Abstract
Understanding the history of forests and their species' demographic responses to past disturbances is important for predicting impacts of future environmental changes. Tropical rainforests of the Guineo-Congolian region in Central Africa are believed to have survived the Pleistocene glacial periods in a few major refugia, essentially centred on mountainous regions close to the Atlantic Ocean. We tested this hypothesis by investigating the phylogeographic structure of a widespread, ancient rainforest tree species, Symphonia globulifera L. f. (Clusiaceae), using plastid DNA sequences (chloroplast DNA [cpDNA], psbA-trnH intergenic spacer) and nuclear microsatellites (simple sequence repeats, SSRs). SSRs identified four gene pools located in Benin, West Cameroon, South Cameroon and Gabon, and São Tomé. This structure was also apparent at cpDNA. Approximate Bayesian Computation detected recent bottlenecks approximately dated to the last glacial maximum in Benin, West Cameroon and São Tomé, and an older bottleneck in South Cameroon and Gabon, suggesting a genetic effect of Pleistocene cycles of forest contraction. CpDNA haplotype distribution indicated wide-ranging long-term persistence of S. globulifera both inside and outside of postulated forest refugia. Pollen flow was four times greater than that of seed in South Cameroon and Gabon, which probably enabled rapid population recovery after bottlenecks. Furthermore, our study suggested ecotypic differentiation—coastal or swamp vs terra firme—in S. globulifera. Comparison with other tree phylogeographic studies in Central Africa highlighted the relevance of species-specific responses to environmental change in forest trees.
Keywords: forest refuge theory, phylogeography, psbA-trnH, SSR, tropical Africa, approximate bayesian computation
Introduction
Forest refugia denote regions where forest taxa are believed to have persisted and evolved through adverse climatic conditions. The general premise of the forest refuge theory, which was developed with reference to the Pleistocene climatic oscillations, is that climate change can lead to isolation of conspecific populations, which will then become genetically differentiated and eventually speciate (Haffer, 1969; Prance, 1982). Forest refuge theory became a popular model for explaining biological diversification in the tropics in the 1980s when palaeoecological and geomorphological data challenged the hypothesis of long-term environmental stability and a resulting slow accumulation of species over time (Prance, 1982; Mayr and O'Hara, 1986). The identification of forest refugia is a promising approach to the conservation of biodiversity under climate change, as refugia represent locations that have avoided regional climate extremes and large-scale disturbance and therefore should have accumulated substantial biodiversity (Millar et al., 2007). Also, genetic approaches to identifying refugia can reveal evolutionary processes that are highly relevant for predicting plant species' response to future climate change (McMahon et al., 2011).
In temperate regions, refuge locations and postglacial migration routes were suggested for many woody taxa based on palaeoecological data and further confirmed by geographical patterns of genetic diversity at neutral markers (for example, Petit et al., 2003; Heuertz et al., 2006). These studies indicated latitudinal and altitudinal range shifts with, typically, glacial persistence in southern refugia followed by northward postglacial recolonization for northern hemisphere thermophilous species (Petit et al., 2003). In tropical areas, refuge theory has been rejected in the Amazonian basin but remains an active hypothesis in tropical Africa and Australia (Ghazoul and Sheil, 2010). Climate-induced altitudinal, but not latitudinal, shifts and postglacial population expansion have recently been suggested in Africa (Born et al., 2011).
The Guineo-Congolian phytogeographic region represents the largest regional centre of endemism for the African rainforest, being subdivided into three subcentres: Upper Guinea (West Africa), Lower Guinea and Congolia (White, 1979; Figure 1). Refuge theory in this region is supported by palaeoecological data, which indicates that the forest was fragmented during cool and dry glacial periods of the Pleistocene and that it expanded into grasslands under warmer and wetter conditions (Maley, 1996; Bonnefille, 2007). As refugia are thought to have led to speciation in isolation, refuge locations embedded in the current rainforest distribution have been proposed based on high species richness or endemism, especially in recently radiated or poorly dispersing groups (Sosef, 1994; Droissart, 2009). A review of palaeoecological and plant and animal species distribution data suggested three upland refugia in Upper Guinea, six in Lower Guinea (one on the Cameroon Volcanic Line (CVL) in West Cameroon, one in Southwest Cameroon, three in Gabon and one in the Mayombe massif in the Republic of Congo, Democratic Republic of Congo and Angola) and one large lowland refuge in the Congo Basin (Maley, 1996; Figure 1). Limitations for identifying refugia based on palaeoecological and/or species distribution data in Central Africa are threefold: (1) the palaeoecological record is still scarce and pollen profiles typically show temporal discontinuities and poor taxonomic resolution (for example, see Bonnefille, 2007). (2) Species richness and endemism may be poor proxies for refugia because the high habitat heterogeneity of upland areas favours high biodiversity and can trigger speciation due to diversifying selection without requiring historical isolation (for example, see Moritz et al., 2000). (3) Species distribution data in many plant and animal groups are inappropriate for indicating refugia because sister taxa diverged before the Pleistocene (Moritz et al., 2000; Plana, 2004).
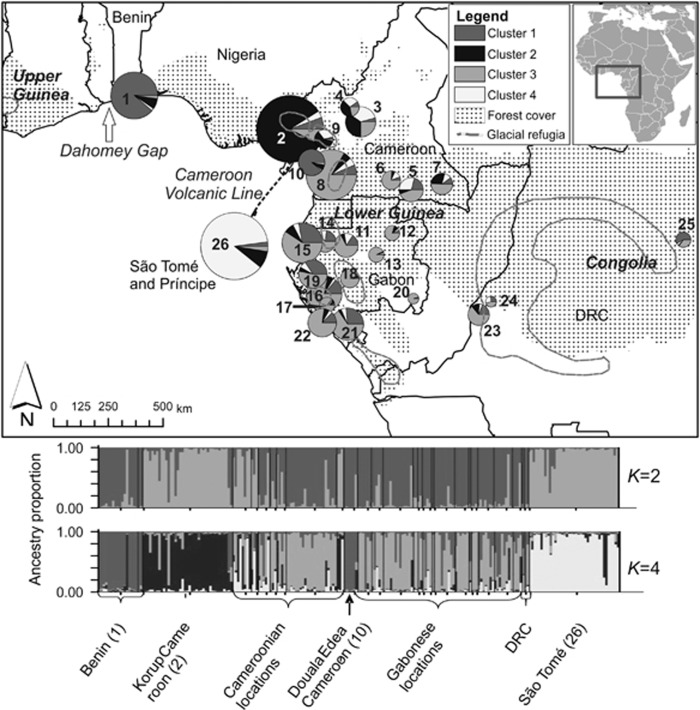
Figure 1.
Above: Map of the geographical distribution of GPs identified by STRUCTURE (K=4 clusters) for S. globulifera in Atlantic Equatorial Africa and postulated refuge locations (areas limited with grey lines) during the last glacial maximum (LGM, about 18 000 years BP) adapted from Maley (1996). Numbers correspond to sampling locations in Table 1. Chart size increases with sample size (ranging from 1 to 39 individuals). Regional names are given in italics. Below: Bar plots showing STRUCTURE ancestry proportions in K=2 and K=4 clusters. Each individual is represented as a line segment, which is vertically partitioned into K coloured components representing the individual's estimated proportions of ancestry in the K clusters. DRC, Democratic Republic of the Congo.
Phylogeographic studies should allow for a broader test of the refuge theory in Africa and have indeed begun providing insights into responses of tree species to past climatic oscillations. For example, the two Cameroonian refuges proposed by Maley (1996) are also supported by the distribution of endemic alleles in Irvingia gabonensis (Lowe et al., 2010) and Santiria trimera (Koffi et al., 2011). In Aucoumea klaineana, genetic diversity is structured in four differentiated gene pools (GPs; identified using nuclear microsatellites), the location of which coincides with refugia postulated from species distribution data in Gabon (Born et al., 2011). However, genetic patterns in three pioneer (Distemonanthus benthamianus, Erythrophleum suaveolens and Milicia excelsa) and one non-pioneer (Greenwayodendron suaveolens) species were not in agreement with postulated refuges but showed a common north–south divide in Lower Guinea, which was suggested to reflect the seasonal inversion near the equator (Daïnou et al., 2010; Dauby et al., 2010; Debout et al., 2011; Duminil et al., 2010). These emerging trends allow formulation of hypotheses on biogeographic scenarios and evolutionary processes operating within species that can be evaluated with multilocus genetic data (preferably using loci with contrasting inheritance) using statistical phylogeographic approaches (Knowles, 2009).
In the present study, we chose an ancient widespread tropical tree species, S. globulifera L. f. (Clusiaceae; originated ca 45–30 Ma ago, see below), typical of mixed humid and freshwater swamp forests in Africa and America, to test the Pleistocene refuge theory and assess population demographic processes in Central Africa. The gradual nature of morphological variation in S. globulifera has prevented its subdivision (Abdul-Salim, 2002; Oyen, 2005) and the species represents an interesting contrast to recently radiated, species-rich families, such as Begoniaceae, Orchidaceae or Rubiaceae, which are typically used to support the refuge theory in Africa (Sosef, 1994; Droissart, 2009). Furthermore, the genus Symphonia has a distinctive pollen morphology providing solid evidence that it was widespread in tropical Africa long before the Pleistocene (reviewed in Dick et al., 2003). Therefore, if this species harboured localized endemic alleles and distinct GPs coinciding with proposed refuges, this would corroborate the refuge theory in Africa and give further support for the relevance of this model for biodiversity organization at the within-species level.
Specifically, we searched for signals of past demographic events in the spatial genetic constitution of S. globulifera in the Guineo-Congolian region using plastid DNA sequences, which essentially inform on colonization processes, and nuclear microsatellites, which provide more robust multi-locus information on the distribution of diversity. We included samples from a comprehensive set of locations through the region with a strong emphasis on Atlantic Equatorial Africa (Lower Guinea), addressing the following questions:
(1) Is the genetic diversity geographically structured in the study region? If so, are cpDNA haplotypes and SSR gene pools structured according to geographic features (mountain or ocean barriers and the forest-savannah zone of the Dahomey gap, see below) or can their distribution be explained in terms of origin from distinct forest refugia sensu Maley (1996)?
(2) Do current GPs or regions harbour signs of a recent demographic bottleneck, such as would be expected if they were still affected by last glacial maximum (LGM) forest fragmentation, or of population expansion, such as would be expected if rapid colonization occurred from refugia? What is the order of divergence of the distinct GPs?
(3) What is the pollen-to-seed dispersal distance ratio (σp/σs) estimated from markers with contrasting inheritance and how does this quantity help to explain the colonization history of the species?
Materials and methods
Study species
S. globulifera L.f. (synonym S. gabonensis [Vesque] Pierre) is a medium-sized tree (25−40 m tall) occurring in Africa from Guinea Bissau to Tanzania and in America from Mexico to Brazil. It has a wide ecological amplitude and grows in forests from sea level to 2600 m altitude (in East Africa) with an annual rainfall of 650–2100 mm and a mean annual temperature of 23–27 °C (Oyen, 2005). It is a late successional species in evergreen mixed humid forests where it regenerates by seeds, requiring shade for germination (Oyen, 2005). The species can grow on a variety of soils (Lescure and Boulet, 1985) and can be locally common along rivers, in swamp forests and on the inner edges of mangroves, where regeneration occurs mostly by root suckers (Van Andel, 2003; Oyen, 2005). The oldest fossil pollen records of Symphonia (fossil taxon Pachydermites diederexi) were found in Nigeria and date to the mid-Eocene (ca 45 Ma, Jan du Chêne and Salami, 1978) and there are records through the Oligocene and Miocene from Angola (Dick et al., 2003). American fossils of the taxon are younger, the oldest dating to the early/mid-Miocene (ca 18–15 Ma, reviewed in Dick et al., 2003). S. globulifera has 16–23 congeners, all endemic to Madagascar where S. globulifera does not occur (Abdul-Salim, 2002). The distribution, palaeoecological and genetic data favour Africa or Madagascar as the geographic origin of the genus (Dick et al., 2003; M Heuertz, OJ Hardy and CW Dick, unpublished data).
S. globulifera has showy red flowers and is pollinated by butterflies, hummingbirds and perching birds in the Neotropics (Bittrich and Amaral, 1996) and by various insects and sun birds in Africa (Oyen, 2005). Seeds are dispersed by bats, tapirs, rodents, primates and deer in the Neotropics and by hornbills, primates and duikers in Africa (Forget et al., 2007). S. globulifera is a predominantly outcrossing species but in some populations from the Neotropics significant levels of biparental inbreeding have been detected (Aldrich and Hamrick, 1998; Degen et al., 2004; da Silva Carneiro et al., 2007). Bird-mediated pollen dispersal has been shown to be limited in the Neotropics (mean 27–54 m, Degen et al., 2004; 154–444 m, da Silva Carneiro et al., 2007). No information on pollen (or seed) dispersal distances are available for the African range.
Plant material and DNA extraction
Leaf or cambium samples (n=251) were collected from S. globulifera individuals in coastal forests in Benin (forest-savannah mosaic of the Dahomey gap, between Upper and Lower Guinea, 19 samples), and almost exclusively in terra firme forests of Cameroon and Gabon (Lower Guinea, 182 samples), the Democratic Republic of Congo (Congolia, 9 samples) and from the Island of São Tomé in the Gulf of Guinea (41 samples, Table 1, Figure 1). The sampling was conducted between 2006 and 2010 jointly with several other species as part of a series of field missions led by the Université Libre de Bruxelles. The forest of each locality or village was prospected for 1 or 2 days, which, in combination with a variable density of S. globulifera (from presumed absence of the species to ca 6 individuals per ha, with diameter at breast height >20 cm), resulted in variable sample sizes per locality. The plant material was immediately dried with silica gel. DNA was extracted using the Invisorb DNA Plant HTS 96 Kit (Invitek, Berlin, Germany).
Table 1. Plant material analysed in Symphonia globulifera.
| No. | Location | Latitude | Longitude | Localities included | n | ncp | dmean (km) | dmax (km) |
|---|---|---|---|---|---|---|---|---|
| 1 | Benin | 6.42585° N | 2.47969° E | Niaouli, Porto Novo, Ahozon, Kraké | 19 | 13 | 29.10 | 72.44 |
| 2 | Cameroon Korup | 5.05685° N | 8.83818° E | Korup National Park and 50 ha plot of the Smithsonian Tropical Research Institute | 39 | 14 | 2.44 | 6.33 |
| 3 | Cameroon CN | 5.34846° N | 11.72144° E | Yoko, Mégan, Nyafianga, Djendjou cliffs | 8 | 40.49 | 116.43 | |
| 4 | Cameroon N | 5.95185° N | 11.28245° E | Mandah, Monkoin | 3 | 30.73 | 42.46 | |
| 5 | Cameroon S1 | 2.57775° N | 13.78582° E | Lélé, Mbalam, Mindourou | 5 | 5 | 70.42 | 103.69 |
| 6 | Cameroon S2 | 2.96657° N | 12.965° E | Dja Biosphere Reserve camp Mamma, Mbouma | 3 | 3 | 51.44 | 77.04 |
| 7 | Cameroon SE | 2.80969° N | 15.03372° E | Ngato, Moloundou, management unit UFA10 | 4 | 3 | 46.23 | 90.36 |
| 8 | Cameroon SW, south-west; | 3.19157° N | 10.52427° E | Akom, Bipindi, Ngovayang massif, management unit UFA 09-028 (EFFA), | 22 | 13 | 23.53 | 80.72 |
| 9 | Cameroon Volcanic Line | 4.64713° N | 10.22126° E | Mount Koupé, Yingui region | 3 | 1 | 63.71 | 93.15 |
| 10 | Cameroon W coast | 3.69213° N | 9.71888° E | RF Douala-Edéa, Limbe | 6 | 1 | 29.58 | 86.23 |
| 11 | Gabon CNE | 0.31051° N | 11.1281° E | Mékié, Ndjolé North | 5 | 31.54 | 77.07 | |
| 12 | Gabon NE1 | 0.81011° N | 13.00449° E | Makokou, Bélinga | 2 | 82.55 | 82.55 | |
| 13 | Gabon NE2 | 0.0697° S | 12.36367° E | Ivindo | 2 | 0.36 | 0.36 | |
| 14 | Gabon Crystal | 0.4726° N | 10.27115° E | Crystal Mountains | 4 | 4 | 0.63 | 1.00 |
| 15 | Gabon coast | 0.41459° N | 9.33374° E | Cap Estérias, Pointe Denis, Pongara | 14 | 14 | 18.07 | 46.16 |
| 16 | Gabon CW | 1.66178° S | 10.23387° E | Niambo-kamba, Mandji W | 11 | 32.91 | 77.10 | |
| 17 | Gabon CS | 2.12277° S | 10.35227° E | Moukalaba-Doudou National Park | 2 | 78.37 | 78.37 | |
| 18 | Gabon Waka | 1.03788° S | 11.27908° E | Waka, Lopé Reserve | 3 | 2 | 39.93 | 59.90 |
| 19 | Gabon W | 0.86243° S | 9.77248° E | Petite Silang, Lake Avanga, Lake Anengue, Enyonga | 7 | 52.97 | 95.51 | |
| 20 | Gabon SE | 1.86288° S | 13.87708° E | Ossélé | 1 | 1 | NC | NC |
| 21 | Gabon SW | 2.90301° S | 11.18371° E | Douano, Eastern slope of the Mayombe massive, Bikamba | 9 | 2 | 36.23 | 72.18 |
| 22 | Gabon Gamba-Doudou | 2.84693° S | 10.1492° E | Gamba, Luango | 7 | 6 | 52.46 | 149.40 |
| 23 | DRC W1 | 2.51542° S | 16.53403° E | MbouMon, Mbanzi | 4 | 4 | 12.78 | 24.57 |
| 24 | DRC W2 | 1.98474° S | 17.0428° E | Boleke | 1 | 1 | NC | NC |
| 25 | DRC N | 0.53791° N | 24.88835° E | Bamboussoko, Yangambi | 2 | 106.84 | 106.84 | |
| 26 | São Tomé | 0.27857° N | 6.56685° E | São Tomé | 39 | 5 | 2.81 | 8.38 |
| Overall sample | 586.74 | 2672.05 |
Abbreviations: CN, centre-north; CNE, centre-north-east; CS, centre-south; CW, centre-west. DRC, Democratic Republic of the Congo; dmean and dmax, mean and maximum pairwise distance between individuals sampled; n, number of individuals sampled and genotyped at nSSR markers; ncp, number of individuals sequenced at psbA-trnH; NC, not calculated; SE, south-east; SW, south-west.
Location, sampling location (in italics and underlined for locations coinciding with refuges sensu Maley (1996); latitude and longitude, central geographic coordinates of sampling locations in decimal degrees. Numbers correspond to locations in Figure 1.
Plastid DNA sequences
A subsample of 94 individuals from 18 sampling locations (Figure 2) was sequenced at the psbA-trnH plastid DNA (cpDNA) intergenic spacer using the psbAF and trnHR primers (Sang et al., 1997). The total PCR volume of 25 μl contained approximately 20 ng of template DNA, 1 × HF PCR reaction buffer, 0.1 μℳ of each primer, 200 μℳ of each deoxyribonucleotide triphosphate (dNTP) and 0.25 U of Phusion polymerase (Finnzymes, Espoo, Finland). The PCR profile was 30 s at 98 °C, 35 cycles of 5 s at 98 °C, 30 s at 50 °C and 45 s at 72 °C, and a final elongation of 3 min at 72 °C. PCR products were purified on filter columns (QIAquick96 kit, Qiagen, Hilden, Germany or MSB HTS PCRapace/C[96]kit, Invitek) or using the enzymes Exonuclease I and Shrimp Alkaline Phosphatase (GE Healthcare, Waukesha, WI, USA) and quantified on agarose gels (1%). Sequencing reactions were performed in both directions using BigDye v.3.1 chemistry (Applied Biosystems, Lennik, Belgium), purified with an ethanol-sodium acetate protocol and analysed on an ABI3100 or an ABI3730 sequencer (Applied Biosystems). Sequences were edited and aligned in CodonCode Aligner 3.0.1. (CodonCode Corporation, Dedham, MA, USA) including base-calling with Phred (Ewing et al., 1998) and alignment with Phrap (Green, 2009). A nucleotide site was considered polymorphic if different variants had at least a Phred quality value of 25, corresponding to an error probability of 3/1000.
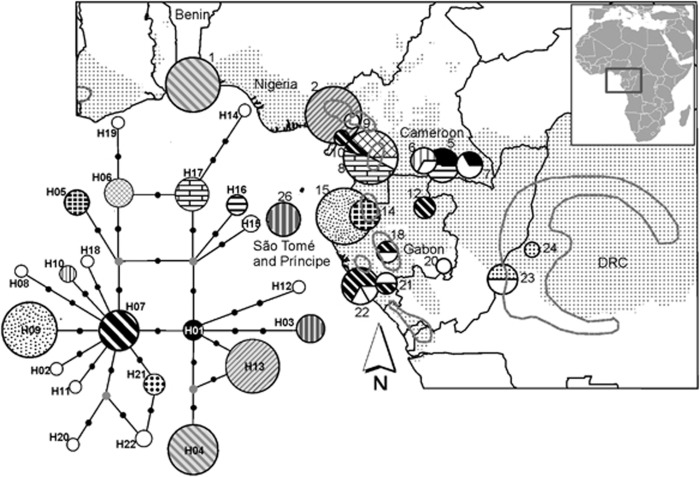
Figure 2.
Distribution of psbA-trnH haplotypes of S. globulifera. Chart size increases with sample size (ranging from 1 to 14 individuals). Grey lines delimit postulated refuge locations during the LGM (about 18 000 years BP) adapted from Maley (1996). A grey background colour indicates haplotypes found in Benin, Cameroon Korup and in São Tomé, to facilitate comparison with the map of SSR GPs (Figure 1). Numbers correspond to sampling locations in Tables 1 and 2. Haplotypes detected in one or two samples in a single population only are marked in white. Lower left: Haplotype network with haplotype numbers in bold (small black circles indicate mutations; small grey circles indicate putative haplotypes not observed in this study).
Microsatellite markers
Five nuclear microsatellites (SSRs) developed for neotropical S. globulifera were optimized for our African samples: SG03 and SG18 (Degen et al., 2004), SGC4 and SG19 (Aldrich et al., 1998), and SG10 (Vinson et al., 2005). Forward primers were fluorescently labelled with IRDye700 or IRDye800 (see Supplementary Information 1). The total PCR volume of 10 μl contained approximately 25 ng of template DNA, 1 × PCR Reaction Buffer (Bioline, London, UK), locus-specific concentrations of MgCl2 (Supplementary Table 1), 0.2 mℳ of each primer, 0.2 mℳ of each dNTP and 1 U of Taq polymerase (Ecotaq, Bioline). The PCR conditions were 3 min at 94 °C, a locus-specific number of cycles for denaturation (30 s at 94 °C), annealing (30 s at locus-specific temperatures) and elongation (45 s at 72 °C), and a final elongation for 7 min at 72 °C (Supplementary Information 1).
PCR products were diluted with a formamide-loading buffer and separated on acrylamide gels on a 4300 DNA analyzer (Li-Cor Biosciences, Lincoln, NE, USA). Microsatellite allele sizes were determined using the genotyping automation software SAGAGT (Li-Cor Biosciences) in comparison with the SequaMark DNA size marker (Invitrogen, San Diego, CA, USA). Length variants that could not unequivocally be distinguished were pooled into the same allele-class.
Plastid DNA data analysis
CpDNA haplotypes were defined taking into account only point mutations (single-nucleotide polymorphisms, SNPs) omitting insertions and deletions (indels), because they contained complex repetitive regions, which were difficult to code and added noise to the haplotype analysis. A haplotype network was constructed using a maximum parsimony method implemented in the median-joining algorithm of the software NETWORK version 4.600 (Fluxus Technology Ltd, Suffolk, UK; Bandelt et al., 1999). The geographical distribution of haplotypes was illustrated in a GIS (Geographic information system) map built in ArcMap9.3.1 (ArcGIS 9, ESRI, Redlands, CA, USA).
The genetic diversity of all samples was estimated as the number of haplotypes and as haplotypic diversity based on unordered (h) or ordered (v) haplotypes (Pons and Petit, 1996). For the latter, a genetic distance matrix was computed among haplotypes, defining the distance between two haplotypes as the number of polymorphic sites differing between them. Differentiation among locations and between pairs of locations with at least three individuals sampled was assessed with SPAGeDi1.2g (Hardy and Vekemans, 2002) using the corresponding statistics, GST for unordered and NST for ordered alleles. Permutation (10 000) of individuals among populations was used to test for deviation of GST or NST from zero. The impact of mutations on among-population differentiation (test for NST>NST[permuted]), that is, the presence of phylogeographic structure, was assessed by 10 000 permutations of pairwise haplotype distances among pairs of haplotypes.
Microsatellite data analysis
To identify the GP composition of the sample, we used the Bayesian clustering method implemented in STRUCTURE 2.2 (Pritchard et al., 2000). We ran an admixture model with correlated allele frequencies between clusters. Ten runs were performed for each number of clusters K=1 to K=10 with a burn-in length of 100 000 and a run length of 200 000 iterations. The K that best described the data was determined from the graph of posterior log likelihood of the data, LnP(D|K), against K, and from delta K (Evanno et al., 2005). For graphical representation of the clustering results, samples were grouped into 26 locations based on geographic proximity (Table 1, Figure 1) and a GIS map was built in ArcMap9.3.1. Clustering results were verified with TESS 2.3.1 (Chen et al., 2007), which infers ancestry proportions of individuals in K user-defined clusters, similar to STRUCTURE, and that includes the sampling location of each individual as prior information. We ran TESS using a conditional auto-regressive admixture model with burn-in of 10 000 and run length of 50 000 iterations, a spatial interaction parameter of 0.99 and a trend degree surface of 1. Ten repetitions were carried out for each K=1 to K=8.
For analyses at the regional level, regions were defined based on the geographic location of GPs. Overall and pairwise genetic differentiation among regions were assessed computing FST (Weir and Cockerham, 1984) using FSTAT 2.9.3.2 (Goudet, 1995) and RST, an analogue of FST accounting for allele size (Slatkin, 1995), using SPAGeDi1.2g. FST and RST were tested against the null expectation of absence of population structure with permutation tests. A phylogeographic signal was tested for by comparing RST to its expectation (RST[permuted]) obtained from 10 000 permutations of allele sizes among alleles using SPAGeDi1.2g. The test is significant if stepwise-like mutations have contributed to differentiation, for example, in the case of ancient isolation (Hardy et al., 2003). For each GP, the expected heterozygosity or gene diversity (HE) and the allelic richness (RS) were computed in FSTAT.
Demographic history analysis
The demographic history of each region was first explored computing the ‘bottleneck' statistic T2 with BOTTLENECK 1.2.02 (Cornuet and Luikart, 1996). T2 represents the deviation of the sample gene diversity from the gene diversity expected for the number of alleles and is positive (heterozygosity excess) in the case of a recent bottleneck and negative in the case of recent population expansion (see Supplementary Information 2). As Approximate Bayesian Computation (ABC) methods have recently been shown to have increased power for bottleneck detection (Peery et al., 2012), we used the ABC framework in DIYABC v1.0.4.46beta (Cornuet et al., 2008; available at http://www1.montpellier.inra.fr/CBGP/diyabc) to compare four demographic scenarios in each region. Scenarios were defined based on the relative past pollen abundance of forest vs savannah species in Central Africa (for example, see Bonnefille, 2007; Ngomanda et al., 2009; Dupont, 2011): (1) null model of constant population size, (2) a recent bottleneck coinciding approximately with the 1000–3000 BP rainforest disturbance, (3) a bottleneck coinciding approximately with the LGM, ca 15 000–22 000 BP and (4) an older bottleneck coinciding approximately with the previous glacial period (ca 130 000–190 000 BP). Initial runs were carried out to explore the parameter space. Parameter values were then drawn from informed broad prior distributions, assuming a generation time of 100 years (Supplementary Information 3), and 106 microsatellite data sets were simulated for each scenario. As summary statistics, we computed the mean number of alleles, mean genetic diversity, mean allele size variance and mean Garza–Williamson's M (Garza and Williamson, 2001). M=k/r is the mean ratio of the number of alleles, k, to the allele size range, r (Garza and Williamson, 2001), and is believed to detect more ancient and severe bottleneck signals than methods based on the deficit of rare alleles such as T2 (Williamson-Natesan, 2005). We estimated the posterior probability of each scenario using logistic regression on the 1% simulated data sets with summary statistics closest to the observed data in each region and compared the 95% highest posterior density intervals (HPD) of scenarios (Cornuet et al., 2008). We then estimated the posterior distribution of parameters from the 1% best simulated data sets using local linear regression and a logit transformation of parameters.
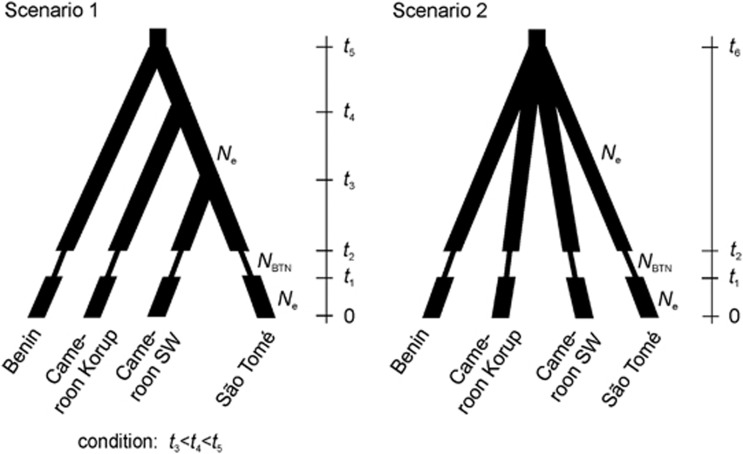
We also used ABC to examine the order of divergence of the four identified GPs/regions (see Results, Figure 3 and Supplementary Information 4). We chose a representative location of each region to correct for a potential bias in coalescence times that may occur due to uneven sampling across regions (more recent coalescence in under-sampled GPs/regions, Städler et al., 2009). Representative locations were defined as >80% STRUCTURE ancestry at the location level and similar sample size: location 1 (Benin) for GP1, 2 (Cameroon Korup) for GP2, 8 (Cameroon SW) for GP3 and 26 (São Tomé) for GP4 (see Table 1 and Figure 1). In each location, all individuals of the location were included in the analysis to account for migration events or uncertainty in STRUCTURE ancestry proportions. We computed the pairwise differentiation statistics RST and (δμ)2 (Goldstein and Pollok, 1997) between these locations with SPAGeDi1.3d and constructed UPGMA trees using Phylip v.3.69 (Felsenstein, 1989). (δμ)2 should retain the phylogenetic signal between GPs better than RST as it is less affected by drift (Hardy et al., 2003), although in our case, the same topology was recovered. We therefore considered two scenarios (topologies) to assess the divergence history in the study area: (1) the (δμ)2-based population tree and (2) simultaneous divergence of all locations (Figure 3). We simulated 106 data sets in each scenario with DIYABC using broad informed parameter priors (Supplementary Information 4). Because we previously detected an LGM or penultimate glacial bottleneck in all regions (see Results) and because geographic features separating regions (CVL, ocean barrier in the Gulf of Guinea) are older than these events, a bottleneck timed between 15 000 and 200 000 years, similarly in all GPs to reduce model complexity, was simulated along each terminal branch after population divergence events (Supplementary Information 4). As summary statistics, we used the pairwise FST, the pairwise shared allele distance, the pairwise (δμ)2, the mean classification index for two samples (Rannala and Mountain, 1997) and the mean genetic diversity per population (for a total of 34 summary statistics). Model choice and the estimation of posterior parameter distributions were done as described above. In addition, the goodness of fit of the models was checked by simulating 1000 data sets from the posterior predictive distribution of parameters, and comparing these to the observed data by means of summary statistics different from those chosen for model fitting: mean number of alleles, mean allele size variance and mean Garza-Williamson's M.
Figure 3.
Two scenarios simulated in DIYABC to assess the population divergence history, (1) (δμ)2-based population tree and (2) simultaneous divergence of all locations. A bottleneck timed approximately between 15 000 and 200 000 years was simulated in terminal branches (for details see Supplementary Information 4).
Pollen- vs seed-mediated gene flow
To estimate the relative contribution of seed dispersal to overall gene dispersal (σs/σg, where σs2 and σg2 are half the mean squared dispersal distances for seed and genes, respectively), we calculated σs/σg=[2Sp(nuclear)/Sp(plastid)]1/2. To assess the pollen-to-seed dispersal distance ratio (σp/σs), we used [(Sp(plastid)/Sp(nuclear))-2]1/2 (formulas assuming hermaphrodite plants and purely maternal inheritance of plastids). The Sp statistics reflect the strength of the spatial genetic structure and were computed from the regression slope of pairwise kinship coefficients between individuals on the logarithm of the spatial distance (Vekemans and Hardy, 2004). These estimates were computed across the South Cameroon and Gabon region, which had sufficient sample size and was closer to demographic equilibrium than the other regions.
Results
CpDNA diversity and structure
In a total of 94 S. globulifera samples from 18 locations from the Guineo-Congolian region, we identified 22 SNPs in the psbA-trnH intergenic region, resolving 22 haplotypes (Supplementary Information 5, Figure 2, GenBank accession numbers JQ996246-JQ996339). The haplotype network presented several reticulations reflecting homoplasy (that is, recurrent mutations at some sites) in the psbA-trnH region (Figure 2, Supplementary Information 5). Neighbouring haplotypes differed by a maximum of three mutations. Several geographic locations presented a single haplotype and most haplotypes (19 out of 22) were endemic to a single location. Endemic haplotypes occurred in proposed refuges (sensu Maley) and in other locations (Figure 2, Table 2 and Supplementary Information 5). Haplotype diversity was assessed in each location considering phylogenetic information using v statistics, or ignoring this information using h statistics. Population ‘Cameroon SW' (8), located on a proposed refuge, was the most diverse location with four haplotypes (hs=0.680 and vs=0.336), whereas ‘Cameroon S2' (6) was the most phylogenetically diverse harbouring two distantly related haplotypes (H10 and H14, hs=0.667 and vs=1.343; Table 2). The south of Lower Guinea (Gabon and Democratic Republic of the Congo) harboured haplotypes that were all closely related to the widespread H07 (maximum distance of three mutations, hs=0.795 and vs=0.604), whereas the north of Lower Guinea (South Cameroon) harboured more phylogenetically diverse haplotypes that were related to H07 or H17 (hs=0.895 and vs=0.850; Table 2, Figure 2). GST among locations was 0.566 and NST was 0.722, significantly larger than NST[permuted] (P<0.001) and indicative of a phylogeographic signal at cpDNA.
Table 2. Genetic diversity for psbA-trnH haplotypes in S. globulifera.
| Location | n | A | AP | hS | vS | |
|---|---|---|---|---|---|---|
| 1 | Benin | 13 | 1 | 1 | 0 | 0 |
| 2 | Cameroon Korup | 14 | 1 | 1 | 0 | 0 |
| 5 | Cameroon S1 | 5 | 2 | 1 | 0.600 | 0.403 |
| 6 | Cameroon S2 | 3 | 2 | 2 | 0.667 | 1.343 |
| 7 | Cameroon SE | 3 | 2 | 1 | 0.667 | 0.448 |
| 8 | Cameroon SW | 13 | 4 | 3 | 0.680 | 0.336 |
| 9 | Cameroon Volcanic Line | 1 | 1 | 1 | NC | NC |
| 10 | Cameroon W coast | 1 | 1 | 0 | NC | NC |
| 12 | Gabon NE1 | 2 | 1 | 0 | 0 | 0 |
| 14 | Gabon Crystal | 4 | 1 | 1 | 0 | 0 |
| 15 | Gabon coast | 14 | 1 | 1 | 0 | 0 |
| 18 | Gabon Waka | 2 | 2 | 1 | 1 | 0.336 |
| 20 | Gabon SE | 1 | 1 | 1 | NC | NC |
| 21 | Gabon SW | 2 | 2 | 1 | 0 | 0.336 |
| 22 | Gabon Gamba-Doudou | 6 | 3 | 2 | 0.600 | 0.336 |
| 23 | DRC W1 | 4 | 2 | 1 | 0.667 | 0.224 |
| 24 | DRC W2 | 1 | 1 | 0 | NC | NC |
| 26 | São Tomé | 5 | 1 | 1 | 0 | 0 |
| North of Lower Guinea | 26 | 10 | 9 | 0.895 | 0.850 | |
| South of Lower Guinea | 36 | 10 | 9 | 0.795 | 0.604 | |
| Overall | 94 | 22 | 22 | 0.968 | 0.968 |
Abbreviations: A, number of haplotypes; AP, number of private or endemic haplotypes; DRC, Democratic Republic of the Congo; hS and vS, gene diversity per location based on unordered and ordered alleles, respectively; n, sample size; NC, not calculated.
Location, sampling location name (in italics and underlined for locations coinciding with refuges sensu Maley (1996). Numbers correspond to locations in Figure 2.
Genetic diversity and structure at SSRs
We identified a total of 111 alleles and an average heterozygosity of HE=0.860 at the five SSRs (for summary statistics per locus and sampling location, see Supplementary Information 1). Log-likelihood values for the admixture model in STRUCTURE increased as a function of the number of clusters K, reaching a plateau for K⩾4 and the delta K vs K graph (Evanno et al., 2005) was bimodal, suggesting K=2 or K=4 as best describing the data (Supplementary Information 6). For K=2, one cluster comprised Benin and the majority of the samples from Cameroon and Gabon, and the other cluster samples from West Cameroon (Korup) and from São Tomé (Figure 1). For K=4, GP1 was essentially represented in Benin (location 1) and in a coastal region close to Douala (location 10) in Cameroon; GP2 was centred on West Cameroon (Korup, location 2); GP3 was centred on South Cameroon (east of the CVL) and Gabon; and GP4 was mostly represented in São Tomé (location 26; Figure 1). No substructure was identified in any of the four GPs. When geographic location of individuals was used as prior information in TESS 2.3.1 (Chen et al., 2007), the same four GPs were identified (Supplementary Information 5) with slightly higher average maximum individual ancestry (87.9%±0.13% (s.d.) vs 83.7%±0.15% (s.d.) in STRUCTURE).
According to the primary location of the four GPs, we defined four main geographic regions; Benin (location 1); West Cameroon (Korup, location 2); South Cameroon (east of the CVL) and Gabon (locations 5–8 and 11–24); and São Tomé (location 26; Figure 1). Sampling locations 3, 4 and 9 of the CVL were not included because they were located in a contact zone between GPs and had small sample sizes (three to eight individuals). Also, location 10 (Cameroon W coast) was not included as this was a small sample (six individuals) from a distinct habitat (river bank) that we suspected to have distinct history (see Discussion). Location 25 was excluded because it had also small sample size (two individuals) and belonged to a distinct phytogeographic region (Congolia). Differentiation among regions was FST=0.135 (P<0.0001) and RST=0.185 (P<0.0001), and there was a phylogeographic signal (RST>RST[permuted], P<0.01) across the study area. Pairwise FST between regions ranged from 0.095 to 0.183 and phylogeographic signals were detected for all pairwise comparisons between Benin, West Cameroon and São Tomé (Table 3). Endemic alleles were found in all regions, 3 in Benin, 2 in West Cameroon, 31 in the geographically extended South Cameroon and Gabon, and 2 in São Tomé. Allelic richness was highest in South Cameroon and Gabon, whereas heterozygosity was similar in all regions (Table 4).
Table 3. Pairwise genetic differentiation between regions in S. globulifera.
| FST/RST | Benin | West Cameroon | South Cameroon, Gabon | São Tomé |
|---|---|---|---|---|
| Benin | 0.109* | 0.095* | 0.183* | |
| West Cameroon | 0.410* | 0.127* | 0.128* | |
| South Cameroon, Gabon | 0.121 | 0.167 | 0.154* | |
| São Tomé | 0.415** | 0.213* | 0.135 |
Above diagonal: FST (test with H1: FST≠0: *P<0.05); below diagonal: RST (test with H1: RST>RST[permuted]: *P<0.05; **P<0.01). Significance values refer to levels after Bonferroni correction.
Table 4. Nuclear genetic diversity, allelic richness and demographic signatures in four geographic regions in S. globulifera.
| RS | HE | Scenario 1: constant-size population | Scenario 2: ca 3000–1000 BP bottleneck | Scenario 3: LGM bottleneck | Scenario 4: penultimate glacial bottleneck | |
|---|---|---|---|---|---|---|
| Benin | 7.86 (0.91) | 0.770 (0.061) | 0.0631 (0.0590, 0.0672) | 0.3081 (0.3001, 0.3161) | 0.5208 (0.5120, 0.5236) | 0.1080 (0.1023, 0.1137) |
| West Cameroon | 8.33 (1.47) | 0.803 (0.041) | 0.0631 (0.0593, 0.0669) | 0.2663 (0.2590, 0.2736) | 0.5783 (0.5703, 0.5864) | 0.0923 (0.0875, 0.0972) |
| South Cameroon and Gabon | 11.36 (2.40) | 0.769 (0.086) | 0.3401 (0.2391, 0.4412) | 0.0032 (0.0020, 0.0043) | 0.1609 (0.1083, 0.2136) | 0.4957 (0.3527, 0.6388) |
| São Tomé | 8.16 (1.27) | 0.772 (0.057) | 0.0001 (0.0000, 0.0001) | 0.1487 (0.1368, 0.1607) | 0.8461 (0.8341, 0.8581) | 0.0051 (0.0040, 0.0062) |
Abbreviation: LGM, last glacial maximum.
RS, allelic richness and its standard error (s.e.) for a sample of 16 individuals per population; HE (SE), gene diversity; Scenario 1–4, posterior probabilities (95% highest posterior density intervals) for four demographic scenarios estimated with DIYABC from the 1% simulations closest to the observed data. The most likely scenario is indicated in bold type.
Demographic history inference
Analyses with the Bottleneck program suggested signals of recent bottlenecks in Benin and West Cameroon (Supplementary Information 2). ABC analyses for population size change indicated that scenario 3 with a bottleneck timed approximately during the LGM had higher posterior probability than alternative scenarios in these regions, and its likelihood (51–58%, based on the 95% HPD) was about twice that of scenario 2 with a more recent bottleneck (Table 4). In São Tomé, the LGM bottleneck was also the most likely scenario, unequivocally (HPD 83–86%). In South Cameroon and Gabon, scenario 4 with a bottleneck during the penultimate glacial was the most likely, although its HPD (35–64%) overlapped with that of scenario 1 of constant population size (24–44%, Table 4). This older bottleneck was not an artefact because of a more extended sampling compared with other regions, as repeating the simulations in a representative location (8) confirmed scenario 4 (probability 90–92%, 95% HPD). Posterior estimates for scaled population sizes (by mutation rate) and bottleneck time events converged, although 95% confidence intervals were wide (Table 5, Supplementary Information 3). They indicated an older bottleneck in South Cameroon and Gabon, as expected from model choice results. The data were insufficient to conclude on any population size differences, either current or during the bottleneck, across regions.
Table 5. Posterior parameter distribution (median and 95% confidence interval) for the most likely demographic scenario in each region (scenario 3 with an LGM bottleneck in Benin, West Cameroon and São Tomé and scenario 4 with a bottleneck during the penultimate glacial in South Cameroon and Gabon).
| Benin | West Cameroon | South Cameroon and Gabon | São Tomé | |
|---|---|---|---|---|
| Neμ | 8.1 (2.5, 27.7) | 11.6 (3.7, 33.6) | 27.1 (11.2, 46.0) | 25.7 (10.9, 44.0) |
| NBTNμ | 0.080 (0.014, 0.304) | 0.114 (0.022, 0.352) | 0.259 (0.110, 0.452) | 0.095 (0.020, 0.295) |
| taμ | 0.042 (0.011, 0.131) | 0.058 (0.017, 0.140) | 0.767 (0.315, 1.210) | 0.084 (0.034, 0.144) |
| tbμ | 0.060 (0.014, 0.185) | 0.082 (0.022, 0.192) | 1.130 (0.462, 1.790) | 0.131 (0.051, 0.204) |
Abbreviation: LGM, last glacial maximum.
Current and bottleneck population sizes, Ne and NBTN, and time of beginning ta and end tb of bottleneck are given, scaled by mutation rate μ.
The two divergence scenarios, (1) (δμ)2-based population tree and (2) simultaneous divergence of all locations (Figure 3), could not be distinguished using DIYABC: their respective probabilities were 0.460 and 0.540, with overlapping HPDs, (0.403,0.518) and (0.482,0.597). The estimates of divergence times scaled by mutation rate converged in both scenarios, but had broad confidence intervals (Supplementary Information 4). In scenario 1, the scaled median time of the oldest divergence event, divergence of Benin, t5μ, was 4.98 (1.26–20.2, 95% CI), which translated to ∼3.32 Ma ago (0.84–13.47 Ma ago, 95% CI) when dividing by the estimated mutation rate and multiplying by generation time. Median estimates for subsequent divergence events t4μ and t3μ lay within the 95% CI of t5μ. In scenario 2, the scaled median time of simultaneous divergence of all locations, t6μ, was 3.01 (0.61–13.7, 95% CI), translating to ∼1.31 Ma ago (0.27–5.98 Ma ago, Supplementary Information 4). Model checking revealed a relatively good fit: comparing our data to data sets simulated from the posterior predictive distribution, only two (Garza-Williamson's M for locations Cameroon SW (8) and São Tomé (26)) of 12 summary statistics lay outside of the 95% CI, but within the 99% CI for scenario 1, and one summary statistic (M for São Tomé (26)) lay between the 95 and the 99% CI for scenario 2. Therefore, the poor model discrimination and poor precision of divergence time estimates can rather be attributed to the information content of the data than to bad choice of model priors.
Pollen- vs seed-mediated gene flow
The relative contribution of seed to overall and to pollen-mediated gene dispersal distances was assessed in the South Cameroon and Gabon region. Significant spatial genetic structure occurred both for cpDNA, Sp(plastid)=0.118, and for SSRs, Sp(nuclear)=0.007, in this region. The ratio of seed to gene dispersal distance, σs/σg, was 0.35. The ratio of pollen to seed dispersal distance, σp/σs, was 3.84, indicating that gene flow via pollen was more wide-ranging than via seeds in South Cameroon and Gabon.
Discussion
Genetic diversity and differentiation of S. globulifera, an ancient tropical rainforest tree
Genetic diversity in African S. globulifera was similar to that in America. Expected heterozygosity, HE, at SSRs was 0.75 (Africa, this study) and 0.78 (Neotropics, Dick and Heuertz, 2008; with three SSRs overlapping with this study). These values are typical for widespread outcrossing long-lived plants growing at late succession stages (Nybom, 2004). Plastid DNA also showed similar polymorphism in both continents at the psbA-trnH fragment with 22 SNPs in Africa and 23 in America (Dick and Heuertz, 2008). This is a high level of variation compared with other species in the same African study region: 2 SNPs in Milicia excelsa (Daïnou et al., 2010) or 14 SNPs in Anthonotha macrophylla s. l. (Leguminosae; M Heuertz and OJ Hardy, unpublished data), a taxon that potentially harbours several species (FJ Breteler, Personnel Communication, 2007). High diversity at the population and species level in S. globulifera suggests high effective population sizes, estimated to ∼25 000 (GP1, Benin) to ∼45 000 (GP3, South Cameroon and Gabon) from scaled Ne estimates. These estimates are somewhat smaller than estimates in the single Pinus taeda GP (∼100 000, Willyard et al., 2007), which has a wide distribution in the SE of the USA and grows at much higher density than S. globulifera. In S. globulifera, consistency with Ne can be attributed to a long history of effective breeding and dispersal strategies ensuring population connectivity across large parts of the range. Genetic differentiation was also similar on both continents in S. globulifera: FST=0.135 among regions/GPs in Africa (this study) vs FST=0.138 among neotropical populations. Our results suggest that the genetic structure found in Africa (ca 0.27–13.47 Ma ago, from ABC) originated probably posterior to the first colonization of America by S. globulifera (15–18 Ma ago, Dick et al., 2003). A joint analysis of sequence variation in a phylogenetic framework should help clarifying the relationships between haplotypes from both continents and provide evidence for the number of colonization events (suggested to be three in Dick et al., 2003) and possibly for the African GP of origin (or region, assuming absence of major range shifts) of the more recent colonization events. The psbA-trnH region might not be the most appropriate for this, as homoplasious point mutations and complex insertion/deletion structure may have erased part of the phylogenetic signal.
Geographic features shape the structure of genetic diversity
Geographic features explain the genetic structure obtained from STRUCTURE and TESS analyses on SSRs in S. globulifera in Atlantic Equatorial Africa: GP1 (Benin) was almost exclusive to coastal forests of the Dahomey gap, a forest-savannah mosaic separating Upper and Lower Guinea, GP2 (West Cameroon) and the large GP3 occurred predominantly on opposite sides of the CVL, and GP4 occurred mostly on São Tomé, which is separated from the African mainland by an ocean barrier within the Bight of Bonny. The same geographic pattern was also apparent in cpDNA data, where locations separated according to these geographical features did not share any haplotypes and, in some cases, were fixed for endemic haplotypes: H04 in Benin, H13 in West Cameroon (Korup) and H03 in São Tomé. This apparent fixation of haplotypes may result from pronounced genetic drift, as expected for cpDNA under ancient divergence; however, it could also reflect uneven sampling effort as cpDNA data have been obtained for only one or very few localities in GP1, GP2 and GP4.
The order of divergence events leading to the recognized SSR GPs could not be distinguished based on a modelling approach using ABC. This result allows the following interpretations: (1) distinct GPs originated from a single ancestral population at similar times ( that is, no hierarchical tree topology) or (2) improved data or an improved model may have led to different conclusions. Notably, we did not consider the possibility of gene flow between adjacent GPs, such as GP2 and GP3 separated by the CVL. Increasing sampling effort in regions where distinct GPs come into contact, increasing the number of markers and improving coalescent models should help to disentangle the population divergence history of S. globulifera in Africa in the future.
S. globulifera from Benin was clearly differentiated from Lower Guinea, supported by SSR divergence considering stepwise mutations (RST), by STRUCTURE and TESS results (GP1) and also by a long deletion (152 bp) at psbA-trnH in the Beninese samples (data not shown). This genetic distinctiveness could have two explanations, related either to geography or to ecology. Based on geography, a Beninese GP differentiated from Lower Guinea can be expected, as has been observed for another widespread forest tree, Milicia excelsa (Daïnou et al., 2010). A biogeographic divide between Upper and Lower Guinea is reflected in both patterns of plant and animal endemism (White, 1979; Mayr and O'Hara, 1986; Linder, 2001) and, for some species, in the population genetic structure (for example, Coffea canephora (Gomez et al., 2009); Santiria trimera (Koffi et al., 2011)). This divide is presumably caused by recurrent connections and separations of both forest blocks since the end of the Pliocene, the latest opening of the Dahomey gap dating to ca 3700 BP (Maley, 1996). In M. excelsa, Beninese populations were interpreted to be of Upper Guinean origin because they showed the strongest pairwise differentiation in the study region, which comprised Benin and Lower Guinea (Daïnou et al., 2010). A similar Upper Guinean origin is also plausible in S. globulifera from Benin. Analysis of plant material from Upper Guinea and from Nigeria would be necessary to test this hypothesis. With regard to ecology, Beninese S. globulifera trees were all sampled in coastal forests. A closer look to STRUCTURE ancestry proportions revealed that nine individuals outside Benin had ancestry ⩾0.9 to GP1 characteristic of Benin (but had local cpDNA haplotypes). Five of these individuals were sampled on the bank of the Sanaga river (location 10), one in the coastal forest of Cap Estérias (location 15), one on the bank of the Ogooué river (location 19) and two in locations that were not in the immediate vicinity of rivers (locations 21 and 25 (at ca 10 km from the Congo river bank)). TESS recovered six of these individuals (from the Sanaga bank and Cap Estérias). These results suggest that there might be ecotype differentiation—coastal or swamp vs terra firme—in S. globulifera, which could be tested using more powerful markers and a more systematic sampling of coastal forests.
The CVL started forming at 30 Ma and represents a 1000 km chain of terrestrial volcanoes close to the Cameroon-Nigeria border and of seamounts in the Gulf of Guinea, including São Tomé (emerged ca 13 Ma, Meyers et al., 1998). In the STRUCTURE analysis for K=4, confirmed by TESS results, GP2 typical of location 2 west of the CVL, GP3 typical of South Cameroon and Gabon, and GP4 typical of São Tomé were found together on the CVL (location 9) or east of the CVL (locations 3 and 4). In these locations, individual ancestry proportions were typically high in one of these three GPs (maximum average ancestry 0.86 for GP2, 0.76 for GP3 and 0.66 for GP4) and just a few individuals were admixed. These results suggest that distinct GPs persisted on the CVL or close to it, or that they came into secondary contact on the CVL. Persistence on the CVL is in agreement with the suggestion that the CVL harboured a Pleistocene forest refuge, based on its distinctive flora and palaeoecological data (Maley and Brenac, 1998). The presence of GP4 (São Tomé) on and close to the CVL suggests either gene flow from São Tomé, or, alternatively, a CVL origin of the São Tomé GP. The latter hypothesis is supported to some extent by the indistinctness of São Tomé and West Cameroon in the STRUCTURE analysis for K=2, suggesting a common origin of these GPs. Colonization of São Tomé along with the CVL has been proposed for Drosophila santomea (Cariou et al., 2001), whereas studies on plants more generally proposed colonization of São Tomé from Lower Guinean forests (Stévart, 2003; Koffi et al., 2011). The (δμ)2-based UPGMA tree in S. globulifera rather supported colonization of São Tomé from the South Cameroon and Gabon region, whereas cpDNA data concurred with both colonization scenarios as H03 from São Tomé was fairly closely related to H01 from South Cameroon (locations 5 and 7, east of the CVL) and to H13 from Korup (location 2, west of the CVL).
Demographic processes within regions/GPs and examination of the forest refuge hypothesis
The forest refuge theory (Haffer, 1969; Prance, 1982) presumes the isolation of populations in refugia during unfavourable climatic conditions of the Pleistocene. The expected effects are population size reductions (bottlenecks) and subsequent population expansions when conditions improve. These demographic changes leave signals in the genetic constitution of a species that can be examined with a variety of analytical tools (Knowles, 2009).
In S. globulifera, we detected bottleneck signals compatible with forest reduction during the LGM in Benin (GP1), in West Cameroon (GP2) and in São Tomé (GP4). These results should be interpreted keeping in mind that the dating of bottlenecks is imprecise as it was based on model choice using ABC, only few competing demographic models were tested, and the scenarios in these models were kept relatively simple. GP2 is located close to the proposed Pleistocene forest refuge on the CVL (Maley, 1996; Maley and Brenac, 1998). This refuge was supported by the discovery of endemic alleles in the rainforest trees Santiria trimera (Koffi et al., 2011) and Irvingia gabonensis (Lowe et al., 2010). Therefore, one interpretation is that S. globulifera in West Cameroon shared this refuge (see previous section) and has not yet recovered from an LGM bottleneck. S. globulifera from Benin and São Tomé also bore signals of an LGM bottleneck. This could be attributed to local persistence in reduced-size populations (for instance in gallery forests), or alternatively, to a recent colonization of Benin from Upper Guinea (see previous section). Our genetic divergence results are compatible with Pleistocene persistence of S. globulifera on São Tomé. The oceanic climate has probably buffered the Pleistocene climatic oscillations on the island, which is a recognized refuge for Pre-Pleistocene lineages of other forest plants (for example, Begonias, Plana et al., 2004).
S. globulifera from South Cameroon and Gabon bore an older bottleneck signal than the other regions. It was detected with DIYABC but not with the BOTTLENECK software. This illustrates that using Garza-Williamson's M as summary statistic in an ABC approach improved the power of bottleneck detection compared with approaches based on heterozygosity (Garza and Williamson, 2001; Peery et al., 2012). The absence of a recent bottleneck signal indicates that GP3 was either not as heavily affected by the LGM as other GPs, or that it recovered better, or both. Instead, allele size ranges conserved signals of (poorly dated) older demographic dynamics, which broadly coincide with late Pleistocene forest contraction–expansion dynamics in Lower Guinea and Congolia inferred from offshore pollen deposits (Dupont, 2011). Most S. globulifera plastid haplotypes in South Cameroon and Gabon had narrow ranges or were endemic at the location level. Endemic haplotypes were found in all locations belonging to the proposed refuges examined (sensu Maley, 1996), as well as outside of them, suggesting wide-ranging long-term persistence of S. globulifera in the region. Narrow haplotype ranges contrast with the absence of substructure at the SSR level (established using STRUCTURE), which suggests much stronger genetic drift for cpDNA than for SSRs. This can be attributed to the lower historical effective population sizes of cpDNA due to the lack of recombination and the exclusively maternal inheritance of cpDNA. The four times further ranging pollen than seed dispersal detected in our study must have contributed to fast Holocene recovery of effective population sizes in South Cameroon and Gabon, whereas, on the other side, we did not find clear evidence for recolonization by seed from major postulated refuges in West Cameroon, SW Cameroon and the Crystal Mountains in Gabon (Maley, 1996). The only haplotype that ranged over hundreds of km, H07, occurred in the proposed refuges of SW Cameroon, and of the Chaillu massif (Waka) and the Gamba-Doudou complex in Gabon, suggesting that it has been widespread before the forest contraction of the LGM. Our limited sampling in the Congo Basin revealed endemic haplotypes but did not allow us to assess the importance of the proposed Congo refuge in terms of diversity or recolonization.
From these observations, we infer that our data supports partly the refuge hypothesis sensu Maley (1996), as endemic cpDNA haplotypes were found in all the proposed refugia examined. Our results indicated that S. globulifera survived unfavourable Pleistocene climatic conditions in additional locations, such as São Tomé and regions in Lower Guinea not previously proposed as refugia. Some of these locations may correspond to floodplain or gallery forests with assured water availability, similarly to the Pleistocene riverine persistence that has been proposed for some Caesalpinioideae species in Gabon (Leal, 2004). In S. globulifera, the survival in numerous, widespread forest fragments as well as high levels of pollen gene flow were probably key to a fast recovery of the species after glacial periods in Lower Guinea.
Conclusions
This study highlights the importance of geographic features in structuring the genetic constitution of the ancient rainforest tree species S. globulifera in the Guineo-Congolian phytogeographic region, in agreement with other population genetic studies of plant or animal taxa (Cariou et al., 2001; Brouat et al., 2004; Daïnou et al., 2010). Recent (LGM) bottlenecks were inferred in Benin, West Cameroon and São Tomé, suggesting that S. globulifera was affected by Pleistocene cycles of forest contraction. In the GP located in South Cameroon and Gabon, wide-ranging LGM persistence of the species enabled rapid post-glacial recovery of population sizes, especially through pollen flow, and only an older bottleneck remained traceable. In this region, S. globulifera did not display SSR substructure, contrarily to other forest tree species (Born et al., 2011; Debout et al., 2011; Daïnou et al., 2010). Furthermore, our study suggested ecotypic differentiation between coastal or swamp forest and terra firme S. globulifera. Our findings underline that species, and even GPs within species, responded individually to past disturbances. Therefore, the basis for making reliable predictions for communities in the face of current climate change should lie in gathering and jointly interpreting solid case studies on past demographic scenarios in species with a wide range of life history traits (Knowles, 2009).
Data archiving
Microsatellite data have been deposited on Dryad (doi:10.5061/dryad.3j300) and cpDNA sequences have been deposited on GenBank (JQ996246-JQ996339).
Acknowledgments
We thank Carmen García-Barriga (INIA-CIFOR, Madrid) for technical assistance in the laboratory and Armand Boudaby, Charlemagne Nguembou, Crépin Djopamdé, Elie Montchowui, Emerand Gassang, Fidel Baya, Gabriel Debout, Jean-François Gillet, Jérôme Duminil, Jérôme Laporte, Michel Arbonnier, Gilbert Todou, Nils Bourland, Paul Zok, Pierre-André Ntchandi-Otimbo, Pierre Agbani, Théophile Ayol, Charles Doumenge, Ingrid Parmentier, Peter Mambo, Gilles Dauby, Kasso Daïnou, Eben-Ezer Ewedje and Guillaume Koffi for their contribution to the sample collection. We also acknowledge the logging companies Pallisco, SFID, Wijma, CEB and CBG, the Missouri Botanical Garden (Central African Program), the CENAREST (Gabon), the Smithsonian Institution (Gabon Biodiversity Program), Patrice Ipandi (ENEF, Gabon), Bonaventure Sonké (Université de Yaoundé I, Cameroon) and Charles Doumenge (CIRAD) for facilitating field work and sample collection and thank Charles Doumenge and Katalin Csilléry for stimulating discussions. KB acknowledges a PhD scholarship (FPI) of the Spanish Ministry of Science and Innovation (MICINN). MH is a ‘Ramón y Cajal' researcher of the MICINN and acknowledges funding from MICINN (JAE-Doc program), from the Belgian Fund for Scientific Research (FRS-FNRS projects 1.2.155.06F, FC66560-1.5.095.08F and F.4.519.10.F) and the Spanish Ministry for Economy and Competitiveness (CGL2012-40129-C02-02/ AFFLORA).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abdul-Salim K. Systematics and Biology of Symphonia L. f. (Clusiaceae) PhD thesis. Harvard University: Boston; 2002. [Google Scholar]
- Aldrich PR, Hamrick JL. Reproductive dominance of pasture trees in a fragmented tropical forest mosaic. Science. 1998;281:103–105. doi: 10.1126/science.281.5373.103. [DOI] [PubMed] [Google Scholar]
- Aldrich PR, Hamrick JL, Chavarriaga P, Kochert G. Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Mol Ecol. 1998;7:933–944. doi: 10.1046/j.1365-294x.1998.00396.x. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bittrich V, Amaral MCE. Pollination biology of Symphonia globulifera (Clusiaceae) Plant Syst Evol. 1996;200:101–110. [Google Scholar]
- Bonnefille R. Tropical rainforest responses to climate change. Praxis Publishing: Chichester.; 2007. Rainforest responses to past climate changes in tropical Africa; pp. 117–170. [Google Scholar]
- Born C, Alvarez N, McKey D, Ossari S, Wickings EJ, Hossaert-McKey M, et al. Insights into the biogeographical history of the Lower Guinea Forest Domain: evidence for the role of refugia in the intraspecific differentiation of Aucoumea klaineana. Mol Ecol. 2011;20:131–142. doi: 10.1111/j.1365-294X.2010.04919.x. [DOI] [PubMed] [Google Scholar]
- Brouat C, McKey D, Douzery EJP. Differentiation in a geographical mosaic of plants coevolving with ants: phylogeny of the Leonardoxa africana complex (Fabaceae: Caesalpinioideae) using amplified fragment length polymorphism markers. Mol Ecol. 2004;13:1157–1171. doi: 10.1111/j.1365-294X.2004.02113.x. [DOI] [PubMed] [Google Scholar]
- Cariou ML, Silvain JF, Daubin V, Da Lage JL, Lachaise D. Divergence between Drosophila santomea and allopatric or sympatric populations of D. yakuba using paralogous amylase genes and migration scenarios along the Cameroon volcanic line. Mol Ecol. 2001;10:649–660. doi: 10.1046/j.1365-294x.2001.01225.x. [DOI] [PubMed] [Google Scholar]
- Chen C, Durand E, Forbes F, François O. Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes. 2007;7:747–756. [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Santos F, Beaumont MA, Robert CP, Marin JM, Balding DJ, et al. Inferring population history with DIYABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics. 2008;24:2713–2719. doi: 10.1093/bioinformatics/btn514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Carneiro F, Sebenn AM, Kanashiro M, Degen B. Low interannual variation of mating system and gene flow of Symphonia globulifera in the Brazilian Amazon. Biotropica. 2007;39:628–636. [Google Scholar]
- Daïnou K, Bizoux JP, Doucet JL, Mahy G, Hardy OJ, Heuertz M. Forest refugia revisited: SSRs and cpDNA sequences support historical isolation in a wide-spread African trees with high colonization capacity, Milicia excelsia (Moraceae) Mol Ecol. 2010;19:4462–4477. doi: 10.1111/j.1365-294X.2010.04831.x. [DOI] [PubMed] [Google Scholar]
- Dauby G, Duminil J, Heuertz M, Hardy OJ. Chloroplast DNA polymorphism and phylogeography of a Central African tree species widespread in mature rainforests: Greenwayodendron suaveolens (Annonaceae) Trop Plant Biol. 2010;3:4–13. [Google Scholar]
- Debout GDG, Doucet JL, Hardy OJ. Population history and gene dispersal inferred from spatial genetic structure of a Central African timber tree, Distemonanthus benthamianus (Caesalpinioideae) Heredity. 2011;106:88–99. doi: 10.1038/hdy.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degen B, Bandou E, Caron H. Limited pollen dispersal and biparental inbreeding in Symphonia globulifera in French Guiana. Heredity. 2004;93:585–591. doi: 10.1038/sj.hdy.6800560. [DOI] [PubMed] [Google Scholar]
- Dick CW, Abdul-Salim K, Bermingham E. Molecular systematic analysis reveals cryptic Tertiary diversification of a widespread tropical rain forest tree. Am Nat. 2003;162:691–703. doi: 10.1086/379795. [DOI] [PubMed] [Google Scholar]
- Dick CW, Heuertz M. The complex biogeographic history of a widespread tropical tree species. Evolution. 2008;62:2760–2774. doi: 10.1111/j.1558-5646.2008.00506.x. [DOI] [PubMed] [Google Scholar]
- Droissart V. Étude taxonomique et biogéographique des plantes endémiques d'Afrique centrale atlantique: le cas des Orchidaceae. PhD thesis. Université libre de Bruxelles: Brussels; 2009. [Google Scholar]
- Duminil J, Heuertz M, Doucet JL, Bourland N, Cruaud C, Gavory F, et al. CpDNA-based species identification and phylogeography: application to African tropical tree species. Mol Ecol. 2010;19:5469–5483. doi: 10.1111/j.1365-294X.2010.04917.x. [DOI] [PubMed] [Google Scholar]
- Dupont L. Orbital scale vegetation change in Africa. Quat Sci Rev. 2011;30:3589–3602. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl M, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP–Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Forget PM, Dennis AJ, Mazer SJ, Jansen PA, Kitamura S, Lambert JE, et al. 2007Seed allometry and disperser assemblages in tropical rainforests: a comparison of four floras on different continentsIn: Dennis AJ, Schupp EW, Green RJ, Westcott DA, (eds)Seed dispersal theory and its application in a changing world 5–36.
- Garza JC, Williamson EG. Detection of reduction in population size using data from microsatellite loci. Mol Ecol. 2001;10:305–318. doi: 10.1046/j.1365-294x.2001.01190.x. [DOI] [PubMed] [Google Scholar]
- Ghazoul J, Sheil D. Tropical Rain Forest Ecology, Diversity, and Conservation. Oxford University Press: Oxford; 2010. [Google Scholar]
- Goldstein DB, Pollock DD. Launching microsatellites: a review of mutation processes and methods of phylogenetic inference. J Heredity. 1997;88:335–342. doi: 10.1093/oxfordjournals.jhered.a023114. [DOI] [PubMed] [Google Scholar]
- Gomez C, Dussert S, Hamon P, Hamon S, Kochko AD, Poncet V. Current genetic differentiation of Coffea canephora Pierre ex A. Froehn in the Guineo-Congolian African zone: cumulative impact of ancient climatic changes and recent human activities. BMC Evol Biol. 2009;9:167. doi: 10.1186/1471-2148-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. Fstat (Version 1.2): a computer program to calculate F-statistics. J Heredity. 1995;86:485–486. [Google Scholar]
- Green P.2009. Phrap, version 1.090518 http://phrap.org .
- Haffer J. Speciation in Amazonian Forest Birds. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Charbonnel N, Fréville H, Heuertz M. Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics. 2003;163:1467–1482. doi: 10.1093/genetics/163.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDI; a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- Heuertz M, Carnevale S, Fineschi S, Sebastiani F, Hausman JF, Paule L, et al. Chloroplast DNA phylogeography of European ashes, Fraxinus sp. (Oleaceae): roles of hybridization and life history traits. Mol Ecol. 2006;15:2131–2140. doi: 10.1111/j.1365-294X.2006.02897.x. [DOI] [PubMed] [Google Scholar]
- Jan du Chêne RE, Salami MB. Palynology and micropaleontology of the Upper Eocene of the well nsukwa-1 (Niger Delta, Nigeria) Archives des Sciences. 1978;13:5–9. [Google Scholar]
- Knowles LL. Statistical phylogeography. Annu Rev Ecol Evol Syst. 2009;40:593–612. [Google Scholar]
- Koffi KG, Hardy JH, Doumenge C, Cruaud C, Heuertz M. Diversity gradients and phylogeographic patterns in Santiria trimera (Burseraceae), a widespread African tree typical of mature rainforests. Am J Bot. 2011;98:254–264. doi: 10.3732/ajb.1000220. [DOI] [PubMed] [Google Scholar]
- Leal ME. The African Rain Forest During the Last Glacial Maximum, an archipelago of Forests in a Sea of Grass. PhD thesis. University of Wageningen; 2004. [Google Scholar]
- Lescure JP, Boulet R. Relationship between soil and vegetation in a Tropical Rain Forest in French Guiana. Biotropica. 1985;17:155–164. [Google Scholar]
- Linder HP. Plant diversity and endemism in sub-Saharan tropical Africa. J Biogeogr. 2001;28:169–182. [Google Scholar]
- Lowe AJ, Harris D, Dormontt E, Dawson IK. Testing putative African tropical refugia using chloroplast and nuclear DNA phylogeography. Trop Plant Biol. 2010;3:50–58. [Google Scholar]
- Maley J. The African rain forest: main characteristics of changes in vegetation and climate from the upper Cretaceous to the Quaternary. Proc Roy Soc Edinburgh. 1996;104B:31–73. [Google Scholar]
- Maley J, Brenac P. Vegetation dynamics, palaeoenvironments and climatic changes in the forests of western Cameroon during the last 28,000 years B.P. Rev Paleobot Palyno. 1998;99:157–187. [Google Scholar]
- Mayr E, O'Hara RJ. The biogeographic evidence supporting the Pleistocene forest refuge hypothesis. Evolution. 1986;40:55–67. doi: 10.1111/j.1558-5646.1986.tb05717.x. [DOI] [PubMed] [Google Scholar]
- McMahon SM, Harrison SP, Armbruster WS, Bartlein PJ, Beale CM, Edwards ME, et al. Improving assessments of climate-change impacts on global biodiversity. Trends Ecol Evol. 2011;26:249–259. doi: 10.1016/j.tree.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Meyers J, Rosendahl B, Harrison C, Ding Z. Deep-imaging seismic and gravity results from offshore Cameroon volcanic line, and speculation of African hotlines. Tectonophysics. 1998;284:31–63. [Google Scholar]
- Millar CI, Stephenson NL, Stephens SL. Climate change and forests of the future: managing in the face of uncertainty. Ecol Appl. 2007;17:2145–2151. doi: 10.1890/06-1715.1. [DOI] [PubMed] [Google Scholar]
- Moritz C, Patton JL, Schneider CJ, Smith TB. Diversification of rainforest faunas: an integrated molecular approach. Annu Rev Ecol Evol Syst. 2000;31:533–563. [Google Scholar]
- Ngomanda A, Neumann K, Schweizer A, Maley J. Seasonality change and the third millennium BP rainforest crisis in southern Cameroon (Central Africa) Quat Res. 2009;71:307–318. [Google Scholar]
- Nybom H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol. 2004;13:1143–1155. doi: 10.1111/j.1365-294X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- Oyen LPA.2005Symphonia globulifera L.f. [Internet] Record from ProtabaseIn: Louppe D, Oteng-Amoako AA, Brink M, (eds)PROTA (Plant Resources of Tropical Africa/Ressources végétales de l'Afrique tropicale) Wageningen: Netherlands; < http://database.prota.org/search.htm > accessed on 29 March 2012.. [Google Scholar]
- Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet-Bëer E, Robinson S, et al. Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol. 2012;21:3403–3418. doi: 10.1111/j.1365-294X.2012.05635.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Aguinagalde I, Beaulieu JL, de Beaulieu JL, Bittkau C, Brewer S, et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Plana V. Mechanisms and tempo of evolution in the African Guineo-Congolian rainforest. Philos T Roy Soc B. 2004;359:1585–1594. doi: 10.1098/rstb.2004.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plana V, Gascoigne A, Forrest LL, Harris D, Pennington RT. Pleistocene and pre-Pleistocene Begonia speciation in Africa. Mol Phylogenet Evol. 2004;31:449–461. doi: 10.1016/j.ympev.2003.08.023. [DOI] [PubMed] [Google Scholar]
- Pons O, Petit RJ. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996;144:1237–1245. doi: 10.1093/genetics/144.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prance GT. Biological Diversification in the Tropics. Colombia University Press: New York; 1982. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Mountain JL. Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA. 1997;94:9197–9201. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae) Am J Bot. 1997;84:1120–1136. [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:1463–1463. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosef M. Refuge begonias: taxonomy, phylogeny and historical biogeography of Begonia sect. Loasibegonia and sect. Scutobegonia in relation to glacial rain forest refuges in Africa. Agricultural University Wageningen Papers. 1994;94:1–306. [Google Scholar]
- Stévart T. Étude taxonomique, écologie et phytogéographique des Orchidaceae en Afrique centrale atlantique. PhD thesis. Université Libre de Bruxelles: Brussels; 2003. [Google Scholar]
- Städler T, Haubold B, Merino C, Stephan W, Pfaffelhuber P. The impact of sampling schemes on the site frequency spectrum in nonequilibrium subdivided populations. Genetics. 2009;182:205–216. doi: 10.1534/genetics.108.094904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Andel TR. Floristic composition and diversity of three swamp forests in northwest Guyana. Plant Ecol. 2003;167:293–317. [Google Scholar]
- Vekemans X, Hardy OJ. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol. 2004;13:921–935. doi: 10.1046/j.1365-294x.2004.02076.x. [DOI] [PubMed] [Google Scholar]
- Vinson CC, Amaral AC, Sampaio I, Ciampi AY. Characterization and isolation of DNA microsatellite primers for the tropical tree, Symphoniaglobulifera Linn. f. Mol Ecol Resources. 2005;5:202–204. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- White F. The Guineo-Congolian region and its relationships to other phytochoria. Bull Jard Bot Nat Belgique. 1979;49:11–55. [Google Scholar]
- Willyard A, Syring J, Gernandt DS, Liston A, Cronn R. Fossil calibration of molecular divergence infers a moderate mutation rate and recent radiations for Pinus. Mol Biol Evol. 2007;24:90–101. doi: 10.1093/molbev/msl131. [DOI] [PubMed] [Google Scholar]
- Williamson-Natesan EG. Comparison of methods for detecting bottlenecks from microsatellite loci. Conserv Genet. 2005;6:551–562. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.