Abstract
X-linked sex-ratio distorters that disrupt spermatogenesis can cause a deficiency in functional Y-bearing sperm and a female-biased sex ratio. Y-linked modifiers that restore a normal sex ratio might be abundant and favored when a X-linked distorter is present. Here we investigated natural variation of Y-linked suppressors of sex-ratio in the Winters systems and the ability of these chromosomes to modulate gene expression in Drosophila simulans. Seventy-eight Y chromosomes of worldwide origin were assayed for their resistance to the X-linked sex-ratio distorter gene Dox. Y chromosome diversity caused males to sire ∼63% to ∼98% female progeny. Genome-wide gene expression analysis revealed hundreds of genes differentially expressed between isogenic males with sensitive (high sex ratio) and resistant (low sex ratio) Y chromosomes from the same population. Although the expression of about 75% of all testis-specific genes remained unchanged across Y chromosomes, a subset of post-meiotic genes was upregulated by resistant Y chromosomes. Conversely, a set of accessory gland-specific genes and mitochondrial genes were downregulated in males with resistant Y chromosomes. The D. simulans Y chromosome also modulated gene expression in XXY females in which the Y-linked protein-coding genes are not transcribed. The data suggest that the Y chromosome might exert its regulatory functions through epigenetic mechanisms that do not require the expression of protein-coding genes. The gene network that modulates sex ratio distortion by the Y chromosome is poorly understood, other than that it might include interactions with mitochondria and enriched for genes expressed in post-meiotic stages of spermatogenesis.
Keywords: sex ratio distortion, Y chromosome, gene expression, spermatogenesis, mitochondria
Introduction
The Drosophila Y chromosome is an unusual molecule. In Drosophila melanogaster the Y chromosome is ∼40 megabases but harbors only ∼14 protein-coding genes. All of the Y-linked genes with known phenotypes are essential for spermatogenesis and male fertility (Carvalho et al., 2009; Krsticevic et al., 2010). The Y chromosome is otherwise composed of multimegabase-long segments of repetitive sequences, including satellite repeats and transposable elements. Because of this feature, the Y chromosome is maintained throughout most of the cell cycle as highly condensed heterochromatin. Expression of the few protein-coding loci requires major chromatin remodeling in the testis (Ashburner et al., 2005). The D. melanogaster Y chromosome also harbors a functional rDNA array that comprises hundreds of active rRNA genes and non-transcribed spacer repeats (Bonaccorsi and Lohe, 1991), the latter constituting the paring sites of the X and Y chromosomes during male meiosis (McKee et al., 1992; Ren et al., 1997). In Drosophila simulans, a sibling species of D. melanogaster, the rDNA array on the Y chromosome consists only of the non-transcribed spacer repeats (Brosseau, 1960; Lohe and Roberts, 1990), which are presumed to be the X-Y pairing site during meiosis in this species. The content of protein-coding genes is conserved in the Y chromosome of D. simulans and D. melanogaster (Koerich et al., 2008).
According to the canonical theory, sex chromosomes have evolved from a homologous pair of autosomes. The X and Y chromosomes are therefore expected to share homologous genes, although homology might be limited at advanced stages of X-Y divergence. X-Y chromosome homology is evident in some Drosophila species, such as Drosophila miranda. In this species, a young pair of sex chromosomes has only recently evolved from autosomes. As there is no recombination in Drosophila males, the neo-Y chromosome has lost genetic diversity and accumulated deleterious mutations, in line with the expected evolutionary trajectory of non-recombining chromosome (Bull, 1983; Rice, 1987; Charlesworth and Charlesworth, 2000; Bachtrog et al., 2008; Kaiser et al., 2011). For most Drosophila species, however, the homology between the X and Y chromosomes is very limited. For example, the Y chromosome from D. melanogaster and D. simulans do not share protein-coding genes with their respective X chromosomes (Carvalho et al., 2009). Hence, it has been suggested that the Y chromosome from these species might have originated from B chromosomes (Hackstein et al., 1996; Carvalho and Clark, 2005). In any circumstance, a consensus is emerging that the Drosophila Y chromosome is not an inert and passive portion of the genome. Instead, recent studies paint a picture of a dynamic chromosome (Carvalho, 2002; Lemos et al., 2008; Carvalho et al., 2009; Kaiser et al., 2011). A survey of the 14 Y-linked genes of D. melanogaster across 12 Drosophila species found only three genes present in all species and seven of them were acquired within the past 63 million years (Koerich et al., 2008).
The notion that the Y chromosome has an active role in the genome has been further supported by studies demonstrating its contribution to diverse phenotypes, in spite of the very few protein-coding genes residing on this chromosome. The Y chromosome has been shown to regulate organismal traits, such as male fertility (Chippindale and Rice, 2001), sex ratio distortion (Carvalho et al., 1997; Montchamp-Moreau et al., 2001), thermal adaptation (Rohmer et al., 2004) and behaviors (Stoltenberg and Hirsch, 1997; Huttunen and Aspi, 2003). The regulatory bases for these phenotypes have been suggested by genome-wide gene expression studies in D. melanogaster (Lemos et al., 2008, 2010; Jiang et al., 2010; Paredes et al., 2011). Transcription of hundreds of genes is altered by Y chromosome variants, which differ in their ability to modulate epigenetic states across the genome (Lemos et al., 2010). These genes are clustered into functional classes related to mitochondria, immunity, as well as chromatin structure and maintenance (Lemos et al., 2008, 2010; Paredes et al., 2011). Variable titration of nuclear proteins by the Y chromosome might cause imbalances in the nucleus and consequently the modulation of gene expression (Platero et al., 1998; Janssen et al., 2000).
Here we studied the ability of polymorphic Y chromosomes of D. simulans to modulate genome-wide gene expression levels and the sex-ratio. Sex-ratio describes a genetic phenomenon in which the offspring sex ratio deviates from the 50% female expectation. At least three sex ratio distortion systems have been uncovered in D. simulans, namely the Paris, the Durham and the Winters systems (Tao et al., 2007b). In the Paris sex-ratio system, the Y chromosome has high frequency of breakage and non-disjunction during meiosis II (Cazemajor et al., 2000). Consequently, the Y-bearing gametes fail to complete spermatogenesis, thus leading to a functional sperm pool dominated by X-bearing sperm and female-biased sex ratio in the offspring (Montchamp-Moreau and Joly, 1997; Montchamp-Moreau, 2006). In the Durham system, in addition to Y-breakage in meiosis II, the autosomal suppressor also has pleiotropic effect on male fertility (YT, unpublished). In the Winters system, the Y chromosome appears to undergo normal disjunction, but post-meiotic spermiogenesis of the Y-bearing gametes is disrupted (Tao et al., 2007a). Distortion is caused by the X-linked gene Dox (Distorter on the X; Tao et al., 2007a). In offspring sired by males in which the autosomal suppressor Nmy (not much yang) is removed, the sex ratio is strongly female biased (Tao et al., 2007b).
In this study we focused on the Winters system. We first identified polymorphic Y chromosomes that vary in their resistance to the Winters sex-ratio distorter gene Dox, resulting in sex ratio ranging from 63% to 98% females. We then investigated genome-wide gene expression in representative males harboring Y chromosomes that are resistant (low sex ratio) and Y chromosomes that are sensitive (high sex ratio) to Dox. A disproportionate number of testis-specific genes were upregulated by the resistant Y, while mitochondria-related and accessory gland-specific genes were downregulated. Similar analyses using XXY females indicated that the expression of protein-coding genes is not required for the Y-linked regulatory functions in D. simulans. In conclusion, the D. simulans Y chromosome shows abundant polymorphic variation that regulates sex-ratio distortion in natural populations. The data raise the prospects that the responsible polymorphisms might not reside exclusively within proteins.
Materials and methods
Drosophila strains
Y chromosomes from 78 isofemale stocks of D. simulans were introgressed into a nearly identical background (SR1227: w Dox; nt; nmy[sim1427]) (Tao et al., 2007b), using a cross scheme (Supplementary Figure S1) in which all chromosomes except the fourth were tracked with phenotypic markers. These stocks include 44 collected in the Carlson Orchards, Harvard, Massachusetts and 34 of global origin (Supplementary Table S1). For gene expression analyses, flies were grown in incubators maintained at 25 °C, 65% of relative humidity and constant light. Newly emerged adults were collected and aged for 2 days at the same rearing condition before they were flash-frozen in liquid nitrogen and stored at −80 °C. Whenever females were analyzed, virgin females were collected within 7 h after eclosion. All strains were expanded in vials; independent sets of biological replicates were frozen for expression analyses. XXY females were obtained by crossing the SR1227/Y chromosome substitution males to the C(1)DX, y w females.
Sex-ratio analysis
The sex ratio of a male genotype was measured by crossing 5 males to 10 virgin SR1227 females at room temperature (∼23 °C), with an average of 2.7 replicates for each of the 78 Y-introgression strains. Offspring were sexed and counted until the nineteenth day after mating. Sex ratio was calculated as the proportion of females.
Gene expression analyses
Microarrays were ∼18 000-feature cDNA arrays spotted with D. melanogaster cDNA PCR products (Lemos et al., 2008). We have recently annotated these probes using the D. simulans genome. Possible biases introduced by using a D. melanogaster platform for a D. simulans genome are minimized because the probed genomes are essentially identical except for the Y chromosomes. The platform has been successfully used to profile gene expression variation in D. simulans when the probed genomes are isogenic for the sequences assayed (Araripe et al., 2010; Sackton et al., 2011). Here the probed genome is the SR1227 isogenic background. Total RNA was extracted from whole flies using TRIzol (Life Technologies, Carlsbad, CA, USA). The synthesis of cDNA and its labeling with fluorescent dyes (Cy3 and Cy5) as well as hybridization reactions were carried out using 3DNA protocols and reagents (Genisphere, Hatfield, PA, USA). Slides were scanned using Axon 4000B scanner (Axon Instruments, Molecular Devices, Sunnyvale, CA, USA) and GenePix Pro 6.0 software (Molecular Devices). We used highly stringent quality control criteria to ensure reliability of foreground intensity reads for both Cy3 and Cy5 channels (Lemos et al., 2008). This procedure screens out poor-quality arrays and ensures that the resulting data are minimally sensitive to the parameters of the normalization method. The final data set consists of 58 high-quality arrays in which >70% of all expressed spots passed our highly stringent quality criteria (Lemos et al., 2008, 2010). We used balanced designs with dye swaps (Supplementary Figure S2). The Y chromosome from Drosophila sechellia might have a significant impact on gene expression and was included. Foreground fluorescence of dye intensities was normalized by the Loess method in Bioconductor/Limma (Smyth and Speed, 2003; Smyth, 2005). The significance of variation in gene expression was assessed with the Bayesian Analysis of Gene Expression Levels (Townsend and Hartl, 2002). Results were checked for consistency using Linear Models in Limma. False discovery rates were empirically estimated by permutation of the data set. The microarray gene expression data reported here can be obtained at the Gene Expression Omnibus database under accession number GSE43665.
Functional categories
Enrichment in Gene Ontology (GO) categories was assessed with modMine (March 2012) (Contrino et al., 2012), and the GO significance was estimated using the Holm–Bonferroni method for multiple hypothesis testing (Holm, 1979). List of genes in various biological categories were obtained from the FlyBase (McQuilton et al., 2012).
Tissue-specific gene expression
As a proxy for the tissue specificity of gene expression in D. simulans, we used the D. melanogaster data available from the FlyAtlas (downloaded on December 2011) (Chintapalli et al., 2007). We filtered the data to include only a non-redundant set of adult tissues (brain, eye, thoracic abdominal ganglion, crop, midgut, hindgut, tubule, testis, accessory gland, salivary gland, adult fat body, heart and trachea). For each Affymetrix probe in the FlyAtlas data set and for each tissue we set the expression level to 0 unless the probe was called as ‘present' in at least two out of four arrays and then averaged over all probes and arrays for each FBgn to calculate an expression level for each gene in each tissue. Genes with expression counts <100 were conservatively set to zero. We calculated a tissue specificity index (TSIi) for tissue i in a set of n tissues as the expression in tissue i over the total expression values across all tissues 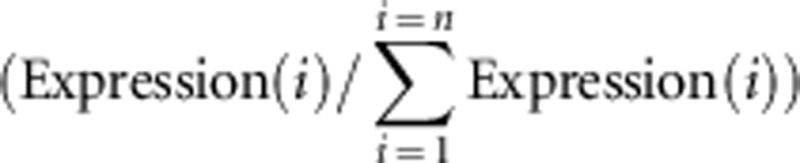 . Genes expressed exclusively in a focal tissue have a TSI equal to 1. We define a gene as tissue specific if this gene has a TSI>0.90 for the focal tissue. We further conservatively require that tissue-specific genes to have expression counts >250 units in the focal tissue.
. Genes expressed exclusively in a focal tissue have a TSI equal to 1. We define a gene as tissue specific if this gene has a TSI>0.90 for the focal tissue. We further conservatively require that tissue-specific genes to have expression counts >250 units in the focal tissue.
Results and discussion
Natural polymorphism in Y-linked suppressors of the Winters sex-ratio system
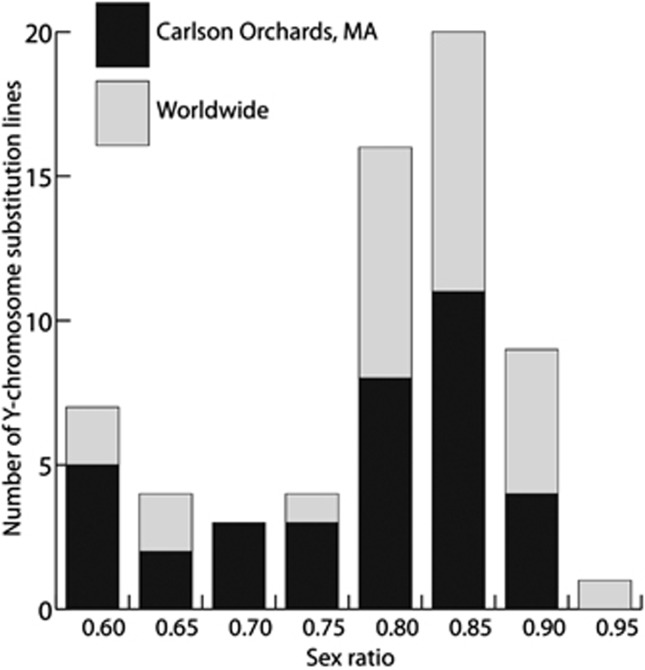
Several studies have shown that Y chromosomes from D. melanogaster or D. simulans are polymorphic for their ability to modulate organismal phenotypes. Here we first investigated whether the Y chromosome in D. simulans could modulate sex ratio distortion of the Winters system. We introgressed a collection of 78 independently derived Y chromosomes into the isogenic D. simulans stock SR1227 (w Dox; nt; nmy[1427]) (Supplementary Figure S1). The SR1227 genotype expresses the sex-ratio distorter Dox located on the X chromosome but lacks the functional suppressor Nmy on the third chromosome. Hence, SR1227 males with their original Y chromosome sire offspring broods with a biased sex ratio of ∼75% (Tao et al., 2007a). Thirty-four Y chromosomes investigated had global origins, while the remaining 44 Y chromosomes were derived from isofemale strains collected in Carlson Orchards, Harvard, Massachusetts (Supplementary Table S1). These Y chromosomes introgressed into the SR1227 genetic background significantly contributed to the large variance of sex ratio in the offspring, which ranged from 63.1% to 98.4% females (analysis of variance with GLM, df=77, P<2.2 × 10−22, Figure 1). Even the Y chromosomes originating from a single Carlson Orchards population displayed a similarly large span of variation (two-sample Kolmogorov–Smirnov test, P=0.7443) (Figure 1). The sample included males producing a highly skewed female-biased sex-ratio (sensitive Y chromosomes) and males displaying a fair rescue of the sex-ratio phenotype (resistant Y chromosomes). These observations are in agreement with previous findings in other sex-ratio systems where the Y chromosome varied in its resistance to sex ratio distortion (Carvalho et al., 1997; Montchamp-Moreau et al., 2001) and support the theoretical predictions of high genetic variances for sex-ratio suppressor on the Y chromosome (Curtsinger and Feldman, 1980; Jaenike, 1999).
Figure 1.
Distribution of sex ratios as regulated by naturally occurring Y chromosomes. Sex ratios were measured in offspring sired by male genotypes isogenic to w Dox; nt; nmy [sim1427], except for the introgressed Y chromosomes. Y chromosomes were independently derived from 78 isofemale D. simulans stocks, among which 44 stocks originated from a single locality in Massachusetts and 34 stocks were of global origins.
Regulation of testis-specific genes in males with resistant Y chromosomes
In order to gain insight into the molecular mechanism through which Y chromosomes modulate the Winters sex-ratio, we performed genome-wide gene expression analysis in four representative Y chromosome-introgression strains. The four strains carried Y chromosomes derived from the Carlson Orchards population and showed contrasting sex ratio distortion. The resistant Y chromosome strains Y71 and Y76 produced 64.0±0.9% (mean±s.e.m.) and 67.1±1.4% females, respectively, while the sensitive Y chromosome strain Y52 and Y56 produced 85.7±1.5% and 82.7±1.7% females, respectively. The progenitor strain (SR1227; Yct) produced a sex ratio of 73.3±2.0% females (Tao et al., 2007a), and was profiled as a control.
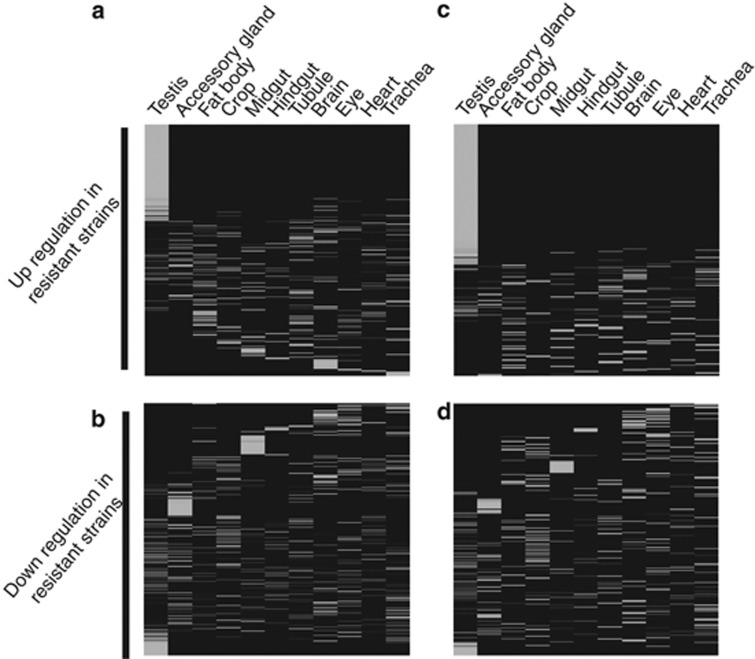
We first contrasted genome-wide gene expression differences between the resistant Y chromosome strains and the more sensitive SR1227 control (Yct). We identified 436 genes downregulated and 409 genes upregulated in the resistant Y chromosome strains (Bayesian Posterior Probability (BPP)>0.99; false discovery rate (FDR)<0.10). Tissue specificity of differentially expressed genes were analyzed using the FlyAtlas (Chintapalli et al., 2007). Among the 409 upregulated genes, we observed a significant enrichment for testis-specific genes (P<10−10, Fisher's exact test; Figure 2a). Conversely, the 436 downregulated genes were not enriched in any single tissue but instead were broadly expressed across multiple tissues (Figure 2b). This result suggests that Y-linked resistance to sex ratio distortion is associated with a general robustness of testis-specific gene expression. Although informative, the contrast between the resistant Y chromosome strains and SR1227 might be confounded by inter-population Y chromosome divergence that might be unrelated to the sex-ratio phenotype (Lemos et al., 2008, 2010). We therefore analyzed a smaller set of 521 differentially expressed genes from the statistical contrast between the two resistant and two sensitive Y chromosome introgression lines (BPP>0.99; FDR<0.10). Among these, 233 genes were upregulated (Figure 2c) and 288 genes were downregulated (Figure 2d) in the resistant Y strains relative to sensitive ones, with a modest but significant correlation in fold change between the two resistant strains (ρ=0.18, P<0.001). Tissue-specific analysis of these genes reinforced the results of the initial analysis: genes consistently upregulated in strains with resistant Y chromosomes were enriched for targets with testis-specific expression (P<10−16, Fisher's exact test; Supplementary Table S2). Indeed, nearly 45% of all the upregulated genes were expressed exclusively in this single tissue (Figure 2c). Conversely, among genes downregulated in the Y resistant lines, we observed that only a few showed testis expression (Figure 2d). The lack of gene ontology classification limits the detection of functional clustering among testis-specific genes in Drosophila (McQuilton et al., 2012). Altogether, our analyses suggest an association between the suppression of sex-ratio by the resistant Y chromosomes and the higher expression of testis-specific genes.
Figure 2.
Disproportionate upregulation of testis-specific genes in males with resistant Y chromosomes. Genes differentially expressed in D. simulans were clustered based on the D. melanogaster FlyAtlas tissue expression data (Chintapalli et al., 2007). Panels show the distribution of differentially expressed genes across 11 adult tissues. Expression levels were normalized for each gene with light and dark denoting high and low transcript abundance, respectively. (a) 409 genes that were upregulated and (b) 436 genes that were downregulated in the resistant Y chromosome males as contrasted to the control SR1227 males. Similarly, (c) 233 genes were upregulated and (d) 288 genes were downregulated in the resistant Y males as contrasted to the sensitive Y males.
Modulation of mitochondria-associated genes in sex-ratio males
Evolutionary theory predicts a disproportional contribution of sex chromosomes (Hamilton, 1967) and cytoplasmic genes (Cosmides and Tooby, 1981) to the control of sex ratio distortion. Intragenomic conflicts over sex ratio control, which is a perpetual tug-of-war among the X, the Y, the autosomes and the cytoplasmic genes, might cause several well-known genomic patterns uncovered in recent years, including meiotic sex chromosome inactivation and out-of-the-X traffic of testis-specific genes (Meiklejohn and Tao, 2010). Interestingly, many of the mitochondrial genes have evolved to function specifically during spermatogenesis in Drosophila (Gallach et al., 2010). This organelle has an essential role in sperm morphogenesis (Noguchi et al., 2011) and motility (Fuller, 1993), although it is not inherited through males in Drosophila (DeLuca and O'Farrell, 2012). Altogether, the system sets the stage in which interactions between mitochondria and Y-linked modifiers of sex-ratio can evolve.
We observed significant enrichment for several functions related to metabolism and the mitochondria among the 436 genes downregulated in the resistant Y chromosome strains relative to the more sensitive SR1227 (Table 1). Specifically, about 7.5% of all downregulated genes (33 genes out of 436) cluster in the following categories: cellular respiration (GO:0045333; P=1.34 × 10−19), respiratory electron transport chain (GO:0022904; P=8.97 × 10−16) and ATP synthesis-coupled electron transport (GO:0042773; P=5.79 × 10−13). Among the genes in the last category are four encoded by the mitochondrial genome (mitochondrial cytochrome c oxidase subunit II (FBgn0013675), mitochondrial cytochrome c oxidase subunit III (FBgn0013676), mitochondrial NADH-ubiquinone oxidoreductase chain 5 (FBgn0013684) and mitochondrial cytochrome b (FBgn0013678)). We observed similar patterns in the set of 288 genes downregulated in males with the resistant Y chromosome relative to males with the sensitive Y chromosome (Table 2). The signature of Y chromosome by mitochondria interaction is indeed stronger when we subset the analysis to the downregulated genes with the highest expression in the testis (Table 3).
Table 1. Gene Ontology (GO) analysis of the downregulated genes identified in the resistant Y chromosome males relative to SR1227.
| GO term | Biological process | P-valuea | No. of genes |
|---|---|---|---|
| GO:0045333 | Cellular respiration | 1.34E−19 | 33 |
| GO:0015980 | Energy derivation by oxidation of organic compounds | 2.91E−18 | 33 |
| GO:0006091 | Generation of precursor metabolites and energy | 1.47E−16 | 35 |
| GO:0022904 | Respiratory electron transport chain | 8.97E−16 | 25 |
| GO:0022900 | Electron transport chain | 5.69E−15 | 25 |
| GO:0042773 | ATP synthesis coupled electron transport | 5.79E−13 | 22 |
| GO:0042775 | Mitochondrial ATP synthesis coupled electron transport | 2.11E−12 | 21 |
| GO:0006119 | Oxidative phosphorylation | 4.40E−12 | 22 |
| GO:0006120 | Mitochondrial electron transport, NADH to ubiquinone | 6.54E−08 | 13 |
| GO:0007602 | Phototransduction | 6.04E−07 | 14 |
| GO:0009583 | Detection of light stimulus | 0.0000028 | 14 |
| GO:0055114 | Oxidation–reduction process | 0.0000149 | 47 |
| GO:0009581 | Detection of external stimulus | 0.0000454 | 14 |
| GO:0009060 | Aerobic respiration | 0.0028969 | 10 |
| GO:0009416 | Response to light stimulus | 0.0078173 | 15 |
| GO:0009314 | Response to radiation | 0.0089795 | 16 |
| GO:0006084 | Acetyl-CoA metabolic process | 0.0341105 | 9 |
Abbreviations: ATP, adenosine triphosphate; NADH, nicotinamide adenine dinucleotide.
Corrected with the Holm–Bonferroni Multiple Hypothesis Test.
Table 2. GO analysis of the downregulated genes identified in the resistant Y chromosome males relative to the sensitive ones from the same population.
| GO term | Biological process | P-valuea | No. of genes |
|---|---|---|---|
| GO:0009583 | Detection of light stimulus | 0.000818 | 10 |
| GO:0045333 | Cellular respiration | 0.006737 | 13 |
| GO:0003012 | Muscle system process | 0.009314 | 6 |
| GO:0006091 | Generation of precursor metabolites and energy | 0.012548 | 15 |
| GO:0015980 | Energy derivation by oxidation of organic compounds | 0.017841 | 13 |
Abbreviation: GO, Gene Ontology.
Corrected with the Holm–Bonferroni Multiple Hypothesis Test.
Table 3. GO analysis of the top 50 highest expressed testis-specific genes identified as downregulated in the resistant Y males contrasted to the sensitive ones.
| GO term | Biological process | P-valuea | No. of genes |
|---|---|---|---|
| GO:0045333 | Cellular respiration | 0.001731 | 7 |
| GO:0015980 | Energy derivation by oxidation of organic compounds | 0.003105 | 7 |
| GO:0006091 | Generation of precursor metabolites and energy | 0.016984 | 7 |
| GO:0042775 | Mitochondrial ATP synthesis coupled electron transport | 0.029857 | 5 |
| GO:0042773 | ATP synthesis coupled electron transport | 0.040518 | 5 |
Abbreviations: ATP, adenosine triphosphate; GO, Gene Ontology.
Corrected with the Holm–Bonferroni Multiple Hypothesis Test.
Abnormal mitochondrial degeneration during spermatogenesis is a remarkable feature of the sex-ratio males of D. simulans (Ramamurthy et al., 1980). Our analyses did not detect the differential regulation of key apoptotic genes, an observation suggesting that the degeneration process is not associated with typical apoptotic pathways. Instead, the regulation of genes associated with ATP biosynthesis might point to inefficiencies of the energetic metabolism due to the mitochondrial degeneration in the Y-bearing spermatids that lead the upregulation of bioenergetic genes to compensate the energetic needs of the ‘ill' cell (Heddi et al., 1999; Epstein et al, 2001). Altogether, the data support the notion that maternally and paternally inherited elements might genetically interact in the etiology of sex ratio distortion.
Genes expressed at post-meiotic stages of spermatogenesis are particularly prone to mis-regulation in sex-ratio males
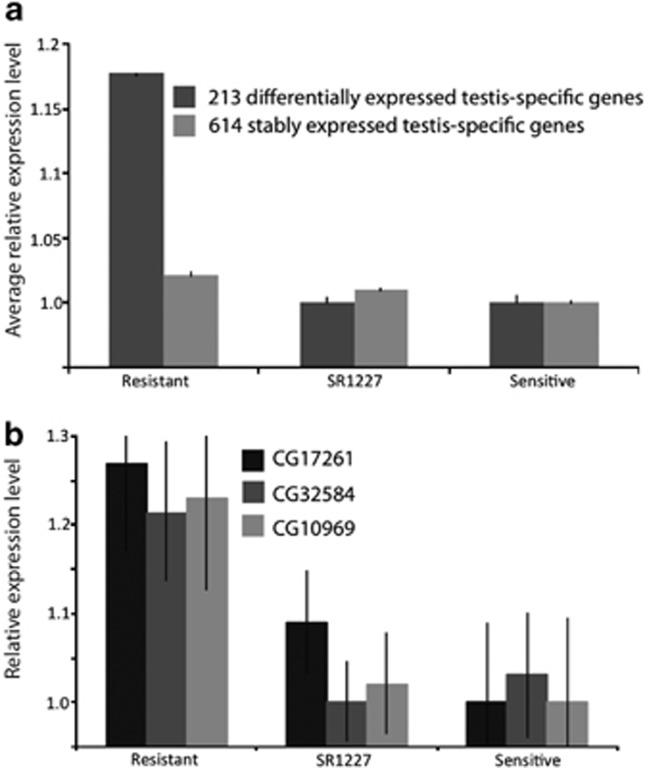
Disturbance of testis-specific gene expression in sex-ratio males might be caused by a general breakdown of spermatogenesis or might be limited only to those genes directly related to the deficiency of the Y-bearing sperm. The testis of SR1227 males does not show any overtly impaired development, but abnormal spermiogenesis of Y-bearing sperm is evident by ultrastructural studies (Tao et al., 2007a), favoring the latter explanation. We narrowed our analyses to testis-specific genes to address this question. We observed 1102 testis-specific genes in the FlyAtlas, 827 of which were detected in our data set. A set of 213 testis-specific genes were differentially expressed across any of the three possible contrasts between the resistant Y, the sensitive Y and the SR1227 control males (Figure 3a). The remaining genes comprise a rather large set (614 out of 827 or 74.3%) of testis-specific genes that were stably expressed across all the three types of strains (Figure 3a; 0.05<BPP<0.95). Most (93%) of the 213 differentially expressed testis-specific genes have no known functional annotations (Figure 3b). Nevertheless, Vibranovski et al. (2009) reported gene expression variation along three time points of spermatogenesis, namely mitotic, meiotic and post-meiotic stages, and identified their peak expression stages. For genes upregulated in males with resistant Y chromosomes, the frequency (12%) of the meiotic genes, which have peak expression levels at meiotic stage, is similar to the frequency (13%) of meiotic genes in the set of background genes that are not differentially expressed (P=0.98). On the other hand, we observed a significant twofold enrichment for post-meiotic genes (P<1.1 × 10−5), whose expression levels peak at the distal region of the testis that contains mostly spermatids at post-meiotic stages (Vibranovski et al., 2009). This indicates that mis-regulation of testis-specific genes in sex-ratio males is not a result of generally abnormal spermatogenesis; rather it appears to be specifically related to the dysgenesis of Y-bearing sperm. This is consistent with morphological observations (Tao et al., 2007a, 2007b).
Figure 3.
Characterization of the 213 testis-specific genes upregulated in the resistant Y chromosome males. (a) The average expression level of the 213 genes is significantly higher in the resistant Y chromosome males as compared with more sensitive Y males (P<0.01, dark gray). The other 614 testis-specific genes stably expressed across different Y introgression males (0.05<P<0.95, light gray). Error bars denote >99% confidence intervals for the average fold differences (±4 s.e.m.). (b) The 213 genes generally lack annotation, as illustrated by three representatives: CG17261 (black), CG32584 (dark gray), and CG10969 (light gray). Error bars denote 95% credible intervals for individual genes.
Accessory gland expression is downregulated in the resistant Y males
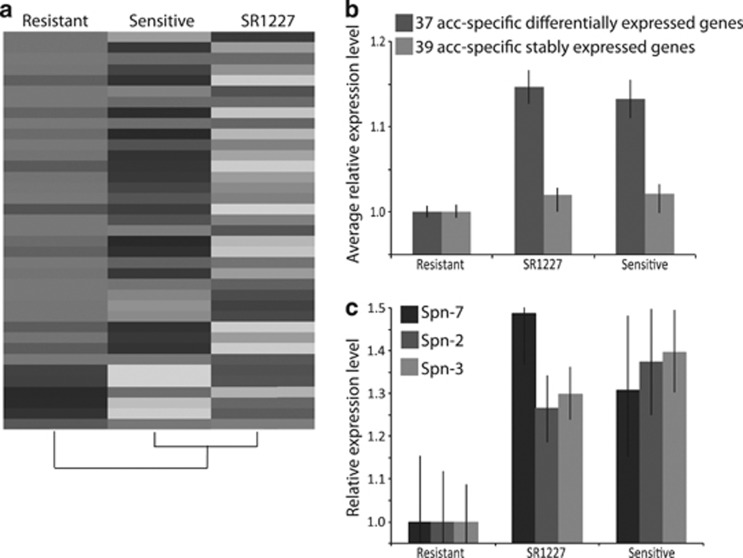
Sex-ratio males are not only weaker in sperm displacement but are also discriminated against by non-virgin females in both Drosophila pseudoobscura and D. simulans (Wu, 1983a, 1983b; Atlan et al., 2004). Males with the Winters sex-ratio also show reduced fertility (Tao et al., 2007b). Furthermore, the Winters system is polymorphic in natural populations with segregating variation that shows evidence of recent selection in both distorters and suppressors (Kingan et al., 2010). Indeed, the invasion of sex-ratio distorters sets the stage for the evolution of genetic suppressors as well as behavioral and physiological mechanisms that might alleviate the fitness cost incurred by a biased sex-ratio (Price et al., 2008). We observed that a cluster of accessory gland-specific genes were significantly downregulated in males with resistant Y chromosomes (P=0.002; Fisher's exact test) (Figures 2b and d). Indeed, the average expression level of 37 accessory gland-specific genes is significantly higher in males expressing more biased sex ratio (Figures 4a and b). Notably, protease inhibitors are accessory gland-specific proteins (Figure 4c) that are able to prevent the premature breakdown of the mating plug and increase sperm vitality (Park and Wolfner, 1995; Chapman, 2001), and that might function to mitigate the lower fitness incurred by sex-ratio males (Tao et al., 2007b). Although higher expression of accessory gland proteins (ACPs) might contribute to partly compensate for impaired testis-specific gene expression in males with skewed sex-ratio (Linklater et al., 2007; Dowling and Simmons, 2012), we cannot formally distinguish between adaptive and non-adaptive mechanisms underlying variation in ACP expression. Similarly, although ejaculate composition might be a mechanism to maintain male fitness in response to the disruption of the testis function in males with highly biased sex ratio, we cannot distinguish direct and indirect effects of Y chromosome on ACP expression. Further population-wide analysis and functional studies are required to ascertain the relationship between Y-linked variation, accessory gland expression and sex ratio distortion. Our expectation is that ongoing natural selection might be currently shaping the dynamics of Y-linked resistance in D. simulans.
Figure 4.
Accessory gland-specific genes were upregulated in the sensitive Y chromosome males. (a) Average fold change in the expression of the 37 accessory gland-specific genes that were differentially expressed across lines (P<0.01). Darker shades denote lower expression level; lighter shades denote higher expression level (Supplementary Figure S3). (b) Average relative expression level of these 37 genes (P<0.01, dark gray), as compared with that of the other 39 accessory gland-specific genes that were stably expressed across genotypes (0.05<P<0.95, light gray). Error bars denote >99% confidence intervals for the average fold differences. (c) Three serine protease inhibitors were upregulated in the sensitive Y chromosome males: serine protease inhibitor 7 (Spn7, black bars), serine protease inhibitor 2 (Spn2, dark gray) and serine protease inhibitor 3 (Spn3, light gray).
Gene expression in XXY female is modulated by the Y chromosome
Natural variants of the D. simulans Y chromosome modulate gene expression and sex-ratio in males. What are the polymorphic Y-linked sequences that account for this polymorphism? Because Drosophila Y-linked protein-coding genes show little to no variation (Zurovcova and Eanes, 1999), it appears unlikely that these proteins might mediate the resistance to Dox. On the other hand, the Y chromosome is mainly composed of non-coding repetitive DNA sequences, and these ‘junk' sequences might have an active role in gene expression regulation (Lemos et al., 2008, 2010). To address the issue, we compared genome-wide gene expression levels between XXY female genotypes that are isogenic except for the Y chromosome. Y-linked proteins are known to have no expression levels in female (Ashburner et al., 2005; Lemos et al., 2010). We contrasted a single resistant Y chromosome with a single sensitive Y chromosome for their effects on XXY female genotypes. We found 683 genes that were differentially expressed between XXY(resistant) and XXY(sensitive) females (BPP>0.99; FDR<0.10), 94 of which were also differentially expressed in males. This is expected in view of the large proportion of testis-specific genes in the male contrast. The 333 genes upregulated in XXY(resistant) show no functional clustering at P<0.01. On the other hand, the 350 genes downregulated in XXY(resistant) show a strong association to cell cycle (50 genes, P<10−7). In terms of gene ontology categories, these genes are enriched for chromosome organization (GO:0051276; 29 genes, P=0.003), DNA replication (GO:0006260; 12 genes, P=0.012) and chromosome segregation (GO:0007059; 14 genes, P=0.034). Examples of these genes include important chromatin components, such as ORC2, ORC5 and ORC6. Similar observations were made before in D. melanogaster (Lemos et al., 2010), which, differ in one important feature from D. simulans in that the latter does not have functional rDNA repeats on its Y chromosome. Hence, neither protein-coding genes nor the rRNA genes appear to be the likely candidates for explaining the Y-linked regulatory variation observed in XXY female genotypes. On the other hand, it is possible that the expression of Y-linked protein-coding genes in males might contribute to the modulation of sex-ratio during spermatogenesis. Finally, non-coding sequences, including the rDNA untranscribed spacer repeats, might be able to provide a previously underappreciated source of regulatory functions of a myriad of organismal traits, including sex-ratio.
Conclusions
Here we have reported abundant Y-linked regulatory variation in D. simulans, some of which might be associated with the Y chromosome's ability to regulate the Winters sex-ratio system. In males with the resistant Y chromosomes, over 200 testis-specific genes were upregulated relative to their expression levels in males with sensitive Y chromosomes. These genes are over-represented by those expressed during the post-meiotic or spermiogenic stage of sperm development, suggesting that the Y-linked regulatory functions are targeting developing Y-bearing spermatid. Further insights into the mechanisms modulating sex-ratio are hindered by the poor functional annotations of testis-specific genes. Our findings nevertheless point to a link between the etiology of sex-ratio and post-meiotic genes and mitochondria. Indeed, mitochondria-associated genes were downregulated in strains with the resistant Y chromosome. These observations concur with recent observations that mitochondrial variants differently regulate testis gene expression in D. melanogaster (Innocenti et al., 2011). In one similar but more extreme case, the mitochondrial type of Brownsville introgressed into the w1118 nuclear background can drastically reduce male fertility (Clancy, 2008). These parallel observations of organismal traits and gene modulations conferred by the Y chromosome and mitochondria might point to a regulatory network of spermatogenesis, including both parties and other nuclear genes. We expect that mitochondria–X chromosome interactions (Rand et al., 2001; Montooth et al., 2010) and mitochondria–Y chromosome interactions may turn out to be critically important in spermatogenesis. Finally, genes specific to the accessory glands were downregulated in males with resistant Y chromosomes, suggesting that this particular tissue might provide protection for spermatogenesis in a stressful environment (that is, sex ratio distortion). The Y chromosome in D. simulans also modulates the expression of hundreds of genes that are broadly expressed across various tissues, but the link between these genes and sex-ratio regulation is elusive. Lastly, the exact identities of the Y-linked genetic elements that regulate sex ratio distortion remain to be discovered.
Data archiving
Expression data deposited in the NCBI's GEO database: GSE43665.
Acknowledgments
YT is supported by NIH R01 HD060679; DLH is supported by NIH R01 GM065169 and GM079536.
The authors declare no conflict of interest
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Araripe LO, Montenegro H, Lemos B, Hartl DL. Fine-scale genetic mapping of a hybrid sterility factor between Drosophila simulans and D. mauritiana: the varied and elusive functions of ‘speciation genes'. BMC Evol Biol. 2010;10:385. doi: 10.1186/1471-2148-10-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA; 2005. [Google Scholar]
- Atlan A, Joly D, Capillon C, Montchamp-Moreau C. Sex-ratio distorter of Drosophila simulans reduces male productivity and sperm competition ability. J Evol Biol. 2004;17:744–751. doi: 10.1111/j.1420-9101.2004.00737.x. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Hom E, Wong KM, Maside X, de Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S, Lohe A. Fine mapping of satellite DNA sequences along the Y chromosome of Drosophila melanogaster: relationships between satellite sequences and fertility factors. Genetics. 1991;129:177–189. doi: 10.1093/genetics/129.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosseau GE. Genetic analysis of the male fertility factors on the Y chromosome of Drosophila melanogaster. Genetics. 1960;45:257–274. doi: 10.1093/genetics/45.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ. Evolution of Sex Determining Mechanisms. Benjamin/Cummings Publishing Company: Menlo Park, CA, USA; 1983. [Google Scholar]
- Carvalho AB. Origin and evolution of the Drosophila Y chromosome. Curr Opin Genet Dev. 2002;12:664–668. doi: 10.1016/s0959-437x(02)00356-8. [DOI] [PubMed] [Google Scholar]
- Carvalho AB, Clark AG. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science. 2005;307:108–110. doi: 10.1126/science.1101675. [DOI] [PubMed] [Google Scholar]
- Carvalho AB, Koerich LB, Clark AG. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genetics. 2009;25:270–277. doi: 10.1016/j.tig.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Vaz SC, Klaczko LB. Polymorphism for Y-linked suppressors of sex-ratio in two natural populations of Drosophila mediopunctata. Genetics. 1997;146:891–902. doi: 10.1093/genetics/146.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazemajor M, Joly D, Montchamp-Moreau C. Sex-ratio meiotic drive in Drosophila simulans is related to equational nondisjunction of the Y chromosome. Genetics. 2000;154:229–236. doi: 10.1093/genetics/154.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T. Seminal fluid-mediated fitness traits in Drosophila. Heredity (Edinb) 2001;87:511–521. doi: 10.1046/j.1365-2540.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Rice WR. Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:5677–5682. doi: 10.1073/pnas.101456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ. Variation in mitochondrial genotype has substantial lifespan effects which may be modulated by nuclear background. Aging Cell. 2008;7:795–804. doi: 10.1111/j.1474-9726.2008.00428.x. [DOI] [PubMed] [Google Scholar]
- Contrino S, Smith RN, Butano D, Carr A, Hu F, Lyne R, et al. modMine: flexible access to modENCODE data. Nucleic Acids Res. 2012;40:D1082–D1088. doi: 10.1093/nar/gkr921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosmides LM, Tooby J. Cytoplasmic inheritance and intragenomic conflict. J Theor Biol. 1981;89:83–129. doi: 10.1016/0022-5193(81)90181-8. [DOI] [PubMed] [Google Scholar]
- Curtsinger JW, Feldman MW. Experimental and theoretical analysis of the ‘sex-ratio' polymorphism in Drosophila pseudoobscura. Genetics. 1980;94:445–466. doi: 10.1093/genetics/94.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca SZ, O'Farrell PH. Barriers to male transmission of mitochondrial DNA in sperm development. Dev Cell. 2012;22:660–668. doi: 10.1016/j.devcel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DK, Simmons LW. Ejaculate economics: testing the effects of male sexual history on the trade-off between sperm and immune function in Australian crickets. PLoS One. 2012;7:e30172. doi: 10.1371/journal.pone.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Waddle JA, Hale W, IV, Dave V, Thornton J, Macatee TL, et al. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M.1993Spermatogenesis The Development of Drosophila melanogasterMartinez-Arias A, Bate M (edsCold Spring Harbor Laboratory Press: New York, NY, USA; 71–147. [Google Scholar]
- Gallach M, Chandrasekaran C, Betran E. Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol Evol. 2010;2:835–850. doi: 10.1093/gbe/evq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstein JH, Hochstenbach R, Hauschteck-Jungen E, Beukeboom LW. Is the Y chromosome of Drosophila an evolved supernumerary chromosome. Bioessays. 1996;18:317–323. doi: 10.1002/bies.950180410. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. Extraordinary sex ratios. A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science. 1967;156:477–488. doi: 10.1126/science.156.3774.477. [DOI] [PubMed] [Google Scholar]
- Heddi A, Stepien G, Benke PJ, Wallace DC. Coordinate induction of energy gene expression in tissues of mitochondrial disease patients. J Biol Chem. 1999;274:22968–22976. doi: 10.1074/jbc.274.33.22968. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Huttunen S, Aspi J. Complex inheritance of male courtship song characters in Drosophila virilis. Behav Genet. 2003;33:17–24. doi: 10.1023/a:1021095331850. [DOI] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH, Dowling DK. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science. 2011;332:845–848. doi: 10.1126/science.1201157. [DOI] [PubMed] [Google Scholar]
- Jaenike J. Suppression of sex-ratio meiotic drive and the maintenance of Y-chromosome polymorphism in Drosophila. Evolution. 1999;53:164–174. doi: 10.1111/j.1558-5646.1999.tb05342.x. [DOI] [PubMed] [Google Scholar]
- Janssen S, Cuvier O, Muller M, Laemmli UK. Specific gain- and loss-of-function phenotypes induced by satellite-specific DNA-binding drugs fed to Drosophila melanogaster. Mol Cell. 2000;6:1013–1024. doi: 10.1016/s1097-2765(00)00100-3. [DOI] [PubMed] [Google Scholar]
- Jiang PP, Hartl DL, Lemos B. Y not a dead end: epistatic interactions between Y-linked regulatory polymorphisms and genetic background affect global gene expression in Drosophila melanogaster. Genetics. 2010;186:109–118. doi: 10.1534/genetics.110.118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Zhou Q, Bachtrog D. Nonrandom gene loss from the Drosophila miranda neo-Y chromosome. Genome Biol Evol. 2011;3:1329–1337. doi: 10.1093/gbe/evr103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingan SB, Garrigan D, Hartl DL. Recurrent selection on the Winters sex-ratio genes in Drosophila simulans. Genetics. 2010;184:253–265. doi: 10.1534/genetics.109.109587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerich LB, Wang X, Clark AG, Carvalho AB. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456:949–951. doi: 10.1038/nature07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsticevic FJ, Santos HL, Januario S, Schrago CG, Carvalho AB. Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster. Genetics. 2010;184:295–307. doi: 10.1534/genetics.109.107516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci USA. 2010;107:15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linklater JR, Wertheim B, Wigby S, Chapman T. Ejaculate depletion patterns evolve in response to experimental manipulation of sex ratio in Drosophila melanogaster. Evolution. 2007;61:2027–2034. doi: 10.1111/j.1558-5646.2007.00157.x. [DOI] [PubMed] [Google Scholar]
- Lohe AR, Roberts PA. An unusual Y chromosome of Drosophila simulans carrying amplified rDNA spacer without rRNA genes. Genetics. 1990;125:399–406. doi: 10.1093/genetics/125.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee BD, Habera L, Vrana JA. Evidence that intergenic spacer repeats of Drosophila melanogaster rRNA genes function as X-Y pairing sites in male meiosis, and a general model for achiasmatic pairing. Genetics. 1992;132:529–544. doi: 10.1093/genetics/132.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P, St, Pierre SE, Thurmond J. FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Tao Y. Genetic conflict and sex chromosome evolution. Trends Ecol Evol. 2010;25:215–223. doi: 10.1016/j.tree.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montchamp-Moreau C. Sex-ratio meiotic drive in Drosophila simulans: cellular mechanism, candidate genes and evolution. Biochem Soc Trans. 2006;34:562–565. doi: 10.1042/BST0340562. [DOI] [PubMed] [Google Scholar]
- Montchamp-Moreau C, Ginhoux V, Atlan A. The Y chromosomes of Drosophila simulans are highly polymorphic for their ability to suppress sex-ratio drive. Evolution. 2001;55:728–737. doi: 10.1554/0014-3820(2001)055[0728:tycods]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Montchamp-Moreau C, Joly D. Abnormal spermiogenesis is associated with the X-linked sex-ratio trait in Drosophila simulans. Heredity. 1997;79:24–30. [Google Scholar]
- Montooth KL, Meiklejohn CD, Abt DN, Rand DM. Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution. 2010;64:3364–3379. doi: 10.1111/j.1558-5646.2010.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Koizumi M, Hayashi S. Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr Biol. 2011;21:805–814. doi: 10.1016/j.cub.2011.04.016. [DOI] [PubMed] [Google Scholar]
- Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: ‘rDNA-sensitive' genes and natural variation. PLoS Genet. 2011;7:e1001376. doi: 10.1371/journal.pgen.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Wolfner MF. Male and female cooperate in the prohormone-like processing of a Drosophila melanogaster seminal fluid protein. Dev Biol. 1995;171:694–702. doi: 10.1006/dbio.1995.1315. [DOI] [PubMed] [Google Scholar]
- Platero JS, Csink AK, Quintanilla A, Henikoff S. Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J Cell Biol. 1998;140:1297–1306. doi: 10.1083/jcb.140.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TA, Hodgson DJ, Lewis Z, Hurst GD, Wedell N. Selfish genetic elements promote polyandry in a fly. Science. 2008;322:1241–1243. doi: 10.1126/science.1163766. [DOI] [PubMed] [Google Scholar]
- Ramamurthy G, Alfert M, Stern C. Ultrastructural studies on spermatogenesis in a sex-ratio-mutant strain of Drosophila simulans. Am J Anat. 1980;157:205–219. doi: 10.1002/aja.1001570208. [DOI] [PubMed] [Google Scholar]
- Rand DM, Clark AG, Kann LM. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics. 2001;159:173–187. doi: 10.1093/genetics/159.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Eisenhour L, Hong C, Lee Y, McKee BD. Roles of rDNA spacer and transcription unit-sequences in X-Y meiotic chromosome pairing in Drosophila melanogaster males. Chromosoma. 1997;106:29–36. doi: 10.1007/s004120050221. [DOI] [PubMed] [Google Scholar]
- Rice WR. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics. 1987;116:161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer C, David JR, Moreteau B, Joly D. Heat induced male sterility in Drosophila melanogaster: adaptive genetic variations among geographic populations and role of the Y chromosome. J Exp Biol. 2004;207:2735–2743. doi: 10.1242/jeb.01087. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Montenegro H, Hartl DL, Lemos B. Interspecific Y chromosome introgressions disrupt testis-specific gene expression and male reproductive phenotypes in Drosophila. Proc Natl Acad Sci USA. 2011;108:17046–17051. doi: 10.1073/pnas.1114690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer: New York, NY, USA; 2005. [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Stoltenberg SF, Hirsch J. Y-chromosome effects on Drosophila geotaxis interact with genetic or cytoplasmic background. Anim Behav. 1997;53:853–864. doi: 10.1006/anbe.1996.0351. [DOI] [PubMed] [Google Scholar]
- Tao Y, Araripe L, Kingan SB, Ke Y, Xiao H, Hartl DL. A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol. 2007a;5:e293. doi: 10.1371/journal.pbio.0050293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Masly JP, Araripe L, Ke Y, Hartl DL. A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biol. 2007b;5:e292. doi: 10.1371/journal.pbio.0050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 2002;3:RESEARCH0071. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long M. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009;5:e1000731. doi: 10.1371/journal.pgen.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI. Virility deficiency and the sex-ratio trait in Drosophila pseudoobscura. I. Sperm displacement and sexual selection. Genetics. 1983a;105:651–662. doi: 10.1093/genetics/105.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI. Virility deficiency and the sex-ratio trait in Drosophila pseudoobscura. II. Multiple mating and overall virility selection. Genetics. 1983b;105:663–679. doi: 10.1093/genetics/105.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurovcova M, Eanes WF. Lack of nucleotide polymorphism in the Y-linked sperm flagellar dynein gene Dhc-Yh3 of Drosophila melanogaster and D. simulans. Genetics. 1999;153:1709–1715. doi: 10.1093/genetics/153.4.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.