Abstract
Antibodies (Abs) against the V3 loop of the human immunodeficiency virus type 1 gp120 envelope glycoprotein were initially considered to mediate only type-specific neutralization of T-cell-line-adapted viruses. However, recent data show that cross-neutralizing V3 Abs also exist, and primary isolates can be efficiently neutralized with anti-V3 monoclonal Abs (MAbs). The neutralizing activities of anti-V3 polyclonal Abs and MAbs may, however, be limited due to antigenic variations of the V3 region, a lack of V3 exposure on the surface of intact virions, or Ab specificity. For clarification of this issue, a panel of 32 human anti-V3 MAbs were screened for neutralization of an SF162-pseudotyped virus in a luciferase assay. MAbs selected with a V3 fusion protein whose V3 region mimics the conformation of the native virus were significantly more potent than MAbs selected with V3 peptides. Seven MAbs were further tested for neutralizing activity against 13 clade B viruses in a single-round peripheral blood mononuclear cell assay. While there was a spectrum of virus sensitivities to the anti-V3 MAbs observed, 12 of the 13 viruses were neutralized by one or more of the anti-V3 MAbs. MAb binding to intact virions correlated significantly with binding to solubilized gp120s and with the potency of neutralization. These results demonstrate that the V3 loop is accessible on the native virus envelope, that the strength of binding of anti-V3 Abs correlates with the potency of neutralization, that V3 epitopes may be shared rather than type specific, and that Abs against the V3 loop, particularly those targeting conformational epitopes, can mediate the neutralization of primary isolates.
The third variable domain (V3) of the human immunodeficiency virus type 1 (HIV-1) gp120 envelope glycoprotein is critical for the formation of syncytia and for virus entry into target cells (24, 55). These functions are mediated by the interaction of the V3 loop with chemokine receptors and are maintained despite the sequence variation that characterizes this region of the virus envelope (18, 51). Indeed, contrary to its name, the V3 loop is characterized by a constant size of 30 to 35 amino acids, a conserved type II β-turn at its tip, a disulfide bond at its base, and a net positive charge (26, 28). Conserved features are also suggested by the structure of the V3 loop discerned by nuclear magnetic resonance studies (47, 52), and conserved elements in the V3 crown and stem are mandatory features for coreceptor interactions (9, 50). All of these structural constraints appear to be imposed by the required interaction of the V3 loop with the coreceptors for HIV-1, CXCR4 or CCR5, and suggest that this region of the virus envelope should induce antibodies (Abs) that are cross-reactive among isolates and inhibitory to virus infectivity.
Initial studies of anti-V3 Abs, induced by brief immunization protocols in animals and tested against a limited number of T-cell-line-adapted (TCLA) strains of the virus, suggested, however, that anti-V3 Abs were type specific and displayed little, if any, cross-reactivity (21, 39). In contrast, anti-V3 monoclonal Abs (MAbs) derived from the cells of HIV-infected subjects displayed broad reactivities with multiple V3 peptides (57) despite sequence diversity in the 11 amino acids spanning the region at the crown of the V3 loop (17). While these MAbs could potently neutralize TCLA strains, most of them displayed weak and sporadic neutralization against most primary isolates (12, 19, 33). Several studies suggested that this could be due to limited exposure of the V3 loop on the surfaces of primary isolates (3, 6, 49). However, studies examining the ability of anti-V3 MAbs to bind to intact virus particles showed that V3 exposure is the rule rather than the exception (34-36). More recent experiments with seven human anti-V3 MAbs and 11 primary isolates revealed a highly significant correlation between the affinity of binding of anti-V3 MAbs to primary isolates and neutralizing potency (15). Within this data set, however, there was significant variation, suggesting that additional factors contribute to the ability of a given Ab to neutralize a particular virus. Hence, it is still unclear how the presence and exposure of the V3 loop affect neutralization sensitivity and how the specificity of anti-V3 Abs contributes to this phenomenon. To address this question, we examined a panel of 32 human anti-V3 MAbs and 13 clade B viruses with diverse sensitivities to neutralization to determine the extent of anti-V3 cross-reactivity among clade B viruses and the nature of the association between virus binding and neutralization.
MATERIALS AND METHODS
Human MAbs.
The 32 human anti-V3 MAbs used for this study, of which 26 were previously described (13-17) and 6 were newly generated, are listed in Table 1. All of the MAbs were generated from HIV-infected individuals by the same cellular method based on the Epstein-Barr virus transformation of peripheral blood mononuclear cells (PBMCs) followed by fusion with heteromyeloma cells, as previously described (11, 16).
TABLE 1.
Human anti-V3 MAbs used for this study
| MAb | Peptide or protein used for selectiona | Reference |
|---|---|---|
| MAbs selected with V3 peptides | ||
| 257 | V3MN | 16 |
| 268 | V3MN | 16 |
| 311 | V3MN | 17 |
| 386 | V3MN | 17 |
| 391/95 | V3MN | 17 |
| 412 | V3MN | 17 |
| 418 | V3MN | 17 |
| 419 | V3MN | 17 |
| 447-52D | V3MN | 17 |
| 453 | V3MN | 17 |
| 504 | V3MN | 17 |
| 537 | V3MN | 17 |
| 782 | V3RF | 13 |
| 838 | V3RF | 13 |
| 908 | V3RF | 13 |
| 1006-15 | V3RF | 13 |
| 1027-15 | V3RF | 13 |
| 1108 | V3987 | 57 |
| MAbs selected with V3-FP or gp120 | ||
| 2182 | V3JR-CSF-FP | 15 |
| 2191 | V3JR-CSF-FP | 15 |
| 2219 | V3JR-CSF-FP | 15 |
| 2412 | V3JR-CSF-FP | 15 |
| 2442 | V3JR-CSF-FP | 15 |
| 2456 | V3JR-CSF-FP | 15 |
| 2424 | V3JR-CSF-FP | This study |
| 2483 | V3JR-CSF-FP | This study |
| 2497 | V3JR-CSF-FP | This study |
| 2557 | V3JR-CSF-FP | This study |
| 2580 | V3JR-CSF-FP | This study |
| 2558 | V392UG037-FP | This study |
| 694/98 | gp120IIIB | 12 |
| 1334 | gp120451 | 14 |
V3JR-CSF-FP is a previously described (22) fusion protein which displays specificity for conformation-sensitive epitopes of V3; V392UG037-FP contains the sequence of a clade A primary isolate (CTRPNNNTRKSVRIGPGQTFYATGDIIGDIRQAHC).
All but three MAbs, namely, 2182, 2557, and 2558, were derived from the cells of subjects from the United States who were presumably infected with clade B viruses. Human MAb 2182 was derived from a clade A virus-infected immigrant from China who is currently living in New York City (15), and MAbs 2557 and 2558 were produced from individuals who were infected with the CRF02_AG virus subtype and are living in Cameroon. The clade A and CRF02_AG env genes were identified by using a heteroduplex mobility assay and by sequencing, respectively (56). The irrelevant human MAb 1418 against parvovirus B19 was used as a negative control (10).
HIV-1 isolates.
Thirteen HIV-1 clade B viruses were used for this study. The isolates IIIB, BaL, SF162, ADA, JR-CSF, JR-FL, US1, and 92US717 were supplied by the AIDS Research and Reference Reagent Program and Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Two primary isolates, P15 and P27 (42), were obtained from D. C. Montefiori, Duke University Medical Center, Durham, N.C. Three chimeric viruses were constructed by subcloning of the KpnI-to-BamHI fragments of the 89.6, aBL-01, and dBR-07 env genes into an NL4-3 plasmid and were provided by Dana Gabuzda, Dana-Farber Cancer Institute, Boston, Mass. (37). All HIV-1 isolates were expanded by two or three cycles of growth on phytohemagglutinin (PHA)- and interleukin-2-stimulated PBMCs, as described previously (29). A single batch of each of the 13 viruses was used for all of the binding and neutralization experiments reported herein to avoid alterations in env sequences due to multiple rounds of expansion.
Binding assays.
The binding of MAbs to intact virions was determined with a capture assay as previously described (15). Briefly, a 96-well plate was coated overnight at 4°C with goat anti-human immunoglobulin G (IgG) Fc Abs (ICN Biomedicals, Aurora, Ohio) at 2.0 μg/ml, and then human MAbs at a saturating level of 10 μg/ml were added for a 1.5-h incubation at 37°C. The plate was blocked with 0.5% bovine serum albumin in phosphate-buffered saline (PBS) containing 10% goat serum and 10 μg of human IgG/ml. The culture supernatant, containing virions at a concentration of 100 ng of p24/ml, was incubated overnight on the plate at room temperature. Viruses captured by immobilized MAbs were lysed with 1% Triton-X in PBS. Between each step of the assay, the plate was washed with PBS containing 0.05% Tween 20, pH 7.4. The p24 in the virus lysate was quantified by using a noncommercial enzyme-linked immunosorbent assay (ELISA) as described previously (34). An irrelevant human MAb, 1418, was used as a negative control.
The relative affinity of MAb binding to intact virions was assessed with the virus capture assay by measuring the amount of MAb required for 50% maximal binding to virions. Plates coated with goat anti-human IgG Fc were incubated with human MAbs at concentrations ranging from 0.0001 to 10.0 μg/ml, followed by the addition of supernatants containing intact virions according to the steps described above for the virus capture assay. The concentration of MAb (in micrograms per milliliter) that gave 50% maximal binding was calculated by linear interpolation with Quattro Pro software (Corel, Ottawa, Canada) when the binding reached the saturation level.
The binding of MAbs to soluble gp120 was determined by ELISA, as described previously (20). Briefly, ELISA plates were coated with sheep anti-C-terminus gp120 antibody (Cliniqa Corporation, Fallbrook, Calif.), followed by incubation with Triton X-100-treated virus preparations adjusted to a concentration of 100 ng of p24/ml. MAbs at a saturating level of 10 μg/ml were added in duplicate, and bound MAbs were detected by using alkaline phosphatase-labeled goat anti-human IgG (γ specific) (Zymed Laboratories, South San Francisco, Calif.). The color was developed by using an amplification system from Life Technologies (Carlsbad, Calif.), and the plates were read at 490 nm. The optical densities (ODs) were normalized to the value given with the negative control, MAb 1418, and are presented as indices (ratios of experimental ODs to mean OD with MAb 1418). Positive binding was defined by using a cutoff value which was calculated as the mean OD with MAb 1418 plus 3 standard deviations.
Production of SF162 pseudovirions.
Pseudotyped virions expressing envelope glycoproteins derived from SF162 (psSF162) were constructed as described previously (15, 40). Briefly, plasmid pNL4-3.Luc.R−E− (5), supplied by the NIH AIDS Research and Reference Reagent Program, and plasmid pLRB826, expressing the env fragment from SF162 (25), were cotransfected into 293 cells by use of FuGENE6 (Roche Diagnostics, Indianapolis, Ind.). Forty-eight hours after transfection, the pseudovirus-containing supernatants were harvested, filtered through a 45-μm-pore-size filter, and stored at −80°C. For measurement of the infectivity of the pseudovirus, a luminescence assay with HOS-CD4/CCR5 cells was used as previously described (15).
Neutralization assays. (i) Luciferase assay.
A single-cycle infectivity assay was used to measure the neutralization of SF162 pseudovirions as described previously (15). Briefly, MAbs at various concentrations and a pseudovirus suspension at a final concentration of 0.5 ng of p24/ml were preincubated for 1.5 h at 37°C. The virus-antibody mixtures were added to HOS-CD4/CCR5 cells (NIH AIDS Research and Reference Reagent Program) which had been seeded 1 day before at 6 × 103 cells per well in a 96-well plate. The cultures were incubated for 3 days at 37°C, washed with PBS, and lysed with lysis buffer (Promega, Madison, Wis.). The cell lysates were transferred to luminometer plates (Corning, Corning, N.Y.), and the luciferase activity (in relative light units) in each well was measured by using Luciferase Substrate (Promega) in a Lumimark Plus system microplate reader (Bio-Rad Laboratories, Hercules, Calif.). The reduction in infectivity was determined by a comparison of the relative light units in the presence and absence of MAbs and is expressed as percent neutralization.
(ii) Single-round PBMC assay.
Single-round PBMC assays were performed according to a previously described method (29). Briefly, each virus was incubated with each MAb at a final concentration of 50 μg/ml. After incubation for 30 min at 37°C, 1.5 × 105 PHA-stimulated PBMCs were added to each well for 2 days. PBMCs were maintained in interleukin-2 medium containing the protease inhibitor indinavir. Cells were then fixed, permeabilized by use of a Cytofix/Cytoperm kit (BD-Pharmingen, San Diego, Calif.), stained with phycoerythrin-conjugated mouse anti-p24 MAb (KC57-RD1; Beckman Coulter), and analyzed in a FACSCalibur flow cytometer (Becton-Dickinson). Typically, 1 to 4% of the PBMCs in control wells were positive for p24 antigen, and 50,000 cells were counted. Data analysis was performed with FlowJo software (Tree Star, San Carlos, Calif.). The percent neutralization was defined as the reduction in the number of p24-positive cells in wells with MAb-treated virus compared with the number in control wells infected without MAb pretreatment. Positive neutralizing activities of the MAbs were determined by using a cutoff based on the 95% confidence level calculated from 13 negative control experiments using irrelevant human antiparvovirus MAb 1418.
Statistical analyses.
t tests, Pearson correlation calculations, and linear regression analyses were performed with GraphPad Prism, version 3.00, for Windows (GraphPad Software, San Diego, Calif.).
RESULTS
Characteristics of 32 human anti-V3 MAbs.
The 32 human anti-V3 MAbs listed in Table 1 were used for this study. They can be divided into two groups, those which were generated by the selection of Ab-producing cells with V3 peptides and those selected with V3 fusion proteins (V3-FPs) or gp120. V3 peptides present only secondary structure, while both V3-FP and gp120 molecules retain the conformation of the V3 loop. All 32 MAbs were tested for binding to recombinant gp120SF162 by ELISA and for neutralization of the SF162-pseudotyped virus by a luciferase assay. Representative data from duplicate experiments are shown in Fig. 1.
FIG. 1.
Neutralization (A and C) and binding (B and D) curves for a panel of 32 human anti-V3 MAbs against psSF162 and gp120SF162 protein. psSF162 is the pNL4-3 luc virus pseudotyped with SF162 env. The human MAbs tested were selected either with V3-FP or gp120 (A and B) or with V3 peptides (C and D). The curves representing MAbs with neutralizing activities above 50% neutralization are shown in red, and those with activities below 50% neutralization are shown in blue. Data for the irrelevant antiparvovirus MAb 1418 (negative control) are shown in green.
There was a distinct difference between the two sets of MAbs when neutralizing activities were compared. Taking the arbitrary, but commonly used, criterion of 50% neutralization, the results show that 11 of the 14 (78%) MAbs selected with either V3-FP or gp120 gave at least this level of neutralization at 1 μg/ml, while only 8 of the 18 (44%) MAbs selected with V3 peptides reached the same level of activity (Fig. 1A and C). Titration of the MAbs tested in the neutralization assays and a regression analysis of the data showed that the geometric mean 50% neutralizing dose for MAbs selected with V3-FP and gp120 (0.06 μg/ml) was significantly lower than that for MAbs selected with V3 peptides (0.40 μg/ml) (P = 0.0005) (Fig. 2A).
FIG. 2.
Neutralization of psSF162 (A) and binding activity of MAbs to gp120SF162 (B). The geometric mean of the 50% neutralization dose and 50% maximal binding values for MAbs selected with V3-FPs or gp120 (•; 0.06 and 0.04 μg of MAb/ml, respectively) were significantly lower than the respective values for MAbs selected with V3 peptides (○; 0.40 and 0.32 μg of MAb/ml).
Similarly, the relative affinity was different between both groups of MAbs. The relative affinity was determined by measuring the MAb concentration required for 50% maximal (half-max) binding to recombinant gp120SF162. The lower the concentration of MAb needed to achieve half-max binding, the higher the relative affinity. As seen in Fig. 1B and D and Fig. 2B, the geometric mean of the half-max binding values was lower for MAbs selected with V3-FP or gp120 (0.04 μg/ml) than for those selected with linear V3 peptides (0.32 μg/ml), with the difference being statistically significant (P = 0.0013).
These results demonstrate that MAbs selected with proteins retaining the V3 loop conformation have more neutralizing activity than those selected with peptides. This observation confirms the critical role played by the conformation of the V3 loop and the need for it to be recognized by Abs that bind avidly, leading to virus neutralization (15).
Relationship between neutralization and affinity.
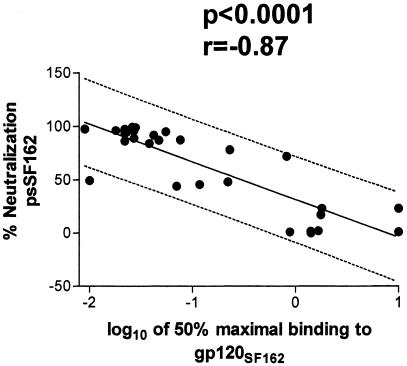
The experiments described above suggest that the neutralizing activities against psSF162 of the 32 MAbs tested correspond to binding activities for soluble gp120 of SF162. A regression analysis of the data showed a significant correlation between the percent neutralization determined with a MAb concentration of 1 μg/ml and the relative affinity of MAbs binding to monomeric gp120SF162 (P < 0.0001; Fig. 3). This analysis indicates that the affinity of anti-V3 MAbs is an important factor in efficient neutralization of psSF162.
FIG. 3.
Linear regression analysis of percent neutralization of psSF162 versus the logarithm of 50% maximal MAb binding to gp120SF162. The best-fit regression line (solid line) and the 95% prediction interval (dashed lines) are shown. The data enclosed by the latter are expected to include 95% of all data points.
Neutralization of clade B viruses.
To extend these studies, we used seven MAbs, including six selected with V3-FP and the best of the MAbs selected with V3MN (MAb 447-52D), to determine the sensitivities of 13 clade B viruses to neutralization. Nine viruses were R5-tropic, one was X4-tropic, and three were dual-tropic; all were produced in PBMCs (Table 2). Human MAb 1418, specific for parvovirus B19, was used as a negative control. For a statistically derived rather than arbitrary value to allow comparative data analysis, significant neutralization was based on the 95% confidence level derived from data collected from 13 experiments with the latter MAb. This established neutralization of >18% as statistically significant.
TABLE 2.
Percent neutralization of B clade viruses in single-round PBMC assays
| Virus | Coreceptor | % Neutralization of indicated virus with MAb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 2182 | 2191 | 2219 | 2412 | 2442 | 2456 | 447-52D | 1418 | ||
| BaL | R5 | 38 | 88 | 91 | 31 | 92 | 75 | 98 | 7 |
| SF162 | R5 | 81 | 96 | 99 | 89 | 93 | 85 | 99 | −20 |
| JR-CSF | R5 | 92 | 90 | 95 | 53 | 93 | 76 | 94 | −3 |
| US1 | R5 | 49 | 41 | 44 | 7 | 33 | 29 | 42 | 12 |
| JR-FL | R5 | 69 | 54 | 35 | −5 | 31 | 1 | 73 | 2 |
| 92US717 | R5 | 1 | 60 | 32 | −5 | 46 | 14 | 80 | 5 |
| P15 | R5 | 14 | 15 | 8 | 21 | 47 | 14 | 38 | 4 |
| P27 | R5 | 34 | 15 | 0 | 10 | 38 | 16 | 33 | 8 |
| 89.6 | R5 and X4 | 11 | 88 | 10 | −7 | 95 | 1 | 98 | 6 |
| IIIB | X4 | 14 | 19 | −1 | 1 | 7 | NT | 98 | 9 |
| ADA | R5 | 5 | 13 | 15 | 15 | 16 | −4 | 45 | 1 |
| dBR-07 | R5 and X4 | 1 | −17 | −15 | −12 | −1 | 6 | 85 | −5 |
| aBL-01 | R5 and X4 | −25 | −14 | −31 | −6 | −8 | 3 | 6 | −11 |
All MAbs were tested at 50 μg/ml. The cutoff value of 18% is based on the 95% confidence level obtained with 13 experiments with the irrelevant MAb 1418. Values of >18% are in bold and represent statistically significant neutralization. NT, not tested.
Of 90 virus-MAb combinations (one combination, IIIB-2456, was not tested), 49 (54%) showed significant neutralization. Each combination was tested in two experiments, and the data from one of these are shown in Table 2. The most cross-neutralizing MAb was 447-52D, which neutralized 12 of 13 viruses, although each of the other MAbs also neutralized 4 or more of the viruses tested. Thus, despite the sequence changes throughout the V3 loop in these viruses (Table 3), anti-V3 MAbs displayed extensive cross-neutralization of primary isolates.
TABLE 3.
V3 sequences of the clade B HIV-1 strains testeda
| Strain | Amino acid sequence |
|---|---|
| Consensus | TRPNNNTRKSIHI..GPGRAFYTTGDIIGDIRQAH |
| BaL | -------------..----------E--------- |
| SF162 | -----------T-..-------A------------ |
| JR-CSF | ---S---------..----------E--------- |
| US1 | I------------..-----I-A--G------R-Y |
| JR-FL | -------------..----------E--------- |
| 92US717 | I-------R--N-..--------------N----- |
| P15 | -X----------M..---A---AR-EV-------- |
| P27 | -E-----I----L..-----WH---Q------K-F |
| 89.6 | --------RRLS-..-------ARRN--------- |
| IIIB | ---------R-R-QR------V-I--K--NM---- |
| ADA | -------------..----------E--------- |
| dBR-07 | --------RG-SM..------LAREQ---N----- |
| aBL-01 | -------KR--TR..----VY------V------- |
Dashes represent identity with the top sequence in the alignment (consensus of clade B); dots represent positions where insertions appear in IIIB.
The viruses tested were chosen to span the spectrum of neutralization sensitivities based on previous experiments, with BaL, SF162, and JR-CSF representing relatively sensitive viruses and ADA, dBR-07, and aBL-01 representing more resistant viruses. In fact, as shown in Table 2, BaL, SF162, and JR-CSF were neutralized by all of the anti-V3 MAbs tested, while ADA and dBR-07 were neutralized by only one MAb and aBL-01 was not neutralized by any of the MAbs tested. The other seven viruses tested fell between the two extremes, being neutralized by two to six of the anti-V3 MAbs. These observations suggest that the V3 domain is functionally accessible in 12 of the 13 viruses tested.
Relative affinity of MAb binding to intact virions.
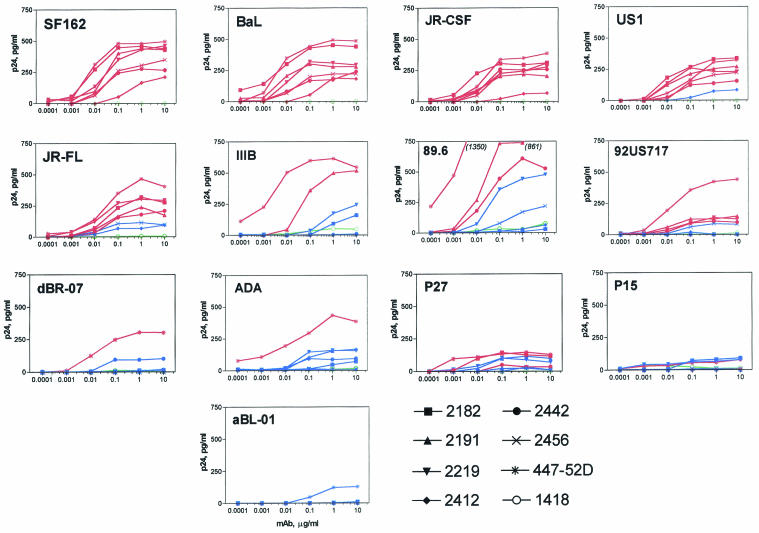
As noted above, 54% of the MAb-virus combinations showed significant neutralization, with some viruses displaying considerably more neutralization sensitivity to anti-V3 MAbs than others. To examine the relationship between MAb binding to virions and neutralization, we performed studies to assess the abilities of MAbs to bind to intact virions. ELISA plates coated with each of the MAbs at concentrations ranging from 0.0001 to 10 μg/ml were used, and the quantity of virus bound was assessed by lysing the bound virus and measuring the levels of released p24, a technique previously used by us and others (2, 4, 34, 38). Representative binding curves for the 90 anti-V3 MAb-virus combinations are shown in Fig. 4.
FIG. 4.
Titration curves for the binding of eight MAbs to 13 intact virions. The MAbs were tested at concentrations ranging from 0.0001 to 10.0 μg/ml and virus was added to all wells at a constant concentration of 100 ng of p24/ml. The amounts of virus bound by MAbs are represented by the picograms of p24 per milliliter released by detergent treatment of bound virus. The binding curves for neutralizing MAbs (as defined in Table 2) are shown in red, those for nonneutralizing MAbs are blue, and those for the negative control MAb 1418 are green.
The binding curves of MAbs which do not neutralize each virus (as defined in Table 2) are shown in blue and fall below those of MAbs with significant neutralizing activities (shown in red). These curves suggest that, in general, virus capture by anti-V3 MAbs corresponds to virus neutralization. A corollary of this is that those viruses whose V3 loops are most exposed and best recognized by anti-V3 Abs are most effectively neutralized by these MAbs.
The various viruses appear to display different thresholds for the level of anti-V3 MAb binding necessary to result in neutralization. Thus, the thresholds for SF162, BaL, and JR-CSF are very low, and consequently all anti-V3 MAbs that bind to them are neutralized. In contrast, viruses such as US1, JR-FL, IIIB, and 89.6 display thresholds below which binding does not result in neutralization. ADA and dBR-07 are distinct in that they are efficiently captured only by MAb 447, and this is the only MAb that is able to effect neutralization of these two viruses. In contrast, the capture of viruses P15, P27, and aBL-01 is relatively weak or at background levels, and neutralization of these viruses is likewise modest or, in the case of aBL-01, undetected.
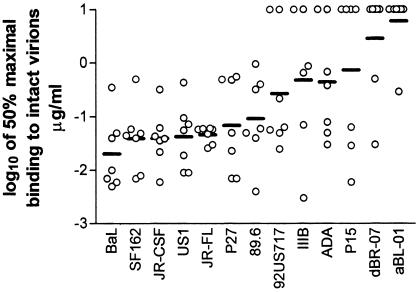
Relative affinities were determined by measuring 50% maximal binding to intact virions; these values are shown in Fig. 5. The lower the half-max value, the higher the relative affinity. The lowest mean relative affinities were for Bal, SF162, JR-CSF, US1, and JR-FL, the same viruses that are most effectively neutralized by the MAbs, again suggesting a correlation between binding and neutralization. A regression analysis showed that relative affinities (half-max values) indeed correlate significantly with percent neutralization (P < 0.0001) (Fig. 6A). This analysis was confirmed when, instead of using half-max binding values, we assessed binding on the basis of virus captured by MAbs under conditions of saturation (10 μg of MAb/ml). Under these conditions, the amount of virus captured by the MAb again significantly correlated with virus neutralization (P < 0.0001) (Fig. 6B). These data confirm and extend previous observations (15).
FIG. 5.
Half-maximal binding of MAbs to intact virions. The logarithms of the half-maximal values (micrograms of MAb/ml) calculated from the binding curves shown in Fig. 4 are shown for each of 13 virus isolates, with mean values shown as solid lines.
FIG. 6.
Linear regression analyses of neutralization and virus binding data for 90 MAb-virus combinations. The data analyzed were taken from Table 2 and Fig. 4 and 5. The analyses show the correlation between percent neutralization and either the log of 50% maximal binding (A) or virus binding at saturation (using 10 μg of MAb/ml) (B). Best-fit regression lines (solid lines) and 95% prediction intervals (dashed lines) are shown.
Binding of MAbs to solubilized gp120s.
The lack of neutralization and binding to particular virions could be due either to the absence of the epitope or to its inaccessibility on the intact particles. To address this issue, we probed detergent-solubilized gp120 molecules from each virus with seven anti-V3 MAbs under saturating conditions (10 μg of MAb/ml) in order to determine the presence of V3 epitopes on the monomeric gp120 molecules. Representative results are shown in Table 4. Each MAb-gp120 combination was tested twice, and MAb binding to soluble gp120 was expressed as an index in order to normalize the results, given the different amounts of gp120 in various virus preparations. Four observations emerged that were relevant to the antigenic variation of the V3 loop and its accessibility on virions to Abs. (i) The gp120s from all viruses tested, except dBR-07, possessed V3 epitopes that were recognized by one or more of the seven anti-V3 MAbs which were derived from individuals infected with heterologous viruses, and even, in the case of MAb 2182, with a virus from a heterologous clade (Table 4). (ii) For 44 of the 90 MAb-virus combinations, binding to soluble gp120 corresponded to neutralization (Tables 2 and 4). (iii) For 38 of the 90 anti-V3 MAb-gp120 combinations tested (42%), there was no recognition of the epitope, attesting to the antigenic variation of the V3 loop. For 33 of these 38 combinations, the absence of binding to gp120 was associated, as expected, with the absence of neutralizing activity for the respective virus. In five of these cases, however, there was no detectable binding of the MAb to gp120, yet neutralization of the corresponding virus did occur (MAb 2182-SF162, MAb 2412-P15, MAb 2442-P15, MAb 2442-P27, and MAb 447-52D-dBR-07) (Tables 2 and 4). In these cases, the MAb would have had to have bound to the virion, in which case binding might have been enhanced by oligomer formation (14) or by changes in the V3 conformation induced by binding to cell surface CD4 (30). (iv) For eight combinations, binding of a MAb to gp120 was noted but resulted in no neutralization, e.g., MAb 2219-P27, MAb 2412-US1, and others (Tables 2 and 4). While representing only 9% of the combinations tested, these may represent instances in which the V3 loop is inaccessible on the virus particles, i.e., it is “cryptic.”
TABLE 4.
Binding of anti-V3 MAbs to solubilized gp120
| Virus | Binding of MAb to indicated virusa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 2182 | 2191 | 2219 | 2412 | 2442 | 2456 | 447-52D | 1418 | |
| BaL | 6.3 | 6.1 | 6.1 | 6.1 | 5.9 | 6.1 | 5.9 | 1.0 |
| SF162 | 1.7 | 18.3 | 12.2 | 12.2 | 12.2 | 7.2 | 18.9 | 1.0 |
| JR-CSF | 11.0 | 11.7 | 12.0 | 11.7 | 9.6 | 11.7 | 11.7 | 1.0 |
| US1 | 13.4 | 13.0 | 12.7 | 12.7 | 9.6 | 12.3 | 12.3 | 1.0 |
| JR-FL | 5.7 | 9.2 | 10.1 | 8.5 | 2.4 | 7.6 | 14.7 | 1.0 |
| 92US717 | 0.5 | 6.8 | 4.2 | 6.6 | 5.1 | 5.7 | 6.8 | 1.0 |
| P15 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 2.7 | 1.0 |
| P27 | 15.0 | 1.4 | 3.5 | 5.0 | 1.4 | 1.4 | 9.2 | 1.0 |
| 89.6 | 0.6 | 2.5 | 1.2 | 1.2 | 5.0 | 1.3 | 7.5 | 1.0 |
| IIIB | 0.7 | 2.1 | 1.4 | 0.7 | 0.7 | NT | 17.8 | 1.0 |
| ADA | 0.7 | 1.3 | 2.0 | 0.7 | 1.3 | 0.7 | 4.0 | 1.0 |
| dBR-07 | 0.7 | 0.7 | 1.3 | 0.7 | 0.7 | 0.7 | 1.3 | 1.0 |
| aBL-01 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 3.1 | 1.0 |
Values shown are index values, expressed as the ratios of mean sample optical densities to mean optical densities with MAb 1418; index values shown in bold are above the cutoff values, which were defined as the means + 3 standard deviations from the negative control experiments using each soluble gp120 preparation and MAb 1418; NT, not tested.
Interestingly, linear V3 sequences did not necessarily predict gp120 binding. For example, most MAbs bound strongly to BaL gp120 but not to ADA gp120, despite identical V3 sequences in these two viruses (Table 3). These data suggest that changes at distant sites in the envelope can affect the conformation and antigenic nature of the V3 loop, an observation which is supported by other work in the literature (32, 48, 54).
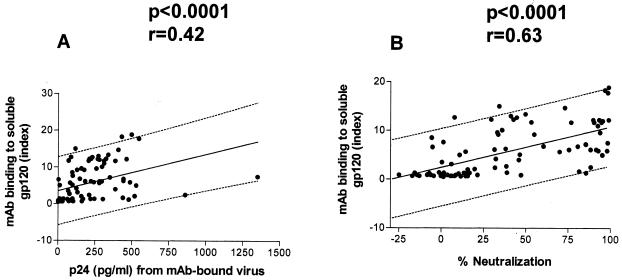
Generally, gp120 binding indices had the highest values for the neutralization-sensitive viruses BaL, SF162, JR-CSF, US1, JR-FL, and 92US717, while the binding indices were lower for the other viruses, which are less sensitive to neutralization. A regression analysis of the data revealed a significant correlation between MAb binding to soluble gp120 and intact virions at saturation, indicating that V3 epitopes present on monomeric gp120 are also present on intact virions and suggesting that their accessibilities to MAbs are similar in the monomeric and oligomeric states (P < 0.0001) (Fig. 7A). Similarly, a significant correlation was found between MAb binding to soluble gp120 and percent neutralization (P < 0.0001) (Fig. 7B). However, the regression data obscure the exceptions noted above, for which anti-V3 MAb binding to gp120 was noted with no consequent neutralization (8 of 90 combinations) and for which no anti-V3 MAb binding to gp120 was noted but neutralization occurred (5 of 90 combinations).
FIG. 7.
Correlation between MAb binding to detergent-solubilized gp120 and capture of intact virions by MAbs under Ab-saturating conditions (A) and between MAb binding to gp120 and percent neutralization at 50 μg of MAb/ml (B). The data analyzed were taken from Tables 2 and 4 and Fig. 4.
DISCUSSION
In this and previous publications, both polyclonal and monoclonal anti-V3 Abs have been shown to neutralize diverse primary isolates of HIV-1 (8, 15, 19, 25, 33), although not all viruses are neutralized by these reagents. The neutralizing activities of anti-V3 Abs may be limited due to antigenic variation of the V3 region, lack of V3 exposure on the surfaces of intact virions, and/or the nature of the Ab specificity. For clarification of this issue, a panel of 32 human anti-V3 MAbs were screened for binding to intact virions, for binding to solubilized gp120 from these viruses, and for neutralization. The results suggest that the V3 loop is similarly exposed on solubilized gp120 and on intact virions, that the V3 loop is accessible in all viruses studied, that the strength of binding of anti-V3 MAbs to intact virions correlates strongly with the potency of neutralizing activity, and that the V3 loops of diverse primary isolates display shared epitopes that can be recognized by anti-V3 MAbs with neutralizing activities.
For this study, the viruses that were selected represented a sampling within the spectrum of neutralization sensitivities and resistances. For example, BaL, SF162, and JR-CSF are well established as viruses that are sensitive to neutralization by various MAbs, including anti-V3 MAbs (29, 44). Other viruses, including ADA, P15, P27, aBL-01, and dBR-07, represent viruses that are relatively resistant to anti-V3 MAbs. The remaining viruses tested fall between these two extremes. The rationale for the use of these viruses was to determine if and how exposure of the V3 loop on the virus surface correlates with neutralization by anti-V3 MAbs. Binding studies revealed that the V3 loop was exposed in all viruses tested, since all could be captured with one or more of the anti-V3 MAbs studied. Each of these viruses, except aBL-01, was also neutralized by at least one MAb, and a highly significant correlation was found between strength of binding to intact virions and potency of neutralization (P < 0.0001) (Fig. 3 and 6).
While all of the viruses examined in this study displayed V3 on their surfaces, data shown above and published previously suggest that the degree of exposure differs with the virus and the conditions studied. Thus, by using saturating conditions in the virus binding assay, in which virions are captured on ELISA plates coated with 10 μg of MAb/ml, we noted large differences in the capture of different viruses. For example, on average, much more BaL was captured by the anti-V3 MAbs than 92US717 (Fig. 4). Nonetheless, the V3 epitopes seemed to be comparably recognized when these same MAbs reacted with the solubilized gp120s from these two viruses (Table 4). These data lead to the conclusions that the relevant V3 epitopes are present within the gp120s of these viruses but that the V3 loop is more accessible on the BaL virus than it is on the 92US717 virus. This view is supported by data in Fig. 5 showing that the average half-max binding of the anti-V3 MAbs is much lower (i.e., the relative affinity is much higher) for BaL than it is for 92US717. Similarly, each MAb tested neutralized BaL more strongly than 92US717 (Table 2). These data reflect the differential exposure of the V3 loop on the intact virions of these two strains, and similar data in the cited tables and figures support the notion that V3 is differentially exposed by different strains of the virus. This concept is extended by data from other studies suggesting that some viruses apparently have cryptic V3 loops (3, 6, 49), although, from the data presented above, this appears to be the exception rather than the rule.
Sensitivity to anti-V3 Abs may also be affected by the mobility of the V3 loop upon interaction of gp120 with CD4. Thus, Mbah et al. (30) showed that, upon treatment with soluble CD4, the exposure of the V3 loop on virions of primary isolates is increased, but this occurs to different degrees with different viruses. Consequently, the sensitivity of viruses to anti-V3 Abs may also be affected by the mobility of the V3 loop during the conformational changes that occur in gp120 upon binding to CD4 (31, 43, 45, 49).
The relative affinities of anti-V3 MAbs were also found to play a profound role in the neutralization process. A highly significant correlation (P < 0.0001) was demonstrated between the percent neutralization of psSF162 tested in a luciferase assay and the half-max values for binding of 32 anti-V3 MAbs with this pseudovirus (Fig. 3). Similarly, a highly significant correlation (P < 0.0001) was found when the percent neutralization in a single-round PBMC assay and half-max virion binding values were analyzed for 90 MAb-virus combinations (Fig. 6). This extends earlier studies in which primary isolates from clades A to F were tested against seven human anti-V3 MAbs for MAb-virion binding and for neutralization (by the GHOST assay and the conventional PHA-blasted PBMC assay), and significant correlations were again found (P < 0.0001 and P < 0.001, respectively) (15). Earlier studies, which were limited to neutralization of TCLA strains and binding to either monomeric gp120 or V3 peptides, also concluded that anti-V3 MAb affinity correlates with its neutralizing potency and further suggested that the dissociation rate (K−1) rather than the association rate (K1) is the principal component which determines neutralizing activity (27, 53). While Abs against other epitopes of the HIV envelope may not be constrained by the same affinity limitations (41), experiments in a multitude of systems consistently support the association between affinity and neutralization for anti-V3 Abs.
In summary, our data indicate that neutralization sensitivity to anti-V3 Abs appears to be affected by the presence or absence of the relevant epitope(s) on the virion envelope, the exposure of the V3 loop on the intact virion, the mobility of the V3 loop during the conformational change induced by CD4, and the affinity of the Ab. Additional factors influencing neutralization sensitivity are the density and/or number of Env oligomers on the surface of a given virus. All of these parameters contribute to the shape of the neutralization curve and the ultimate outcome of Ab-virus interactions. These multiple parameters, and most probably several others, contribute to the complex equation that determines neutralization sensitivity or resistance. Thus, viruses such as BaL, SF162, and JR-CSF, which according to the data shown in Fig. 4 have a very low threshold for resisting Ab-mediated neutralization, appear to have well-exposed V3 loops for which the anti-V3 MAbs have high affinities (Fig. 5); in contrast, viruses such as BR07, ADA, and aBL01 have higher thresholds as a consequence, at least in part, of a poorly exposed V3 loop and low-affinity interactions with anti-V3 MAbs. The existence of a threshold was suggested previously in a study showing that anti-V3 MAb neutralization of TCLA HIV-1 was incremental rather than all or nothing and that each MAb binding an Env oligomer reduced infectivity (46). Recently, Franti et al. also described a threshold effect when studying the interaction between JF-CSF and the anti-gp120 MAbs b12, 447-52D, and 2G12 (M. Franti, S. Frost, M. Guyader, K. Delgado, D. R. Burton, and P. Poignard, Abstr. AIDS Vaccine 2003, abstr. 118, 2003). The data included in Fig. 4, however, indicate for the first time that the levels of these thresholds will vary with each individual strain of virus.
The affinities of the anti-V3 MAbs tested here for psSF162 and 13 clade B viruses grown in PBMCs also varied by more than 3 orders of magnitude (Fig. 1, 4, and 5); similarly, profound differences were described previously in the affinities of several of these MAbs for the V3-FP and V3 peptide of JR-CSF (15). Clearly, HIV-infected subjects make a broad range of anti-V3 Abs, both with respect to affinity and specificity (57). With the single, but important, exception of MAb 447, which was selected with the V3 peptide of MN, the most avid anti-V3 MAbs were selected with V3-FPs, which maintain the native conformation of the V3 loop (22). These data have important implications for vaccine design, as they establish the fact that the human Ab repertoire includes anti-V3 Abs with broad and potent cross-neutralizing activities and they identify the characteristics of the types of anti-V3 Abs which will have the greatest neutralizing efficacies and therefore the highest probabilities of blocking a virus inoculum. These characteristics include broad immunochemical cross-reactivity (Table 4) (35, 57), broad neutralizing activity (Table 2) (15), and a high affinity for gp120 and intact virus particles (Table 4) (15). Antigens used to select MAbs with these characteristics, such as V3-FP, provide a template for the design of immunogens that will focus the immune response on the V3 loop and should induce high-affinity, broadly reactive Abs to conformational epitopes on V3. The apparent requirement for conformational aspects of V3 revealed by our studies may also provide an explanation for the disappointing results with previously used V3 immunogens (1, 7, 23) which lacked the appropriate conformational aspects of the V3 loop.
Acknowledgments
This study was supported in part by grants from the NIH (HL59725, AI36085, and AI 47053) and the Immunology Core of the NYU Center for AIDS Research (NIH grant AI27742) and by research funds from the Department of Veterans Affairs.
We are grateful to Dana Gabuzda and David Montefiori for providing several HIV-1 isolates used in this study.
REFERENCES
- 1.Ahlers, J. D., C. D. Pendleton, N. Dunlop, A. Minassian, P. L. Nara, and J. A. Berzofsky. 1993. Construction of an HIV-1 peptide vaccine containing a multideterminant helper peptide linked to a V3 loop peptide 18 inducing strong neutralizing antibody responses in mice of multiple MHC haplotypes after two immunizations. J. Immunol. 150:5647-5665. [PubMed] [Google Scholar]
- 2.Bastiani, L., S. Laal, M. Kim, and S. Zolla-Pazner. 1997. Host cell-dependent alterations in envelope components of human immunodeficiency virus type 1 virions. J. Virol. 71:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bou-Habib, D. C., G. Roderiquez, T. Oravecz, P. W. Berman, P. Lusso, and M. A. Norcross. 1994. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J. Virol. 68:6006-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavacini, L. A., J. E. Peterson, E. Nappi, M. Duval, R. Goldstein, K. Mayer, and M. R. Posner. 1999. Minimal incidence of serum antibodies reactive with intact primary isolate virions in human immunodeficiency virus type 1-infected individuals. J. Virol. 73:9638-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, B. K., K. Saksela, R. Andino, and D. Baltimore. 1994. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 68:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C.-H., L. Jin, C. Zhu, S. Holz-Smith, and T. J. Matthews. 2001. Induction and characterization of neutralizing antibodies against a human immunodeficiency virus type 1 primary isolate. J. Virol. 75:6700-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conley, A. J., P. Conard, S. Bondy, C. A. Dolan, J. Hannah, W. J. Leanza, S. Marburg, M. Rivetna, V. K. Rusiecki, E. E. Sugg, F. Van Middlesworth, S. A. Warne, J. T. Ulrich, J. A. Rudbach, R. L. Tolman, and E. A. Emini. 1994. Immunogenicity of synthetic HIV-1 gp120 V3-loop peptide-conjugate immunogens. Vaccine 12:445-451. [DOI] [PubMed] [Google Scholar]
- 8.Conley, A. J., M. K. Gorny, J. A. Kessler II, L. J. Boots, M. Ossorio-Castro, S. Koenig, D. W. Lineberger, E. A. Emini, C. Williams, and S. Zolla-Pazner. 1994. Neutralization of primary human immunodeficiency virus type 1 virus isolates by the broadly reactive anti-V3 monoclonal antibody 447-52D. J. Virol. 68:6994-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gigler, A., S. Dorsch, A. Hemauer, C. Williams, S. Kim, N. S. Young, S. Zolla-Pazner, H. Wolf, M. K. Gorny, and S. Modrow. 1999. Generation of neutralizing human monoclonal antibodies against parvovirus B19 proteins. J. Virol. 73:1974-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorny, M. K. 1994. Production of human monoclonal antibodies via fusion of Epstein-Barr virus-transformed lymphocytes with heteromyeloma, p. 276-281. In J. E. Celis (ed.), Cell biology: a laboratory handbook, vol. 2. Academic Press, New York, N.Y.
- 12.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J.-Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorny, M. K., T. C. VanCott, C. Hioe, Z. R. Israel, N. L. Michael, A. J. Conley, C. Williams, J. A. Kessler II, P. Chigurupati, S. Burda, and S. Zolla-Pazner. 1997. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 159:5114-5122. [PubMed] [Google Scholar]
- 14.Gorny, M. K., T. C. VanCott, C. Williams, K. Revesz, and S. Zolla-Pazner. 2000. Effects of oligomerization on the epitopes of the human immunodeficiency virus type 1 envelope glycoproteins. Virology 267:220-228. [DOI] [PubMed] [Google Scholar]
- 15.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. P. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 76:9035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorny, M. K., J.-Y. Xu, V. Gianakakos, S. Karwowska, C. Williams, H. W. Sheppard, C. V. Hanson, and S. Zolla-Pazner. 1991. Production of site-selected neutralizing human monoclonal antibodies against the third variable domain of the HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 88:3238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorny, M. K., J.-Y. Xu, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150:635-643. [PubMed] [Google Scholar]
- 18.Hill, C. M., H. Deng, D. Unutmaz, V. N. Kewalramani, L. Bastiani, M. K. Gorny, S. Zolla-Pazner, and D. R. Littman. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 71:6296-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hioe, C. E., S. Xu, P. Chigurupati, S. Burda, C. Williams, M. K. Gorny, and S. Zolla-Pazner. 1997. Neutralization of HIV-1 primary isolates by polyclonal and monoclonal human antibodies. Int. Immunol. 9:1281-1290. [DOI] [PubMed] [Google Scholar]
- 20.Israel, Z. R., M. K. Gorny, C. Palmer, J. A. McKeating, and S. Zolla-Pazner. 1997. Prevalence of a V2 epitope in clade B primary isolates and its recognition by sera from HIV-1 infected individuals. AIDS 11:128-130. [PubMed] [Google Scholar]
- 21.Javaherian, K., A. J. Langlois, C. McDanal, K. L. Ross, L. I. Eckler, C. L. Jellis, A. T. Profy, J. R. Rusche, D. P. Bolognesi, S. D. Putney, and T. J. Matthews. 1989. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc. Natl. Acad. Sci. USA 86:6768-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayman, S. C., Z. Wu, K. Revesz, H. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller, P. M., B. A. Arnold, A. R. Shaw, R. L. Tolman, F. Van Middlesworth, S. Bondy, V. K. Rusiecki, S. Koenig, S. Zolla-Pazner, P. Conard, E. A. Emini, and A. J. Conley. 1993. Identification of HIV vaccine candidate peptides by screening random phage epitope libraries. Virology 193:709-716. [DOI] [PubMed] [Google Scholar]
- 24.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 25.Krachmarov, C. P., S. C. Kayman, W. J. Honnen, O. Trochev, and A. Pinter. 2001. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1737-1748. [DOI] [PubMed] [Google Scholar]
- 26.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langedijk, J. P., N. K. Back, E. Kinney-Thomas, C. Bruck, M. Francotte, J. Goudsmit, and R. H. Meloen. 1992. Comparison and fine mapping of both high and low neutralizing monoclonal antibodies against the principal neutralization domain of HIV-1. Arch. Virol. 126:129-146. [DOI] [PubMed] [Google Scholar]
- 28.LaRosa, G. J., K. Weinhold, A. T. Profy, A. J. Langlois, G. R. Dreesman, R. N. Boswell, P. Shadduck, D. P. Bolognesi, T. J. Matthews, E. A. Emini, and S. D. Putney. 1991. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant: further clarifications. Science 253:1146. [DOI] [PubMed] [Google Scholar]
- 29.Mascola, J. R., M. K. Louder, C. Winter, R. Prabhakara, S. C. De Rosa, D. C. Douek, B. J. Hill, D. Gabuzda, and M. Roederer. 2002. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J. Virol. 76:4810-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mbah, H. A., S. Burda, M. K. Gorny, C. Williams, K. Revesz, S. Zolla-Pazner, and P. N. Nyambi. 2001. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J. Virol. 75:7785-7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKeating, J. A., J. Cordell, C. J. Dean, and P. Balfe. 1992. Synergistic interaction between ligands binding to the CD4 binding site and V3 domain of human immunodeficiency virus type 1 gp120. Virology 191:732-742. [DOI] [PubMed] [Google Scholar]
- 32.McKeating, J. A., J. Gow, J. Goudsmit, L. H. Pearl, C. Mulder, and R. A. Weiss. 1989. Characterization of HIV-1 neutralization escape mutants. AIDS 3:777-784. [DOI] [PubMed] [Google Scholar]
- 33.Moore, J. P., A. Trkola, B. Korber, L. J. Boots, J. A. Kessler II, F. E. McCutchan, J. Mascola, D. D. Ho, J. Robinson, and A. J. Conley. 1995. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J. Virol. 69:122-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyambi, P. N., M. K. Gorny, L. Bastiani, G. van der Groen, C. Williams, and S. Zolla-Pazner. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 72:9384-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyambi, P. N., H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, A. Nadas, and S. Zolla-Pazner. 2000. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J. Virol. 74:7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nyambi, P. N., A. Nadas, H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, and S. Zolla-Pazner. 2000. Immunoreactivity of intact virions of human immunodeficiency virus type 1 (HIV-1) reveals the existence of fewer HIV-1 immunotypes than genotypes. J. Virol. 74:10670-10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohagen, A., A. Devitt, K. J. Kunstman, P. R. Gorry, P. P. Rose, B. Korber, J. Taylor, R. Levy, R. L. Murphy, S. M. Wolinsky, and D. Gabuzda. 2003. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J. Virol. 77:12336-12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orentas, R. J., and J. E. Hildreth. 1993. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Res. Hum. Retrovir. 9:1157-1165. [DOI] [PubMed] [Google Scholar]
- 39.Palker, T. J., M. E. Clark, A. J. Langlois, T. J. Matthews, K. J. Weinhold, R. R. Randall, D. P. Bolognesi, and B. F. Haynes. 1988. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc. Natl. Acad. Sci. USA 85:1932-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, E. J., M. K. Gorny, S. Zolla-Pazner, and G. V. Quinnan, Jr. 2000. A global neutralization resistance phenotype of human immunodeficiency virus type 1 is determined by distinct mechanisms mediating enhanced infectivity and conformational change of the envelope complex. J. Virol. 74:4183-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parren, P. W., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilgrim, A. K., G. Pantaleo, O. J. Cohen, L. M. Fink, J. Y. Zhou, J. T. Zhou, D. P. Bolognesi, A. S. Fauci, and D. C. Montefiori. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J. Infect. Dis. 176:924-932. [DOI] [PubMed] [Google Scholar]
- 43.Pinter, A., W. J. Honnen, and S. A. Tilley. 1993. Conformational changes affecting the V3 and CD4-binding domains of human immunodeficiency virus type 1 gp120 associated with env processing and with binding of ligands to these sites. J. Virol. 67:5692-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schonning, K., B. Jansson, S. Olofsson, and J. E. Hansen. 1996. Rapid selection for an N-linked oligosaccharide by monoclonal antibodies directed against the V3 loop of human immunodeficiency virus type 1. J. Gen. Virol. 77:753-758. [DOI] [PubMed] [Google Scholar]
- 47.Sharon, M., N. Kessler, R. Levy, S. Zolla-Pazner, M. Gorlach, and J. Anglister. 2003. Alternative conformations of HIV-1 V3 loops mimic β hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure (Cambridge) 11:225-236. [DOI] [PubMed] [Google Scholar]
- 48.Sirko, D. A., and G. D. Ehrlich. 1996. Genotypic and phenotypic characterization of a neutralization-resistant breakthrough population of HIV-1. Virology 218:238-242. [DOI] [PubMed] [Google Scholar]
- 49.Stamatatos, L., and C. Cheng-Mayer. 1995. Structural modulations of the envelope gp120 glycoprotein of human immunodeficiency virus type 1 upon oligomerization and differential V3 loop epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor binding. J. Virol. 69:6191-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suphaphiphat, P., A. Thitithanyanont, S. Paca-Uccaralertkun, M. Essex, and T. H. Lee. 2003. Effect of amino acid substitution of the V3 and bridging sheet residues in human immunodeficiency virus type 1 subtype C gp120 on CCR5 utilization. J. Virol. 77:3832-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 52.Tugarinov, V., A. Zvi, R. Levy, Y. Hayek, S. Matsushita, and J. Anglister. 2000. NMR structure of an anti-gp120 antibody complex with a V3 peptide reveals a surface important for co-receptor binding. Struct. Fold Des. 8:385-395. [DOI] [PubMed] [Google Scholar]
- 53.VanCott, T. C., F. R. Bethke, V. R. Polonis, M. K. Gorny, S. Zolla-Pazner, R. R. Redfield, and D. L. Birx. 1994. Dissociation rate of antibody-gp120 binding interactions is predictive of V3-mediated neutralization of HIV-1. J. Immunol. 153:449-459. [PubMed] [Google Scholar]
- 54.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 55.Wyatt, R., N. Sullivan, M. Thali, H. Repke, D. Ho, J. Robinson, M. Posner, and J. Sodroski. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong, P., S. Burda, M. Urbanski, H. Kenfack, M. Tongo, L. Heyndrickx, A. Nanfack, J. Shang, L. Agyingi, S. Zolla-Pazner, L. Zekeng, and P. Nyambi. 2002. HIV type 1 group M clades infecting subjects from rural villages in equatorial rain forests of Cameroon. J. Acquir. Immune Defic. Syndr. 31:495-505. [DOI] [PubMed] [Google Scholar]
- 57.Zolla-Pazner, S., M. K. Gorny, P. N. Nyambi, T. C. VanCott, and A. Nádas. 1999. Immunotyping of human immunodeficiency virus type 1 (HIV): an approach to immunologic classification of HIV. J. Virol. 73:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]