and dialyzed against buffer containing 20 mM HEPES, 1 mM DTT, 1 mM EDTA, 0.5 M NaCl, and 20% glycerol.
A protocol very similar to that for the His6-IN purification was used to purify His6-INI1 from bacterial cells harboring the construct pQE32-INI1, except that wash buffer containing 20 mM imidazole was used instead of wash buffer containing 40 mM imidazole and the protein was eluted using a continuous gradient of 20 to 800 mM imidazole in elution buffer. The peak fractions were pooled and dialyzed against the same buffer as that used during His6-IN purification.
Both His6-IN and His6-INI1 proteins were quantitated on an SDS-PAGE gel against a BSA standard. While the His6-IN preparations were purified to near homogeneity with no background bands, His6-INI1 was partially pure. The His6-INI1 proteins separated on the SDS-PAGE gel were stained with silver stain, and the bands corresponding to His6-INI1 were quantitated against a BSA standard.
RT assay.
The effect of the S6 transdominant mutant on retroviral particle production was monitored by carrying out RT assays as follows. Culture supernatants (1 ml) from cells transfected with HIV-1R3B, HIV-2, SIVmac, HTLV-1, and Moloney murine leukemia virus (Mo-MuLV) were precipitated by adding 0.5 ml of 30% polyethylene glycol 8000-0.4 M NaCl overnight at 4°C. The precipitated samples were centrifuged at 8,000 × g for 45 min at 4°C, and each pellet was resuspended in 25 μl of solution B (0.9% Triton X-100, 440 mM KCl) and 100 μl of solution A (25 mM Tris [pH 7.8], 0.25 mM EDTA, 0.025% Triton X-100, 50% glycerol, 10 mM DTT, 100 mM KCl). A 5-μl volume of each sample was then incubated with 45 μl of RT reaction cocktail for 1 h at 37°C. The reaction cocktails for all viruses, except for Mo-MuLV, included 50 mM Tris (pH 8.0), 20 mM DTT, 60 mM NaCl, 5 μM oligo(dT), 10 mM poly(rA), 10 mM dTTP, 10 μCi of [32P]dTTP, 10 mM MgCl2, and 0.0005% NP-40. The reaction cocktail for Mo-MuLV consisted of 60 mM Tris (pH 8.3), 24 mM DTT, 0.7 mM MnCl2, 75 mM NaCl, 6 mM oligo(dT), 12 mM poly(rA), 10 μCi of [32P]dTTP, 12 μM dTTP, and 0.0006% NP-40. The reactions were carried out at 37°C for 60 min. After the incubation, 10 μl of each reaction mixture was spotted in triplicate onto DE82 paper (Whatman), washed three times for 30 min each in 2× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with gentle agitation, rinsed in 100% ethanol, and dried under infrared light for 1 h. The DE82 squares containing the RT reaction spots were then cut out from the sheet and placed into scintillation fluid, and the radioactivity was counted in a scintillation counter.
Preparation of concentrated virus.
Culture supernatants from the producer cells were harvested and passed through a low-protein-binding syringe filter (Fisher no. 09-740-37F) to remove cellular particulates. The supernatants were concentrated by sucrose density centrifugation and then treated with subtilisin as previously described (35). Briefly, the concentrated virus was resuspended in 1× phosphate-buffered saline (PBS), an equal volume of 2× subtilisin buffer (2× subtilisin buffer contains 40 mM Tris-Cl [pH 8.0] and 2 mM CaCl2) containing 2 mg of subtilisin per ml (final concentration of subtilisin, 1 mg/ml) was added to the solution, and the mixture was digested overnight (∼18 h) at 37°C. After digestion, PMSF was added to a final concentration of 5 μg/ml to inactivate the subtilisin. The subtilisin-treated virus was centrifuged over 10 ml of a 20% sucrose solution with 5 μg of PMSF per ml. The 10 ml of 20% sucrose solution was carefully layered to form a “pad” under the virus sample, and the remaining space in the centrifuge tube was filled with 1× PBS containing 5 μg of PMSF per ml. The preparations were centrifuged in an SW28Ti rotor at 25,000 rpm for 3 h. After the centrifugation, the PBS and the top one-third (by volume) of the sucrose were carefully removed. The remaining trace of sucrose solution was removed using a fresh pipette tip. The pellet was then gently washed in 1 ml of PBS to remove the remaining sucrose. The resulting concentrated and subtilisin-treated virions were resuspended in lysis buffer (25 mM Tris-Cl [pH 7.5], 50 mM KCl, 0.025% Triton X-100, 50% glycerol). An equal volume of 2× loading buffer (2× loading buffer contains 0.25 M Tris-Cl [pH 6.8], 8% SDS, 40% glycerol, 3.6% β-ME, and 0.005% bromophenol blue) was then added to the samples. The virion proteins were separated by SDS-PAGE and subjected to immunoblot analysis using anti-INI1 antibodies, anti-p24 antibodies to detect the levels of HIV-1 capsid, anti-p30 antibodies to detect the Mo-MuLV capsid, and anti-gp41 antibodies to confirm the complete subtilisin digestion in virion samples.
Estimation of the number of particles present in various retroviral preparations.
The number of particles present in the virion preparations was estimated by quantitative protein staining of SDS-PAGE gels. Serial dilutions of each virus preparation, starting at 1 μg of total protein, were analyzed by SDS-PAGE and stained with SYPRO orange (Molecular Probes, Eugene, Oreg.) as recommended by the manufacturer. The fluorescence intensities of the CA bands were compared to those of a serial dilution series of a BSA stock solution (Sigma-Aldrich, St. Louis, Mo.) that was precisely quantitated by amino acid analysis. SYPRO orange staining fluorescence was observed and captured on a VersaDoc 3000 imaging system (Bio-Rad, Inc., Hercules, Calif.) as recommended by the manufacturer, and intensities were analyzed and concentrations were calculated by linear regression using Quantity One software (Bio-Rad).
Stoichiometric analysis of IN-INI1 in the virions.
The stoichiometry of virion-associated INI1/hSNF5 protein with reference to IN was determined by analyzing the autoradiography images in a 440cf Kodak Digital Science image station. Briefly, the subtilisin-treated virions were separated by SDS-PAGE, immunoblotted using anti-IN and anti-INI1 antibodies sequentially, and detected by the enhanced chemiluminiscence method. The resulting autoradiography films were scanned in a Kodak Digital Science image station, and the relative densities of the bands were determined by using the Kodak Digital Science 1D image analysis software. The relative band intensities of the IN and INI1 proteins present in the virions were normalized against a standard of purified and quantitated His6-IN and partially purified and quantitated His6-INI1 proteins with the assumption that the antibodies recognize the native proteins present in the virions as efficiently as they recognize the recombinant proteins.
Transfections.
293T cells were cotransfected with 20 μg of pCGN-S6 expressing the INI1 transdominant mutant, S6 (51), along with 5 μg of the full-length molecular clones of HIV-1R3B (39), HIV-2 (44), Mo-MuLV (47), SIVmac(AIDS repository no. 133), or HTLV-1 (AIDS repository no. 2817) by using a calcium phosphate transfection method (Specialty Media S-001) as recommended by the manufacturer.
RESULTS
Stoichiometry of INI1/hSNF5 in the HIV-1 virions and its absence in the microvesicular fractions.
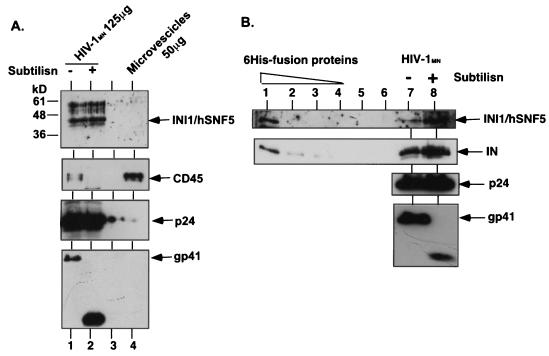
We previously demonstrated that purified HIV-1 virion particles treated with subtilisin incorporate INI1/hSNF5 protein (51). We further demonstrated that coexpression of hemagglutinin (HA)-tagged INI1/hSNF5 in the producer cells leads to the specific incorporation of HA-tagged protein into the purified and subtilisin-treated virions (51). It has been reported that even after two sucrose density gradient centrifugations and subtilisin treatment, the HIV-1 virion preparations may be contaminated with a very small amount (<5%) of microvesicle-associated proteins (34, 36). Therefore, to conclusively demonstrate that the presence of INI1/hSNF5 in the virion preparations is not due to microvesicular contamination, we examined and compared the proteins present in purified HIV-1MN virion preparations to those present in the purified microvesicular fractions. For this purpose, total proteins from the purified fractions of microvesicles (∼50 μg of total proteins) along with purified HIV-1MN virions (125 μg of total protein before digestion) treated with subtilisin and purified on sucrose density gradients were separated by SDS-PAGE and subjected to immunoblot analysis using affinity-purified anti-INI1/hSNF5 antibodies (Fig. 1A). While a clear distinct band corresponding to INI1/hSNF5 was apparent in both subtilisin-treated and untreated HIV-1 virions, no band corresponding to INI1/hSNF5 was detected in the microvesicular fractions, even after longer exposure in repeated experiments (Fig. 1A). These results demonstrate that purified microvesicles are devoid of INI1/hSNF5.
FIG. 1.
INI1/hSNF5 in the virions. (A) Absence of INI1/hSNF5 in the purified microvesicles. Lanes 1, untreated purified HIV-1MN virions; 2, subtilisin-treated purified HIV-1MN virions; 3, empty; 4, purified banded microvesicular fraction. Top panel, immunoblot probed with affinity-purified anti-INI1 antibody; second panel from top, immunoblot probed with anti-CD45 antibody, third panel from top, immunoblot probed with anti-p24 antibody; bottom panel, immunoblot probed with anti-gp41 antibody. (B) Stoichiometric analysis of the IN-to-INI1/hSNF5 ratio in the virions. Lanes: 1 to 4, fivefold dilutions of normalization standards of partially purified His6-INI1/hSNF5 (top panel) and His6-IN (second panel); 5 and 6, empty; 7, untreated HIV-1MN virions; 8, subtilisin-treated HIV-1MN virion. Top panel, immunoblot probed with affinity-purified anti-INI1 antibody; second panel from top, immunoblot probed with monoclonal anti-IN antibody; third panel from top, immunoblot probed with anti-p24 antibody; bottom panel, immunoblot probed with anti-gp41 antibody.
To determine the amount of microvesicular contamination in the batch of HIV-1MN virions used in our experiments, we analyzed the presence of cellular proteins specific to the microvesicular fraction. It has been previously reported that one of the surface proteins, CD45, also called leukocyte common antigen, is specifically enriched in the microvesicular fractions but not in the HIV-1 virion fractions (19). Therefore, we probed the above blot containing the microvesicles and HIV-1 with CD45 antibodies. As illustrated in Fig. 1A (second panel from top), the results indicated that while microvesicular fractions harbor abundant CD45, HIV-1 virions that were not treated with subtilisin contained very small amounts of CD45, which was subsequently eliminated on subtilisin digestion. Nevertheless, the virion preparations, although containing 10-fold less microvesicular contamination, exhibited the presence of a distinct INI1/hSNF5 band and the virions retained very similar amounts of INI1/hSNF5 regardless of the subtilisin treatment. Taken together, these results indicate that INI1/hSNF5 is clearly absent from microvesicular fractions and that the INI1/hSNF5 protein detected in the virion preparations is not due to microvesicular contamination.
Owing to the low avidity of anti-INI1 antibodies available, large amounts of virion preparations were needed to detect INI1/hSNF5 in the above experiments, making it appear as though a very small amount of the protein is present in the virions. Therefore, to ascertain the exact amount of INI1/hSNF5 present in the virions, we determined the stoichiometry of IN to INI1/hSNF5 in these preparations. Since INI1/hSNF5 is likely to be incorporated by its association with HIV-1 IN, we surmised that the INI1/hSNF5 protein will be present in the HIV-1 virions in molar amounts comparable to that of IN. We first purified recombinant His6-tagged INI1/hSNF5 protein expressed in bacteria and quantitated the full-length His6-INI1/hSNF5 protein by comparing serial dilutions of the protein against a BSA standard. The serial dilutions of quantitated protein were loaded onto an SDS-PAGE gel along with subtilisin-treated purified virion fractions. In the same lanes that contained His6-INI1/hSNF5, we also included purified fractions of equimolar amounts of recombinant His6-HIV-1 IN protein to determine the relative ratios of INI1 to IN within the virions. These proteins were subjected to sequential immunoblot analyses using affinity-purified anti-INI1/hSNF5 antibody as well as anti-IN, anti-p24, and anti-gp41 antibodies (Fig. 1B). To avoid variation from experiment to experiment and to be able to cross-compare the relative quantities of proteins in question within the same batch of purified virions, the blot was stripped and reprobed with different antibodies. The intensities of bands in the autoradiogram were subjected to quantitation using the Kodak Digital Science image station. Table 1 illustrates our estimation of the amounts of IN and INI1 proteins present in the virion preparations compared to that of the standard. The results of this analysis indicated that molar ratio of IN to INI1/hSNF5 within the virions is approximately 2:1, indicating that a dimer of IN might bind to a monomer of INI1/hSNF5. Given that there are about 100 IN molecules per HIV-1 particle, we estimate that there are approximately 50 molecules of INI1/hSNF5 per virus particle.
TABLE 1.
Stoichiometry of IN to INI1/hSNF5 in the HIV-1 virions
| Protein | Amt of protein standard (lane 1, Fig. 1B) | Mean intensity of standarda | Mean intensity of band 1b | Amt of protein in band 1 | Mean intensity of band 2b | Amt of protein in band 2 | Average intensity of bands | Average amt of protein |
|---|---|---|---|---|---|---|---|---|
| IN | 8.8 ng | 5.0769 | 7.1433 | 12.38 ng | 9.6827 | 16.78 ng | 8.4130 | 14.58 ng |
| INI1/hSNF5 | 12.5 ng | 3.57 | 1.9905 | 6.97 ng | 4.5164 | 15.81 ng | 3.2535 | 11.39 ng |
| IN-INI1 ratio (ng) | 1.77:1 | 1.1:1 | 1.3:1 | |||||
| IN-INI1 molar ratio | 2.3:1 | 1.44:1 | (1.87 ± 0.43):1 |
INI1 is specifically incorporated into HIV-1 but not into other related lentivirus particles.
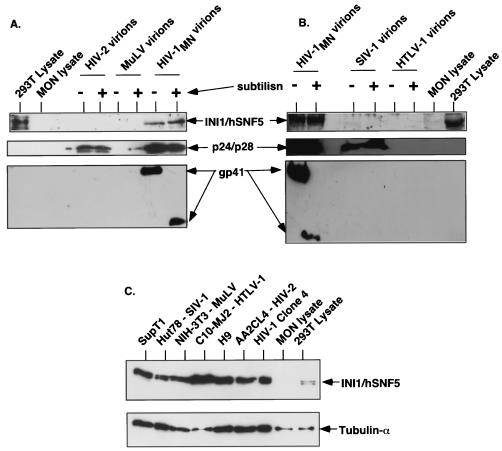
Once we confirmed the virion-specific association of INI1/hSNF5 in the particles and the lack of INI1 in the microvesicular fraction, we tested the specificity of incorporation of INI1 into various retrovirus particles. INI1/hSNF5 is a highly conserved protein and is documented to be present in all the eukaryotic species examined thus far. The conservation among mammalian proteins is very high, with the primary amino acid sequences of the human and mouse proteins being nearly 100% identical, except for one amino acid change. We surmised that if INI1/hSNF5 is incorporated into other retrovirus particles, we should be able to detect it by using the antibodies raised against human protein. To determine the specificity of incorporation of INI1/hSNF5 into various retroviruses, we generated purified virion preparations of primate lentiviruses including HIV-1, HIV-2, and SIVmac, as well as nonlentiviruses such as HTLV-1 and Mo-MuLV. As before, the virions were prepared by banding the culture supernatants from the producer cells on a sucrose gradient to eliminate the microvesicular fractions and treated with subtilisin to eliminate the contaminating proteins. These preparations were further purified through a sucrose cushion. We estimated the approximate number of particles present in each virion preparation, to ensure the presence of equivalent loads, by separating samples by SDS-PAGE, staining these preparations with SYPRO orange dye, and determining the amount of capsid molecule present per microgram of total protein. The approximate numbers of particles present in these preparations were then calculated. As indicated in the Table 2, these preparations contained comparable numbers of virion particles. Total virion proteins were then separated by SDS-PAGE and subjected to immunoblot analysis using affinity-purified anti-INI1/hSNF5 antibodies. The results of these analyses illustrated that while a strong and distinct band corresponding to INI1/hSNF5 was observed in lanes containing HIV-1 virions, no bands of similar size were apparent in lanes containing HIV-2, SIVmac, HTLV-1, or Mo-MuLV particles (Fig. 2). These results demonstrate that incorporation of INI1/hSNF5 is specific to HIV-1. The immunoblots were further probed with anti-p24 (HIV-1), anti-p30 (Mo-MuLV), anti-gp70, or anti-gp41 antibodies to ensure comparable amounts of loading in the subtilisin-treated and untreated samples and to ensure complete digestion with subtilisin.
TABLE 2.
Estimation of the number of virion particles present in various retroviral preparations
| Virus | Amt (μg) of capsid/100 μg of proteina | No. of Gag molecules | No. of particlesb |
|---|---|---|---|
| HIV-1 | 10 | 2.4 × 1014 | 1 × 1011 |
| HIV-2 | 6 | 1.2 × 1014 | 6 × 1010 |
| HTLV-1 | 37 | 9.2 × 1014 | 5 × 1011 |
| Mo-MuLV | 17 | 3.4 × 1014 | 2 × 1011 |
| SIV | 2.5 | 9.6 × 1013 | 2.6 × 1010 |
Total protein.
Based on 2,000 Gag molecules/virion (estimates for various retroviruses range from 1,200 to 5,000).
FIG. 2.
Analysis of INI1/hSNF5 incorporation into various retrovirus particles. (A and B) About 125 μg each of purified and subtilisin-treated samples and the untreated controls were separated by SDS-PAGE and subjected to immunoblot analysis. The blot was sequentially probed with affinity-purified anti-INI1 antibodies (top panel), anti-p24 antibodies (middle panel), and anti-gp41 antibodies (bottom panel). Panels include HIV-2, Mo-MuLV, and HIV-1 virions (A) and HIV-1, SIV, and HTLV-1 (B) virion preparations, along with total-cell lysates from 293T (INI1+/+) and MON (INI1−/−) cells as controls. Note the cross-reactivity of anti-p24 antibodies to capsid protein from HIV-1, HIV-2, and SIV-1 (middle panels). (C) Presence of INI1/hSNF5 in the producer cells. The lanes contain total-cell lysates as noted above the panel. The top panel represents the immunoblot probed with affinity-purified anti-INI1 antibody, and the bottom panel represents the immunoblot probed with anti-tubulin antibody as a loading control.
Since the virions used in this study were produced in different mammalian cell lines, the lack of incorporation of INI1/hSNF5 could be due to the lack of protein expression in these cells. To determine if endogenous INI1/hSNF5 is expressed in sufficient amounts in these cell lines, we prepared lysates from H9, AA2CL4, Hut78, C10-MJ2, and NIH 3T3 cell lines, which were used as producers for HIV-1, HIV-2, SIV-1, HTLV-1, and Mo-MuLV, respectively, and subjected relatively equal amounts of the total proteins from these cells to immunoblot analysis using affinity-purified anti-INI1/hSNF5 antibodies. As an internal control for the protein load, the blot was stripped and reprobed with anti-tubulin antibodies. The results indicated that the producer cell lines used for the preparation of various retroviruses express sufficient quantities of INI1/hSNF5 (Fig. 2C). They also indicate that lack of INI1/hSNF5 incorporation is not due to the lack of expression of INI1/hSNF5 protein in the producer cell lines.
Two-hybrid analysis to determine the protein-protein interaction of various retroviral INs with INI1/hSNF5.
Specific incorporation of INI1/hSNF5 into HIV-1 virions, despite the presence of endogenous INI1/hSNF5 in the producer cells, suggests that the incorporation is not due to a nonspecific scooping effect but, rather, is due to a specific, HIV-1-mediated mechanism. One possible mechanism by which this restricted incorporation is achieved would be by the recruitment of INI1/hSNF5 protein into HIV-1 virions on the basis of its ability to specifically interact with HIV-1 IN but not with other retroviral INs. Another cellular protein, cyclophilin A (CyPA), also has been demonstrated to undergo specific interactions only with HIV-1 but not with other related retroviral capsid proteins (20, 45). Consistent with this property, CyPA is incorporated only into certain clades of HIV-1 and SIVCPZ and not into other retroviruses (6). To determine the specificity of interaction of INI1 with retroviral INs, we first examined the interaction of various INs with INI1/hSNF5 by using the yeast two-hybrid system. Combinations of plasmids expressing LexADB and GAL4AC fusions of IN from HIV-1 (IN-1), HIV-2 (IN-2), SIVmac (IN-S), and HTLV-1 (IN-T) and INI1/hSNF5 were coexpressed in yeast strain CTY10-5d carrying the lacZ gene as a reporter for the two-hybrid analysis. As negative controls, plasmids expressing various LexADB fusion proteins were transformed with plasmids expressing GAL4AC alone and plasmids expressing all of the GAL4AC fusions were transformed with plasmids expressing LexADB alone. The transformants were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) to detect positive interactions between the various retroviral INs and INI1/hSNF5.
A compilation of the results obtained from multiple yeast transformations is presented in Table 3. The results demonstrate that coexpression of IN-1 and INI1/hSNF5 gave the strongest reaction when expressed as either of the two fusion proteins. The expression of LexADB fusions of IN-1 and IN-T along with GAL4AC resulted in white colonies (Table 3). However, the remaining IN proteins gave a low level of background activity. Both IN-2 and IN-S also demonstrated a weaker interaction with INI1/hSNF5. This interaction, however, was orientation dependent and was observed only when IN-2 and IN-S were fused to LexADB and not when they were fused to GAL4AC (Table 3). Interestingly, IN-T, which is not closely related to IN-1, did not show any interaction with INI1/hSNF5 either as a LexADB fusion or as GAL4AC fusion. The lack of interaction of IN-T with INI1/hSNF5 is not due to lack of protein expression or stability, since its fusion protein exhibited homomeric interactions (data not shown). These results indicate that INI1/hSNF5 interacts strongly with HIV-1 IN but only weakly with HIV-2 and SIV INs in the yeast two-hybrid system.
TABLE 3.
Interaction of various retroviral integrases with INI1/hSNF5
| LexADB fusion | GAL4AC fusion | β-Gal staininga |
|---|---|---|
| LexADB | +GAL4AC | − |
| LexADB-IN-1 | +GAL4AC | − |
| LexADB-IN-2 | +GAL4AC | ± |
| LexADB-IN-S | +GAL4AC | ± |
| LexADB-IN-T | +GAL4AC | − |
| LexADB | +GAL4AC-INI1/hSNF5 | − |
| LexADB-IN-1 | +GAL4AC-INI1/hSNF5 | +++ |
| LexADB-IN-2 | +GAL4AC-INI1/hSNF5 | + |
| LexADB-IN-S | +GAL4AC-INI1/hSNF5 | + |
| LexADB-IN-T | +GAL4AC-INI1/hSNF5 | − |
| LexADB-INI1/hSNF5 | +GAL4AC | − |
| LexADB-INI1/hSNF5 | +GAL4AC-IN-1 | +++ |
| LexADB-INI1/hSNF5 | +GAL4AC-IN-2 | ± |
| LexADB-INI1/hSNF5 | +GAL4AC-IN-S | ± |
| LexADB-INI1/hSNF5 | +GAL4AC-IN-T | ± |
| LexADB | +GAL4AC-IN-1 | − |
| LexADB | +GAL4AC-IN-2 | − |
| LexADB | +GAL4AC-IN-S | − |
| LexADB | +GAL4AC-IN-T | − |
+++, strong interaction (blue stain visible within 30 min); +, weak interaction (blue stain visible within 2 to 3 h); ±, insignificant interaction (blue stain visible after 6 to 8 h); −, negative interaction (no blue stain visible).
In vitro binding reaction to determine the direct protein-protein interactions of INs with INI1/hSNF5.
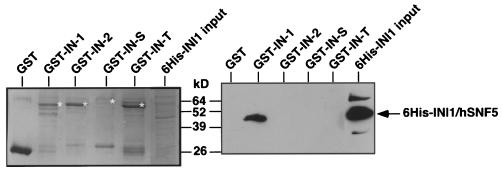
The weak interaction observed between IN-2, IN-S, and INI1/hSNF5 in one of the directions in the yeast two-hybrid system could have been due to the presence of bridging proteins. To clearly determine if these retroviral INs have the ability to directly interact with INI1/hSNF5, we carried out in vitro interaction assays using GST fusions of various INs and the His6-tagged bacterially expressed INI1/hSNF5 protein to corroborate the findings in the yeast two-hybrid system. We expressed various INs as fusions to GST and tested to see if they interacted with His6-INI1/hSNF5 in solution. The results of these in vitro interaction studies are illustrated in Fig. 3. The results of our analysis indicated that only the GST-IN-1 protein showed a distinct and strong interaction with His6-INI1/hSNF5 (Fig. 3). None of the other proteins such as GST (a negative control), GST-IN-2, GST-IN-S, or GST-IN-T displayed any interaction with INI1/hSNF5. The combined results from both the in vitro binding assays and the yeast two-hybrid analysis indicate that INI1/hSNF5 specifically interacts with HIV-1 IN but not with other retroviral INs. These results suggest that the weak interaction exhibited by IN-2 and IN-S in the two-hybrid system could be indirect and due to the presence of bridging proteins.
FIG. 3.
In vitro binding analysis to detect the IN-INI1 interaction. GST and GST fusions of various INs were incubated with bacterial lysates expressing His6-INI1/hSNF5. After the binding reaction, the samples were split in half and were separated on two identical SDS-PAGE gels. One of the gels was stained with Coomassie blue to visualize the amounts of GST fusion proteins (left panel), and the other gel was immunoblotted using anti-His6 antibody to determine the interaction (right panel). The labels at the top of the gels illustrate the proteins present in the binding reaction. The position of His6-INI1/hSNF5 protein detected in the Western analysis is marked. The asterisks next to the bands in the left panel indicate the position of the GST fusion proteins.
Specificity of inhibition of retroviral particle production by the S6 transdominant mutant.
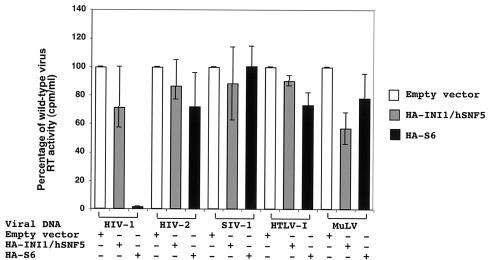
Specific incorporation of INI1/hSNF5 into HIV-1 virions but not into other retrovirus particles is consistent with the observation that interaction of INI1/hSNF5 is restricted to HIV-1 IN. We previously demonstrated that a transdominant mutant of INI1/hSNF5, S6, strongly inhibits HIV-1 particle production (51). Furthermore, analysis of interaction-defective mutants suggests that the effect is due to the specific protein-protein interactions of S6 with IN-1 within the context of Gag-Pol. However, there is a possibility that S6 is inhibiting the assembly and particle production of HIV-1 by affecting the general cellular pathways needed for this process in an as yet undefined manner. To determine whether the S6 protein inhibits viral replication by blocking certain steps of the viral life cycle in a general manner (a generic antiviral effect), we tested the effect of this transdominant mutant on particle production of HIV-1R3B, HIV-2, SIVmac, HTLV-1, and Mo-MuLV. Full-length DNAs from the molecular clones of each of these viruses were transfected into 293T cells in the presence or absence of full-length HA-INI1 or HA-S6. Culture supernatants were obtained 48 h posttransfection, and viral particle production was monitored by assaying for RT activity. By producing viruses in a nonpermissive cell line such as 293T, we could eliminate the possibility of viral spread and examine effects of the transdominant mutant on a single round of viral production of each of the retroviruses.
The RT assay was carried out under buffer conditions that were optimal for each virus. For example, while the RT assay of HIV-1, HIV-2, HTLV-1, and SIV-1 were carried out in the presence of Mg2+, the assay for Mo-MuLV was carried out in the presence of Mn2+. In each assay, the amount of virus produced in the presence of INI1 or S6 was expressed as a percentage of the amount of the control virus produced in the absence of INI1 or S6. We found that while the amount of HIV-1 was drastically reduced in the presence of S6, as previously reported (51), none of the other viruses were similarly affected (Fig. 4). We also found that overexpression of INI1/hSNF5 does not have significant effect on particle production of any of the retroviruses tested. These results indicated that the interaction between S6 and the retroviral INs is a necessary mechanism by which S6 inhibits viral particle production.
FIG. 4.
Effect of the S6 transdominant mutant on the particle production of various retroviruses. 293T cells were cotransfected with full-length molecular clones of HIV-1, HIV-2, SIV-1, HTLV-1, and Mo-MuLV, along with pCGN-S6 (expressing HA-S6), pCGN-INI1 (expressing HA-INI1), or an empty vector. The culture supernatants were assayed for RT activity to determine the amount of virus produced in the supernatant. The RT activity in the culture supernatants of the cells transfected with full-length molecular clones of each virus along with an empty vector was taken as 100%. The figure is a graphic representation of the percentage of RT activity in the culture supernatants of the cells transfected with full-length retroviral clones in the presence or absence of INI1 or S6 compared to that of the empty-vector control.
DISCUSSION
In this study we have examined the specificity and stoichiometry of the incorporation of INI1/hSNF5 into HIV-1 virion particles and determined the specificity of its interaction with various retroviral INs. Comparison of purified microvesicular fractions to purified and subtilisin-treated virions indicate that while INI1/hSNF5 is completely absent from microvesicles, it is present in both subtilisin-treated and untreated samples. In addition to the band corresponding to the expected size of INI1, an unidentified high-molecular-weight band was apparent in the virions. It is not clear if this band represents a modified form of INI1. Despite the lack of INI1/hSNF5 in the microvesicular fractions, we generally needed large quantities of purified virions to clearly detect this protein, owing to the poor qualities of the anti-INI1 antibodies. However, stoichiometric analysis using purified His6-INI1 proteins suggests that approximately one molecule of INI1/hSNF5 is present for every two molecules of IN encapsidated in the virions. Considering that there are only about 100 molecules of IN per particle, this amounts to about 50 molecules of INI1/hSNF5 per particle, explaining the difficulty in detecting the protein readily by immunoblot analysis. It is interesting that the interaction of capsid with CyPA allows the uptake of CyPA into HIV-1 virions, in roughly a 10:1 capsid-to-CyPA ratio (20, 45).
We further evaluated the specificity of incorporation of INI1/hSNF5 into various primate lentiviruses as well as Mo-MuLV. It appears that INI1/hSNF5 incorporation is restricted to HIV-1 and that even closely related retroviruses such as HIV-2 and SIV-1 do not incorporate INI1/hSNF5 despite the abundant presence of the protein in the producer cells. This observation suggests that a virus-specific mechanism is involved in this process. Our finding that INI1/hSNF5 interacts strongly and specifically with only HIV-1 IN in both the yeast two-hybrid system and the in vitro binding reactions lends support to the idea that INI1/hSNF5 is recruited to HIV-1 virions by its direct interaction with IN.
Although yeast two-hybrid studies indicate that the IN-S and IN-T proteins exhibit weak interactions with INI1/hSNF5 in one of the two orientations, the in vitro results indicate that none of the retroviral INs (other than that of the HIV-1) were able to interact with INI1/hNSF5, suggesting that the INI1 interaction is very specific to HIV-1 IN. The specific interaction of INI1 with IN-1 raises the question whether there are different interacting host proteins for other retroviral INs. In addition, it raises the question why HIV-1 IN has evolved to interact with INI1/hSNF5 while other retroviruses do not incorporate this protein. It is possible that other retroviruses interact with cellular proteins that function in the same pathway as INI1/hSNF5. Alternatively, it is possible that the interaction with INI1/hSNF5 confers specific abilities to HIV-1 that are not present in other retroviruses or provides a function that is not required by other retroviruses. Isolation of host interacting proteins for other retroviral INs may give us a better understanding of in vivo integration and host-virus interactions.
It is intriguing that although CyPA is required for normal HIV-1 replication kinetics, it is incorporated only into certain clades of HIV-1 and SIVCPZ and not into other retroviruses (7). Different lentiviral groups seem to be distinguishable by their differential incorporation of CyPA and their CyPA requirement for infectivity. While HIV-1 group M incorporates CyPA and requires it for infectivity, HIV-2 and SIVcpz do not incorporate CyPA and do not require it for infectivity and HIV-1 group O incorporates CyPA but does not require it for infectivity. Another protein that is demonstrated to interact with Gag protein during assembly is TSG101 (and its yeast homologue Vps23), which interacts with the p6 protein containing the late domain. This protein functions in the endosomal sorting pathway (3, 4), and it is hypothesized that it may be a receptor for ubiquitinated proteins (such as monoubiquitinated Gag) that function to select cargo proteins for incorporation into multivesicular bodies (MVB). It is interesting that while all the retroviruses appear to use various components of the MVB, there appear to be both specificity and uniqueness with which each of the retroviruses uses these pathways. While TSG101 directly interacts with the HIV-1 late domain p6, it appears not to interact with Mo-MuLV Gag (14). On the other hand, Vps4, another component of the MVB, appears to directly bind to the MuLV late domain (21). Therefore, it is possible that, similar to the specificity exhibited by the Gag-interacting proteins, the retroviral INs could interact with various proteins functioning in the same pathway as that of INI1/hSNF5 and that the viruses might exhibit differential abilities to incorporate these proteins.
In this study, we also have demonstrated that the ability of retroviral INs to interact with INI1/hSNF5 correlates with the specificity of inhibition of particle production by the INI1/hSNF5 transdominant mutant, S6. We found that while S6 strongly inhibits particle production of HIV-1 in 293T cells, it does not inhibit particle production of other retroviruses including HIV-2, SIV-1, HTLV-1, and Mo-MuLV in the same cells. This result clearly indicates that S6 does not inhibit HIV-1 replication by blocking certain cellular pathways necessary for particle production of retroviruses but, rather, exerts its effect by a direct interaction with HIV-1 IN. We think that this highly specific and potent inhibition is of tremendous value in developing strategies to control HIV-1 replication. Isolation of low-molecular-weight compounds that precisely mimic the specificity and potency of S6 may be therapeutically useful in preventing the reemergence of viruses from latently infected pools. Finally, inhibition of HIV-1 particle production by S6 precludes the use of lentiviruses for gene therapy strategies to deliver S6 into hematopoietic stem cells. Based on our results, we propose that either the use of non-HIV-1-based lentiviral vectors, such as those derived from HIV-2, or chimeric HIV-1 harboring IN sequences from other retroviruses will be valuable in achieving this goal.
Acknowledgments
We thank Vinayaka R. Prasad for critically reading the manuscript and for providing the HIV-2 and HTLV-1 clones. We thank D. Helland and A. Engelman for providing the hybridoma and S. Le Grice for providing the His6-IN expression plasmid. We also thank Pheroze Joshi and Vinayaka Prasad for help with RT assays.
This work was supported by National Institutes of Health grant R01 AI30051-01 to G.V.K. and in part by federal funds from the National Cancer Institute, National Institutes of Health, under contract NO1-CO-12400 to D.O. M.S. acknowledges support from institutional training grant T32-AI07501.
REFERENCES
- 1.Andrake, M. D., and A. M. Skalka. 1995. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J. Biol. Chem. 270:29299-29306. [DOI] [PubMed] [Google Scholar]
- 2.Asante-Appiah, E., and A. M. Skalka. 1999. HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv. Virus Res. 52:351-369. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248-258. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, N., and P. Woodman. 2001. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276:11735-11742. [DOI] [PubMed] [Google Scholar]
- 5.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 6.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 70:4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, P. 1997. Integration, p. 161-203. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Bukovsky, A., and H. Gottlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, S.-W. G., K. P. Davies, E. Yung, R. J. Beltran, J. Yu, and G. V. Kalpana. 1999. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 22:102-105. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 12.Condra, J. H., M. D. Miller, D. J. Hazuda, and E. A. Emini. 2002. Potential new therapies for the treatment of HIV-1 infection. Annu. Rev. Med. 53:541-555. [DOI] [PubMed] [Google Scholar]
- 13.Craig, E., Z.-K. Zhang, K. P. Davies, and G. V. Kalpana. 2002. A masked NES in INI1/hSNF5 mediates hCRM1/Exportin1-dependent nuclear export. EMBO J. 21:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman, A., F. D. Bushman, and R. Craigie. 1993. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 12:3269-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esposito, D., and R. Craigie. 1999. HIV integrase structure and function. Adv. Virus Res. 52:319-333. [DOI] [PubMed] [Google Scholar]
- 19.Esser, M. T., D. R. Graham, L. V. Coren, C. M. Trubey, J. Bess, J. W., L. O. Arthur, D. E. Ott, and J. D. Lifson. 2001. Differential incorporation of CD45, CD80 (B7-1), CD86 (B7-2), and major histocompatibility complex class I and II molecules into human immunodeficiency virus type 1 virions and microvesicles: implications for viral pathogenesis and immune regulation. J. Virol. 75:6173-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 21.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 22.Gulick, R. M. 2003. New antiretroviral drugs. Clin. Microbiol. Infect. 9:186-193. [DOI] [PubMed] [Google Scholar]
- 23.Guyader, M., M. Emerman, L. Montagnier, and K. Peden. 1989. VPX mutants of HIV-2 are infectious in established cell lines but display a severe defect in peripheral blood lymphocytes. EMBO J. 8:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins, T. M., A. Engelman, R. Ghirlando, and R. Craigie. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 271:7712-7718. [DOI] [PubMed] [Google Scholar]
- 25.Kalpana, G. V., and S. P. Goff. 1993. Genetic analysis of homomeric interactions of human immunodeficiency virus type 1 integrase using the yeast two-hybrid system. Proc. Natl. Acad. Sci. USA 90:10593-10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 27.Kestler, H., T. Kodama, D. Ringler, M. Marthas, N. Pedersen, A. Lackner, D. Regier, P. Sehgal, M. Daniel, N. King, and R. Desrosiers. 1990. Induction of AIDS in Rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 248:1109-1112. [DOI] [PubMed] [Google Scholar]
- 28.Lai, L., H. Liu, X. Wu, and J. C. Kappes. 2001. Moloney murine leukemia virus integrase protein augments viral DNA synthesis in infected cells. J. Virol. 75:11365-11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leavitt, A. D., G. Robles, N. Alesandro, and H. E. Varmus. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, D., J. W. Kim, T. Seo, S. G. Hwang, E. J. Choi, and J. Choe. 2002. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 277:22330-22337. [DOI] [PubMed] [Google Scholar]
- 31.Lee, D., H. Sohn, G. V. Kalpana, and J. Choe. 1999. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature 399:487-491. [DOI] [PubMed] [Google Scholar]
- 32.Limon, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 76:12078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morozov, A., E. Yung, and G. Kalpana. 1998. Structure-function analysis of integrase interactor 1/hSNF5L1 reveals differential properties of two repeat motifs present in the highly conserved region. Proc. Natl. Acad. Sci. USA 95:1120-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder, Jr., Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 35.Ott, D. E., L. V. Coren, B. P. Kane, L. K. Busch, D. G. Johnson, R. C. Sowder, Jr., E. N. Chertova, L. O. Arthur, and L. E. Henderson. 1996. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 70:7734-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott, D. E., L. V. Coren, R. C. Sowder, Jr., J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phelan, M. L., S. Sif, G. J. NarIikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 38.Pluymers, W., E. De Clercq, and Z. Debyser. 2001. HIV-1 integration as a target for antiretroviral therapy: a review. Curr. Drug Targets Infect. Disord. 1:133-149. [DOI] [PubMed] [Google Scholar]
- 39.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Joseph, E. R. Doran, J. A. Rafalski, E. A. Whitwhoen, K. Baumeister, L. Ivanoff, S. R. Petteway, M. L. Pearson, J. A. Lautenberger, T. S. Papas, J. Ghrayeb, N. T. Chang, R. C. Gallo, and F. Wong-Staal. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 40.Rosen, C. A., J. G. Sodroski, and W. A. Haseltine. 1985. Location of cis-acting regulatory sequences in the human T-cell leukemia virus type I long terminal repeat. Proc. Natl. Acad. Sci. USA 82:6502-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozenblatt-Rosen, O., T. Rozovskaia, D. Burakov, Y. Sedkov, S. Tillib, J. Blechman, T. Nakamura, C. M. Croce, A. Mazo, and E. Canaani. 1998. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc. Natl. Acad. Sci. USA 95:4152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saksena, N. K., and D. N. Haddad. 2003. Viral reservoirs an impediment to HAART: new strategies to eliminate HIV-1. Curr. Drug Targets Infect. Disord. 3:179-206. [DOI] [PubMed] [Google Scholar]
- 43.Shin, C.-G., B. Taddeo, W. A. Haseltine, and C. M. Farnet. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 64:2421-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song, Q., G. Yang, S. P. Goff, and V. R. Prasad. 1992. Mutagenesis of the Glu-89 residue in human immunodeficiency virus type 1 (HIV-1) and HIV-2 reverse transcriptases: effects on nucleoside analog resistance. J. Virol. 66:7568-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 46.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 47.Wang, M. Q., and S. P. Goff. 2003. Defects in virion production caused by mutations affecting the C-terminal portion of the Moloney murine leukemia virus capsid protein. J. Virol. 77:3339-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yung, E., M. Sorin, A. Pal, E. Craig, A. Morozov, O. Delattre, J. Kappes, D. Ott, and G. V. Kalpana. 2001. Inhibition of HIV-1 virion production by a transdominant mutant of integrase interactor 1. Nat. Med. 7:920-926. [DOI] [PubMed] [Google Scholar]