Abstract
Glacial and postglacial processes are known to be important determinants of contemporary population structuring for many species. In Europe, refugia in the Italian, Balkan and Iberian peninsulas are believed to be the main sources of species colonising northern Europe after the glacial retreat; however, there is increasing evidence of small, cryptic refugia existing north of these for many cold-tolerant species. This study examined the glacial history of Atlantic salmon in western Europe using two independent classes of molecular markers, microsatellites (nuclear) and mitochondrial DNA variation. Alongside the well-documented refuge in the Iberian Peninsula, evidence for a cryptic refuge in northwest France is also presented. Critically, methods utilised to estimate divergence times between the refugia indicated that salmon in these two regions had diverged a long time before the last glacial maximum; coalescence analysis (as implemented in the program IMa2) estimated divergence times at around 60 000 years before present. Through the examination of haplotype frequencies, previously glaciated areas of northwest Europe, that is, Britain and Ireland, appear to have been colonised from salmon expanding out of both refugia, with the southwest of England being the primary contact zone and exhibiting the highest genetic diversity.
Keywords: phylogeography, refugia, microsatellite, haplotype, ND1 gene
Introduction
Genetic differentiation within a species is governed by both contemporary and historical processes. The Pleistocene glaciations, which lasted approximately two million years and terminated around 10 000 years ago with a period of global warming (Andersen and Borns, 1994), are deemed to be one of the most important historical events involved in shaping the large-scale population structuring observed in many species today (Bernatchez and Wilson, 1998; Taberlet et al., 1998). During the major glaciations of the Pleistocene, polar ice sheets expanded considerably, extirpating many populations and compressing species' ranges to their southerly limits, thereby restricting populations to refugial locations (Hewitt, 1999). Following the onset of glacial retreat, populations of species at the northern edge of their range would have expanded into new areas of suitable habitat (Hewitt, 1999). Such rapid expansion from a few founder individuals can lead to a reduction in genetic diversity at the leading edge of recolonisation (Hewitt, 1996); however, a secondary increase in the diversity of resulting hybrid populations has also been observed as recolonisers from different refugia come into contact (Petit et al., 2003). Owing to the genetic differentiation that occurred between refugial populations (due to drift and differential selection), it is possible, through phylogeographic exploration, to trace the movement of species from refugial areas into the previously glaciated regions, which now constitute their native range (for example, Makhrov and Bolotov, 2006).
For most European species, ancestry from the Pleistocene period can be traced back to one or more of the three main refugia in the Iberian, Italian or Balkan Peninsulas (Taberlet et al., 1998; Hewitt, 1999). More recently, however, several studies (Provan et al., 2005; Hoarau et al., 2007; Olsen et al., 2010; Panova et al., 2011) have identified the area comprising the Brittany peninsula in northwest France, the Channel River and Hurd Deep (Lericolais et al., 2003), the undersea relic of the Channel River located just off the northwest coast of Brittany, as a glacial refuge for marine/coastal organisms within northwest continental Europe. At this time, the English Channel was largely dry land with numerous rivers, including the well-documented Channel River, running through the area (Lericolais et al., 2003); modelling studies by Sarnthein (2001) suggest that sea temperatures in the region of the English Channel at this time were 1–2 °C in winter, rising to 5–6 °C in summer. In the case of Atlantic salmon, their current range extends only to the historical Iberian Peninsula refuge (MacCrimmon and Gots, 1979), which is widely accepted as a glacial refuge for Atlantic salmon during the Pleistocene glaciations (Consuegra et al., 2002). However, there is no evidence for their extension into the Italian or Balkan regions at that time and, until now, no evidence of a refuge for this species in northwest France.
Within Europe, Atlantic salmon appear to have differentiated into Atlantic and Baltic sublineages before the last glacial period (Verspoor et al., 1999; Nilsson et al., 2001); hence, it is expected that the two lineages would have differing phylogeographic histories. Extensive phylogeographic exploration in the Baltic region and around Russia to the White and Barents Seas has proved successful in elucidating likely glacial refugia for Atlantic salmon in freshwater lakes present in the Baltic region and their subsequent colonisation routes across the region (Nilsson et al., 2001; Asplund et al., 2004; Säisä et al., 2005; Tonteri et al., 2005). In contrast, the phylogeographic history of Atlantic salmon in northwest Europe (northern Spain, France, Britain and Ireland) remains speculative (Verspoor et al., 1999; Nilsson et al., 2001). Hence, the aim of this study was to elucidate the phylogeographic history of Atlantic salmon in this region.
With the exception of the recently published population analysis of southern European populations (Griffiths et al., 2010), in most Europe-wide evolutionary studies of Atlantic salmon conducted to date, samples of wild populations from the west and northwest, that is, Spain, France, Britain and Ireland, have been largely lacking. This is particularly marked for France and England—a maximum of five samples from across the region were included in studies by Bourke et al. (1997) and Verspoor et al. (1999). In the only studies to focus directly on this region, Payne et al. (1971) and Child et al. (1976) described two lineages based on polymorphisms at the transferrin locus. The so-called ‘Boreal' lineage radiated from a glacial lake in the southern North Sea and was proposed to occupy most of northern Europe, including the Baltic, while the ‘Celtic' race was confined to the southwest of England (UKSouthwest). Much has been achieved in other areas of Europe since these early studies. For example, although no evidence for a Boreal–Celtic divide has since been observed (Ståhl, 1987; Bourke et al., 1997; Verspoor et al., 1999; Consuegra et al., 2002), due to the high genetic diversity consistently reported for Britain and Ireland (Verspoor et al., 1999; Nilsson et al., 2001; Consuegra et al., 2002), this region has been proposed as a contact zone for salmon expanding out of multiple refugia. Sound evidence of a refuge for Atlantic salmon in this region comes in the form of bone relics recovered from a cave in the Iberian Peninsula, which demonstrates the presence of Atlantic salmon in this area for at least the past 40 000 years, that is, during the last glacial period (Consuegra et al., 2002). As proposed for other freshwater fish (Hanfling et al., 2002), it is possible that Atlantic salmon existed north of this refuge, in northwest France or southern England (UKSouth) (Verspoor, 1986), in line with Payne et al.'s (1971) ‘Celtic' hypothesis. In addition, a glacial lake present in the southern North Sea has been proposed as a potential refuge for Atlantic salmon colonising west into the Atlantic and North Sea drainages, east into the Baltic (Verspoor et al., 1999; Säisä et al., 2005) and north into the White and Barents Seas (Verspoor et al., 1999; Asplund et al., 2004; Tonteri et al., 2005), in line with Payne et al.'s (1971) ‘Boreal' hypothesis. However, the allozyme studies of Verspoor (1986), Ståhl (1987) and Bourke et al. (1997) have been brought into question by the revelation that a commonly used isoenzyme (ME-2, mMEP-2*) is likely to be acting under selection pressures (Verspoor and Jordan, 1989; Bourke et al., 1997) and as such is not a suitable marker for phylogeographic studies. This may also be the case for transferrin (Verspoor and Jordan, 1989) as used by Payne et al. (1971) and Child et al. (1976) in the initial investigations.
In recent years, the use of multiple classes of molecular markers has proved highly effective in elucidating phylogeographic histories of freshwater fish (Hanfling et al., 2002; Koskinen et al., 2002). Specifically for Atlantic salmon, the differing yet complementary properties of microsatellite and mitochondrial DNA (mtDNA) have led to robust inferences of phylogeographic histories in the Baltic Seas and northern Europe (Säisä et al., 2005; Tonteri et al., 2005). Our aim, therefore, was to utilise these markers to resolve the phylogeographic history of Atlantic salmon in northwest Europe, by identifying refugial locations and determining colonisation pathways. Specifically, the null hypothesis (H0) to be tested was that salmon colonising northwest Europe expanded out of a single refuge located in the Iberian Peninsula, with alternate hypotheses of salmon residing in additional glacial refugia located north of the Iberian peninsula (in northwest France and/or UKSouth; H1A) and/or in the southern North Sea (H1B). Subsequently, we aim to reconstruct the colonisation pathways of Atlantic salmon into Britain and Ireland from these potential source location(s).
Materials and Methods
Sample collection
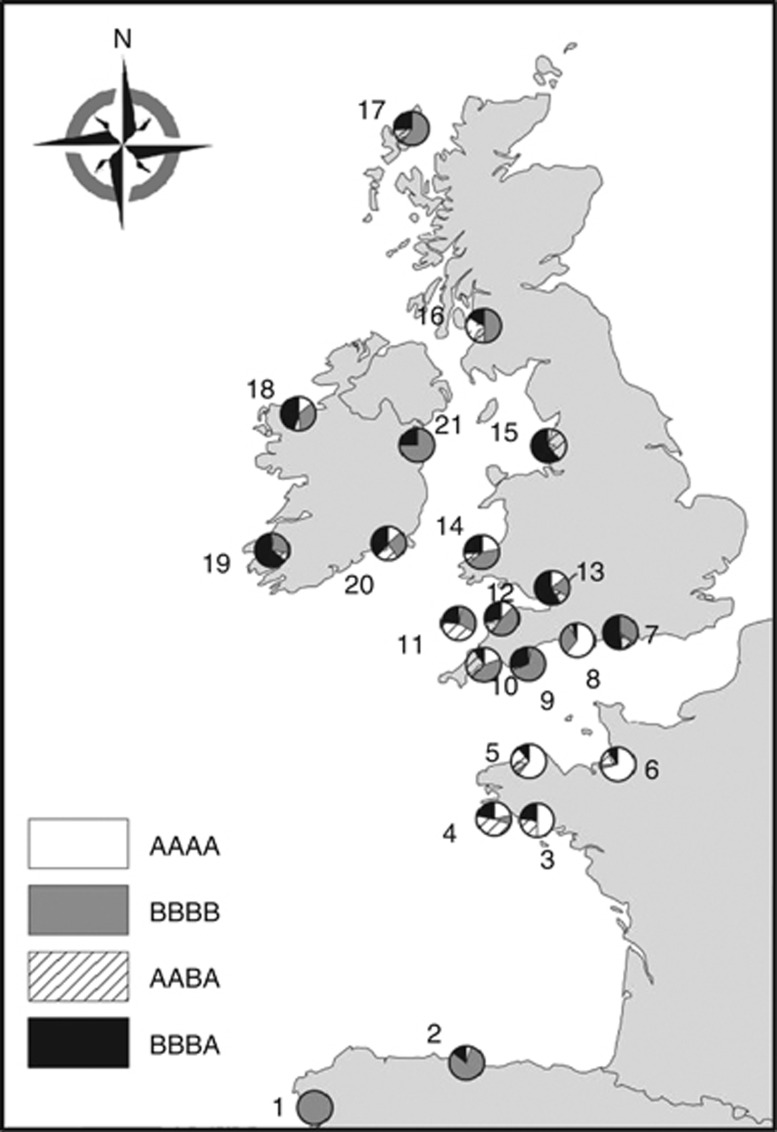
In this study, 723 specimens of Atlantic salmon were collected from 21 rivers that drain into the east coast of the Atlantic Ocean, including the English Channel, the Irish Sea and the Bay of Biscay (Figure 1 and Table 1). The majority of sampling was carried out in 2004 and 2005 during routine juvenile salmon abundance surveys and targeted 1+ parr. The exceptions are the French specimens, which were sampled as scales from rod-caught adult salmon in 2005.
Figure 1.
Sampling locations and ND1 haplotype frequencies. Population ID follows Table 1; composite haplotypes derived from the restriction enzymes HaeIII, HinfI, AvaII and RsaI.
Table 1. Genetic diversity indices for microsatellite data, mtDNA RFLP data and mtDNA RFLP haplotype frequencies.
| River | ID | N |
Microsatellite diversity
|
mtDNA RFLP diversity
|
mtDNA RFLP haplotypes
|
Regional group | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ar | Ho | He | Haplotype diversity | Nucleotide diversity | AAAA | AABA | BBBB | BBBA | ||||
| Ulla | 1 | 40 | 8.43 | 0.70 | 0.72 | 0.000 | 0.00000 | 0.00 | 0.00 | 1.00 | 0.00 | Spain/Spain |
| Sella | 2 | 41 | 11.87 | 0.81 | 0.81 | 0.337 | 0.00883 | 0.05 | 0.00 | 0.80 | 0.15 | Spain/Spain |
| Scorff | 3 | 30 | 11.26 | 0.80 | 0.79 | 0.646 | 0.01774 | 0.50 | 0.27 | 0.00 | 0.23 | France/France |
| Ellé | 4 | 46 | 11.64 | 0.82 | 0.78 | 0.684 | 0.01949 | 0.22 | 0.48 | 0.09 | 0.22 | France/France |
| Léguer | 5 | 28 | 12.62 | 0.81 | 0.79 | 0.577 | 0.01531 | 0.61 | 0.25 | 0.04 | 0.11 | France/France |
| Sée | 6 | 31 | 11.12 | 0.81 | 0.80 | 0.475 | 0.01368 | 0.71 | 0.16 | 0.03 | 0.10 | France/France |
| Itchen | 7 | 29 | 7.79 | 0.74 | 0.75 | 0.616 | 0.01336 | 0.00 | 0.14 | 0.34 | 0.52 | UKSouth/UK |
| Avon | 8 | 23 | 7.00 | 0.73 | 0.81 | 0.557 | 0.02841 | 0.61 | 0.04 | 0.30 | 0.04 | UKSouth/France |
| Dart | 9 | 38 | 10.96 | 0.83 | 0.88 | 0.495 | 0.01016 | 0.03 | 0.03 | 0.66 | 0.29 | UKSw/Spain |
| Fowey | 10 | 36 | 11.48 | 0.81 | 0.82 | 0.708 | 0.02697 | 0.19 | 0.31 | 0.42 | 0.08 | UKSw/UK |
| Camel | 11 | 30 | 12.68 | 0.83 | 0.84 | 0.690 | 0.02170 | 0.03 | 0.43 | 0.30 | 0.23 | UKSw/UK |
| Taw | 12 | 32 | 12.57 | 0.84 | 0.87 | 0.692 | 0.02186 | 0.13 | 0.13 | 0.47 | 0.28 | UKSw/UK |
| Usk | 13 | 30 | 12.58 | 0.84 | 0.86 | 0.632 | 0.01869 | 0.13 | 0.10 | 0.20 | 0.57 | UKSw/UK |
| Teifi | 14 | 27 | 13.38 | 0.84 | 0.85 | 0.732 | 0.02600 | 0.22 | 0.11 | 0.41 | 0.26 | UKSw/UK |
| Ribble | 15 | 25 | 11.29 | 0.82 | 0.84 | 0.530 | 0.01498 | 0.04 | 0.36 | 0.00 | 0.60 | UKNw/UK |
| Ayr | 16 | 32 | 11.36 | 0.83 | 0.83 | 0.627 | 0.02054 | 0.00 | 0.34 | 0.50 | 0.16 | UKNw/UK |
| Creed | 17 | 35 | 11.90 | 0.83 | 0.83 | 0.570 | 0.01402 | 0.00 | 0.14 | 0.60 | 0.26 | UKNw/UK |
| Moy | 18 | 29 | 11.74 | 0.82 | 0.85 | 0.680 | 0.02004 | 0.14 | 0.07 | 0.34 | 0.45 | UKNw/UK |
| Laune | 19 | 47 | 12.11 | 0.83 | 0.82 | 0.531 | 0.01144 | 0.02 | 0.09 | 0.26 | 0.64 | UKNw/UK |
| Barrow | 20 | 37 | 13.33 | 0.86 | 0.86 | 0.739 | 0.02261 | 0.14 | 0.22 | 0.27 | 0.38 | UKNw/UK |
| Boyne | 21 | 36 | 11.57 | 0.82 | 0.80 | 0.386 | 0.00526 | 0.00 | 0.00 | 0.75 | 0.25 | UKNw/Spain |
Abbreviations: Ar, allelic richness; He, expected heterozygosity; Ho, observed heterozygosity; ID, population ID for Figure 1; mtDNA, mitochondrial DNA; N, number of salmon; Nw, northwest; RFLP, restriction fragment length polymorphism; Sw, south west.
mtDNA RFLP composite haplotypes were derived from restriction enzymes AvaII, HaeIII, HinfI and RsaI. Two regional assignments are listed for each population: the first regional assignment classifies a population according to the microsatellite analysis, and the second is based on the mtDNA analysis.
DNA analysis: microsatellites
Genomic DNA was extracted from scales or fin clips of Atlantic salmon according to a Chelex resin protocol (Estoup et al., 1996). Genotypes were determined at 12 microsatellite loci: Ssa157a, SsaD144b (King et al., 2005), Ssa197, Ssa202, Ssa171 (O'Reilly et al., 1996), SSsp2210, SSsp1605, SSsp2201, SSspG7 (Paterson et al., 2004), Ssa289 (McConnell et al., 1995) and SSOSL417, SSOSL85 (Slettan et al., 1996).
Microsatellite analysis followed the protocol of Griffiths et al. (2010). Polymerase chain reactions (PCRs) were carried out in 10 μl reaction volumes and standard PCR reagents were used in a mixture containing 50–100 ng DNA, 0.5 μℳ of each primer, 1.5 mℳ MgCl2, 200 μℳ of each dNTP, 1 × reaction buffer and 0.5 U of Taq DNA polymerase (Bioline, London, UK). The PCR amplification profile consisted of a single denaturing step lasting 3 min at 94 °C, followed by 30 iterations of: 94 °C for 30 s, an annealing step of 30 s at 53–58 °C, depending on the locus being amplified (see Supplementary Information and Supplementary Table 1), and 72 °C for 30 s, with a single final elongation step of 72 °C for 10 min. Primer annealing temperatures for each locus are given in the Supplementary Information and Supplementary Table 1; see Finnegan and Stevens (2008) for additional details.
Size determination of the labelled PCR products was performed using a Beckman-Coulter CEQ 8000 automated DNA sequencer with an internal size standard, according to the manufacturer's instructions. The raw data were analysed with the platform's associated fragment analysis software (Beckman-Coulter, High Wycombe, UK).
DNA analysis: mtDNA RFLPs
The NADH dehydrogenase subunit I (ND1) region of mtDNA was amplified using the primers of Hall and Nawrocki (1995), as modified by Nilsson et al. (2001). The 30 μl reaction volume contained 0.25 U of Taq DNA polymerase (Bioline), 1 × reaction buffer, 0.2 mℳ dNTP, 2 mℳ MgCl2 and 0.5 μℳ of each primer and approximately 50 ng of purified template DNA. PCR amplification consisted of an initial denaturation step of 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 58 °C for 45 s and 72 °C for 60 s, concluding with a final extension phase of 5 min at 72 °C.
Amplified DNA was digested with the restriction enzymes HaeIII, HinfI, AvaII and RsaI in individual 10 μl reactions; all enzymes were supplied by Promega at a concentration of 10 U μl−1. Individual reactions comprised 0.3 μl enzyme, 0.5 μl corresponding buffer mix, 3.2 μl dH2O and 6 μl of amplified DNA, and were incubated at 37 °C overnight. Fragments were separated by electrophoresis and variant fragment patterns, that is, composite haplotypes, were determined following the original ND1 haplotype designation system of Nielsen et al. (1996), and see also Nilsson et al. (2001).
DNA analysis: mtDNA ND1 sequencing
On the basis of the haplotypes identified by mtDNA restriction fragment length polymorphism (RFLP) analysis (see above), a subset of samples that included representatives of all haplotypes identified was subjected to additional ND1 sequencing. This analysis identified additional within-haplotype variation, visualised using a haplotype median-joining network constructed using the program 𝒩ℰ𝒯𝒲𝒪ℛ𝒦 v.4.6.1.1 (available at: www.fluxus-engineering.com/sharenet.htm; Bandelt et al., 1999); in turn, this allowed us to use the software IMa2 (Hey, 2010) to investigate divergence times between haplotype groups.
Primers ND1-F and ND1-R (Nilssen et al., 2001) were used to amplify a 1410 bp region of the mtDNA that includes the ND1 gene. PCR reactions were carried out in a total volume of 25 μl consisting of 1 × HotStar Taq Plus Master Mix (Qiagen, Manchester, UK), 0.2 μℳ of each forward and reverse primer and approximately 50 ng of purified template DNA. After an initial denaturing step at 94 °C for 2 min 30 s, amplification proceeded for 35 cycles at 94 °C for 30 s, 58 °C for 30 s, 72 °C for 60 s and a final extension at 72 °C for 10 min.
PCR products (10 μl) were purified using 0.25 U each of Exonuclease I (New England Biolabs, Hitchin, UK) and Antarctic Phosphotase (USB) incubated at 37 °C for 45 min and 80 °C for 15 min. Sequencing was carried out on a 3130xl Genetic Analyzer (Applied Biosystems, Paisley, UK) using Big Dye (v.1.1) sequencing chemistry.
Microsatellite data analyses
Allele number and allelic richness (Ar) (allele number corrected for sample size) were calculated for all loci within populations using FSTAT v.2.9.3 (Goudet, 1995). Deviations from Hardy–Weinberg expectations and the occurrence of linkage disequilibrium between loci was tested for using the default parameters in 𝒢ℰ𝒩ℰ𝒫𝒪𝒫 v.3.4 (Raymond and Rousset, 1995) with sequential Bonferroni corrections applied for multiple tests across populations (Rice, 1989). To identify null alleles and mis-scoring, all data were run through the program Mℐ𝒞ℛ𝒪-Cℋℰ𝒞𝒦ℰℛ (Van Oosterhout et al., 2004).
FSTAT v.2.9.3 was used to calculate FST values between populations and their significance. Genetic distances between populations were estimated according to Nei's (1987) DA distance, from which a phylogenetic tree was constructed using the neighbour-joining method as implemented in 𝒫𝒪𝒲ℰℛℳ𝒜ℛ𝒦ℰℛ v.3.25 (Liu and Muse, 2005). Strength of support for each node was assessed with 1000 bootstrap replicates and a consensus tree was obtained using the CONSENSE program in 𝒫ℋ𝒴ℒℐ𝒫 v.3.67 (Felsenstein, 1993).
The optimal number of regional groups was explored using STRUCTURE v.2.3.3 (Pritchard et al., 2000). A burn-in period of 30 000 steps was set, followed by 1 000 000 Markov chain Monte Carlo replicates; allele frequencies were set as ‘weakly correlated' as recommended by Pritchard et al. (2000). The number of groups to be simulated (K) was set as 1 to 10, with 10 replicates for each group. The optimal number of groups was estimated using the method of Evanno et al. (2005).
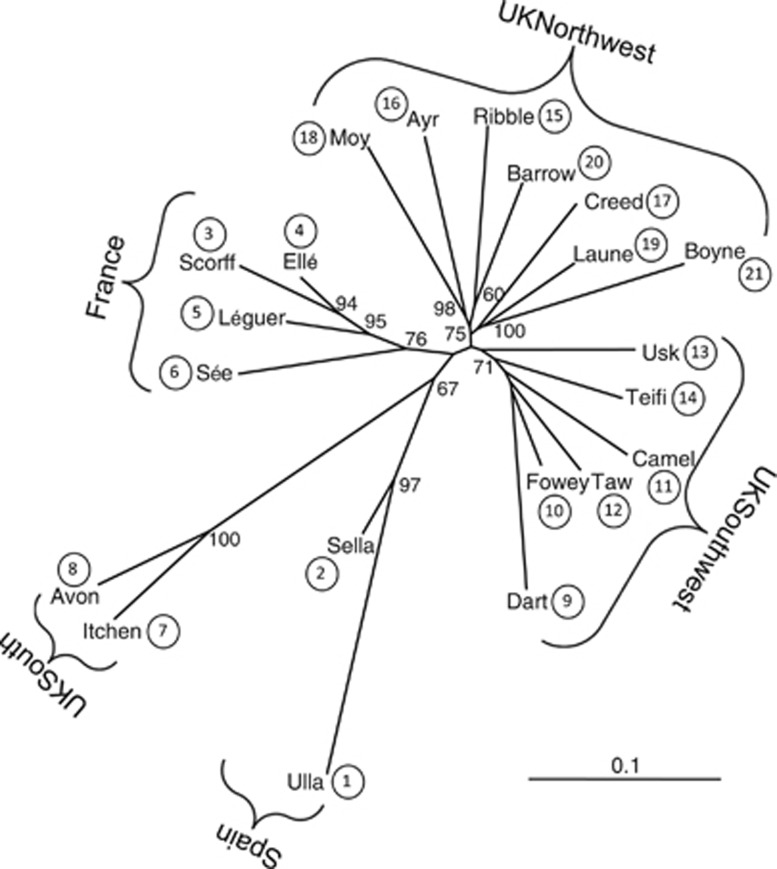
An analysis of molecular variance, as implemented in 𝒜ℛℒℰ𝒬𝒰ℐ𝒩 v.3.5 (Excoffier and Lischer, 2010), was used to examine how genetic variation was partitioned among different hierarchical levels in Atlantic salmon in northwest Europe. Three hierarchical levels were defined: (i) between regional groups; (ii) between populations within regional groups; and (iii) between individuals within populations. Regional groups were defined based on the outcome of the STRUCTURE analysis and the phylogenetic tree (Figure 2), which resolved five broad regions (Spain, France, UKSouth, UKSouthwest, northwest of England (UKNorthwest)/Ireland; see Results). Differences in the average values of Ar, observed heterozygosity (Ho) and expected heterozygosity (He) among these regions were also assessed statistically using FSTAT v.2.9.3.
Figure 2.
Neighbour-joining phylogenetic tree generated from microsatellite data. Bootstrap values >50% are shown.
It has previously been suggested that, taking into account the mutation rate of microsatellite loci and the generation time of salmon, stepwise mutations will only have had a significant effect on differentiation between populations if they diverged before the last glacial maximum (for full details see Tonteri et al., 2005). Therefore, an allele size permutation test (Hardy et al., 2003) implemented in SPAGeDi v.2.1 (Hardy and Vekemans, 2002) was used to assess if RST>FST for populations within and between the five regions identified using STRUCTURE. A significant result (RST>FST) would be indicative of colonisation from multiple sources (Hardy et al., 2003), in this study the French and Spanish refugia. Confidence intervals were calculated with 10 000 permutations of allele sizes among alleles within each locus. The locus Ssa171 was excluded from this analysis as both di- and tetranucleotide repeats are observed at this locus; therefore, it could not have mutated through a stepwise process alone.
PCR–RFLP mtDNA data analyses
The sizes of restriction fragments (cross-referenced with the RFLP scoring system of Nielsen et al., 1996) were used to generate a restriction site binary matrix of presence/absence of fragment bands for each enzyme, and a binary representation of the composite haplotypes of each individual using ℛℰ𝒜𝒫 v.4.0 (McElroy et al., 1992). These were used in subsequent programs in the ℛℰ𝒜𝒫 v.4.0 package to calculate haplotype and nucleotide diversity within populations and a matrix of pairwise distances between populations (Nei, 1987) based on population haplotype frequencies. Subsequently, an unweighted pair-group method using arithmetic averages phylogenetic tree was constructed in 𝒫ℋ𝒴ℒℐ𝒫 v.3.67. Mann–Whitney U-tests were used to statistically investigate the difference of haplotype and nucleotide diversity between the three broad geographic regions depicted in the phylogenetic tree (Spain, France and United Kingdom/Ireland; see Results). Hierarchical diversity was assessed with an analysis of molecular variance implemented in 𝒜ℛℒℰ𝒬𝒰ℐ𝒩 v.3.5 as for the microsatellite analysis (see above), except with the regions defined as Spain, France and Britain/Ireland (see above).
ND1 sequence analysis
Sequences were edited in the ABI program AutoAssembler v.2.0 and aligned in Clustal X (Thompson et al., 1997). A principal coordinates analysis, based on the population pairwise matrix of FST values, was constructed using GenAlEx v.6 (Peakall and Smouse, 2006) to identify population groupings for subsequent estimation of divergence times. In addition, FS (Fu, 1997) was used to test if populations were at drift-migration equilibrium using ARLEQUIN v.3.5.
Timing of divergence between groups of populations was estimated using the isolation with migration (IM) model of population divergence using the program IMa2 (Hey, 2010), with the HKY model of sequence evolution. Analyses were run three times using 40 Markov chain Monte Carlo chains with geometric heating. Following a burn-in of 1 × 106 generations, posterior probabilities of parameters were calculated from 25 × 106 generations of data, sampling every 100 generations. Parameter estimates were adjusted to demographic scales assuming an average salmon generation time of 4 years and a mutation rate of 5.7 × 10−9 substitutions per site per year (Doiron et al., 2002), with upper and lower limits around this rate set at 1 × 10−8 and 1 × 10−9. This mutation rate represented the average of the two mutation rates estimated by Doiron et al. (2002) for ND1 between two charr species and rainbow trout (Oncorhynchus mykiss). Critically for this study, Doiron et al. (2002) looked at mutation rates across 13 mtDNA genes in a range of other salmonid species, including Atlantic salmon, and concluded that the relatively constant mutational differences observed suggested that comparable estimates of divergence would be obtained among salmonid species, regardless of the mitochondrial genes studied.
As demonstrated by Ho and Larson (2006), however, extrapolating mutation rates between species and across evolutionary timescales is seldom straightforward and recent rates of molecular evolution for intraspecific comparisons or newly emerging species may be far more rapid than those estimated from more ancient divergence events. In light of this, additional runs of IMa2 were undertaken with rate parameters derived from divergences between other groups of salmonid fish, including one based on a considerably more recent divergence event (Jacobsen et al., 2012). The additional rates explored comprised one based on divergence of the ND4 gene between Oncorhynchus species (9.7 × 10−9; Wilson and Turner, 2009) and the other a whole mtDNA genome-derived rate based on recent divergences between whitefish species (Coregonus spp.; 1.537 × 10−8; Jacobsen et al., 2012). A recent, comprehensive study of salmonid evolution and interspecific divergence times (Crête-Lafrenière et al., 2012) estimated the rate of mitochondrial molecular divergence within Salmonidae at 0.31% per million years (that is, 3.1 × 10–9; confidence interval (CI): 0.27–0.36% per MY), reaffirming the suitability of the rate parameters used in this study. Until such time as we are reliably able to estimate differential intraspecific rates of mtDNA evolution in salmonids, we have employed rates for which we have reliable supporting information.
Results
Microsatellite analyses
Genetic diversity indices are summarised in Table 1 (see also Supplementary Information and Supplementary Table 2). After sequential Bonferroni corrections were applied, significant deviations from Hardy–Weinberg expectations were observed on 11 occasions; these involved nine populations and six loci (Supplementary Information and Supplementary Table 2). No locus was consistently implicated in departure from Hardy–Weinberg expectations across different samples; thus, data from all 12 loci were retained for subsequent analysis. Of the 1386 tests of linkage disequilibrium between loci, only 26 (that is, <2%) were found to be significant after correction. The majority were single observations, which only occurred in one population. Hence, linkage disequilibrium of loci was deemed to be negligible. The results from Mℐ𝒞ℛ𝒪𝒞ℋℰ𝒞𝒦ℰℛ indicated that no locus consistently showed evidence for null alleles nor error due to stuttering across samples.
Global FST was estimated at 0.0436, with pairwise estimates ranging from 0.001 (Ellé–Léguer; Ellé–Scorff) to 0.156 (Ulla–Itchen). All were significant at P<0.05, except the two lowest values; these involved populations from northwest France that shared the same river mouth and comprised samples of adults ascending the rivers (Supplementary Information and Supplementary Table 3). The neighbour-joining population tree depicted five broad groups of populations reflecting geographical regions (Figure 2). Bootstrap analysis indicated very strong support for the group of Spanish populations (96.2%) and reasonably strong support for the group of French populations (75.2%). Within the United Kingdom and Ireland, the clustering of populations into three geographic regions was clearly depicted: UKSouth, UKSouthwest and UKNorthwest/Ireland. Bootstrap support was very strong for the two UKSouth populations branching together (100%), but less so for the grouping of the UKSouthwest (39.4%) and UKNorthwest/Ireland (26.5%) populations, although within these groups there was stronger support at some branches (Figure 2).
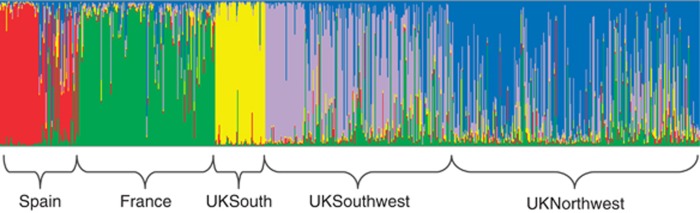
The clustering analysis, as implemented in STRUCTURE, defined an optimum of five regions (that is, k=5), corresponding well with geographic origins (Figure 3). Complementing the neighbour-joining population tree, individuals clustered into the five regions of Spain, France, UKSouth, UKSouthwest and UKNorthwest/Ireland. Similarly, Spain, France and UKSouth all formed very distinct clusters, while the UKSouthwest and UKNorthwest clusters were less distinct and showed considerable overlap.
Figure 3.
Output from 𝒮𝒯ℛ𝒰𝒞𝒯𝒰ℛℰ analysis indicating admixture of individuals in each cluster, defined as k=5. Estimation of the membership fraction to each of five inferred clusters (red, yellow, blue, green and purple) based on microsatellite data; each vertical bar represents one individual and horizontal brackets indicate the geographical regions from which individual fish were sampled.
Hierarchical analysis demonstrated that 3% of variation lay among these five regional groups, 2.2% lay among populations within these regions and the remaining 94.8% lay within populations. Genetic diversity, expressed as average allelic richness and heterozygosity, was lowest in the UKSouth regional group; this was significantly lower (P<0.05) compared with all other regional groups, except Spain (which was close to being significant; Ar: P=0.057; He: P=0.038) (Table 2).
Table 2. Regional genetic diversity indices.
|
Microsatellite diversity
|
mtDNA RFLP diversity
|
|||
|---|---|---|---|---|
| Ar | He | Haplotype diversity | Nucleotide diversity | |
| Spain | 10.151 | 0.771 | 0.166 | 0.0044 |
| France | 11.661 | 0.834 | 0.586 | 0.0166 |
| UKSouth | 7.396 | 0.767 | 0.575 | 0.0209 |
| UKSouthwest | 12.276 | 0.86 | 0.647 | 0.0209 |
| UKNorthwest | 11.9 | 0.856 | 0.571 | 0.0156 |
Abbreviations: Ar, allelic richness; Ho, observed heterozygosity; He, expected heterozygosity.
The allele size permutation test revealed that, on a global scale, multilocus RST values were significantly higher than FST values (FST=0.0436, RST=0.0635, P<0.01; Supplementary Information and Supplementary Figure 1). This suggests that stepwise mutations of microsatellite repeat regions have contributed to the genetic differentiation of the five broad regions and is consistent with the hypothesis that northwest Europe was colonised by Atlantic salmon from different refugia. A significant result was also produced in the comparison of Spain and northwest France (FST=0.0511, RST=0.0985, P<0.05), indicating that these two regions were colonised from, or acted as, different refugia. To test the competing hypotheses of colonisation of the United Kingdom and Ireland, populations from the three clusters comprising this region (UKSouth, UKSouthwest and UKNorthwest/Ireland) were compared. No significant contribution of stepwise mutations to genetic differentiation was observed, suggesting that the three regions were colonised from the same refugial population(s). However, when the UK and Ireland populations were then compared with the potential source/refugial populations of Spain and France, both comparisons were found to be significant (France vs United Kingdom: FST=0.0352, RST=0.0457, P<0.05; Spain vs United Kingdom: FST=0.0448, RST=0.0613, P<0.05). This could be interpreted in two ways: (i) there was a third refuge in UKSouth, from which salmon colonised the whole of the United Kingdom and Ireland; (ii) the United Kingdom and Ireland were a contact zone for Atlantic salmon colonising out of the different refugia in Spain and France, the test being significant because the UK and Ireland populations are comprised of salmon descended from both refugia.
mtDNA RFLP analysis
RFLP analysis of the ND1 region of mtDNA revealed a total of four composite haplotypes (AAAA, AABA, BBBA and BBBB), all of which have been observed previously in populations from northwest Europe (Nielsen et al., 1996; Verspoor et al., 1999; Nilsson et al., 2001). The majority of populations exhibited varying frequencies of all four haplotypes, with only one population fixed for a single haplotype (Ulla; BBBB). All four haplotypes are thought to have been widely distributed before the last glacial period (Nilsson et al., 2001), but due to their frequency distributions, the AAAA and AABA haplotypes have been previously termed ‘Baltic' haplotypes, while the BBBB and BBBA haplotypes were termed ‘Atlantic' haplotypes (Nilsson et al., 2001); for continuation we will also refer to this nomenclature in this study, although it is important to remember that this does not reflect their true origins, rather the nomenclature simply reflects their comparative dominance within the two regions. The Spanish populations were dominated by ‘Atlantic' haplotypes (BBBB; BBBA; 97.5%), while the French populations were dominated by ‘Baltic' haplotypes (AAAA; AABA; 80%). Overall, the United Kingdom was largely dominated by the ‘Atlantic' haplotypes; however, southern UK populations exhibited a higher proportion of ‘Baltic' haplotypes compared with northern UK populations. Genetic diversity, expressed as haplotype and nucleotide diversity, was highest in the United Kingdom, specifically the UKSouthwest group and lowest in Spain (Table 2); a significant reduction in genetic diversity was observed in Spain compared with the United Kingdom (P<0.05). All other comparisons were nonsignificant.
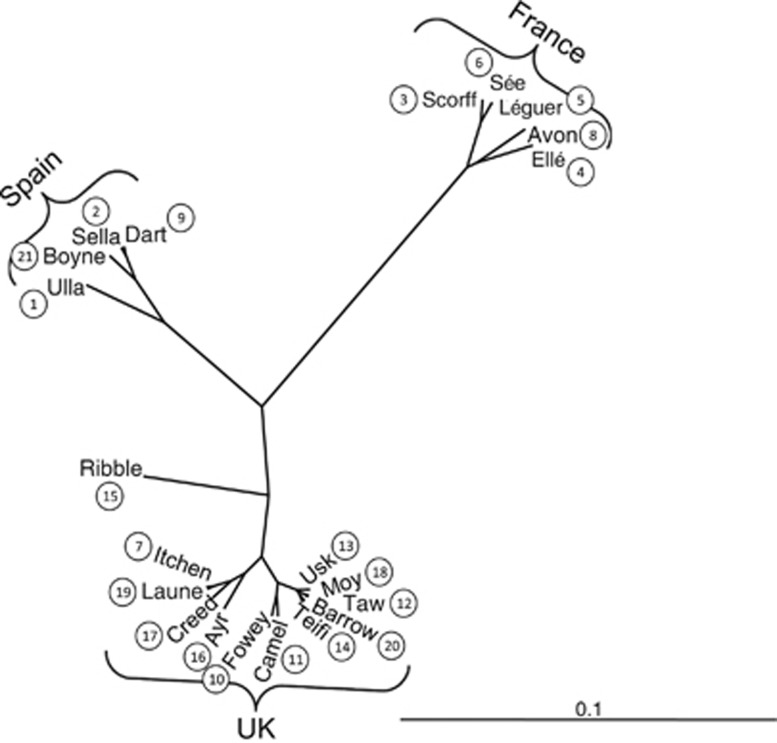
The unweighted pair-group method using arithmetic averages population tree, constructed from Nei's (1987) DA distance (Figure 4), revealed the division of populations into three clusters: one dominated by French populations exhibiting high frequencies of the ‘Baltic' haplotypes, which also contained the geographically proximate Avon; one largely Spanish clade dominated by high frequencies of the ‘Atlantic' haplotypes, with two rivers from other regions (Dart and Boyne), which show similarly high frequencies of the ‘Atlantic' haplotypes; and a third clade containing the majority of UK and Irish samples, which have a more equal distribution of both the ‘Atlantic' and ‘Baltic' haplotypes. Hierarchical analysis for these three broad regions revealed that 34.37% of genetic diversity lay among the three regions, 8.74% lay among populations within each region and 56.89% resided within populations.
Figure 4.
Unweighted pair-group method using arithmetic averages phylogenetic tree generated from mtDNA RFLP data.
ND1 sequence analysis
A 1155 bp region of the mtDNA sequence was obtained from 182 salmon. In total, 22 bases were polymorphic and 20 haplotypes were identified (EMBL accession numbers: HF586486–HF586505). The number of haplotypes per population ranged from one to eight (average=3.85), with two Spanish populations being fixed for a single haplotype; nine haplotypes were found only in a single individual (Supplementary Information, Supplementary Figure 2 and Supplementary Table 4).
Principal coordinates analysis of pairwise FST values of the sequence data reiterated the three groupings defined by mtDNA RFLP analysis (Figure 4): Spain plus the River Dart; France plus the River Avon; and the remaining British and Irish populations (Supplementary Information and Supplementary Figure 3). All subsequent results are based on these three groupings. Fu's Fs was −1.158 (P=0.294) and 0.270 (P=0.518) for the French and Spanish groups, respectively, but was significantly negative for the British and Irish group (−10.589, P=0.0004). A negative value indicates evidence for a history of population expansion.
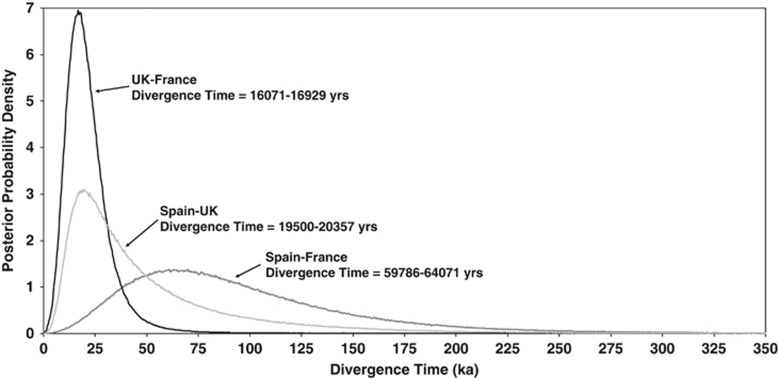
Parameter estimates were consistent across the three runs in the IM analysis (Supplementary Information and Supplementary Table 5). Divergence time estimates between the French and British group and between the Spanish and British group were ∼16 000–20 000 years. The divergence time estimate for the Spanish and French group was much longer, at around 63 000 years (Figure 5). Estimates using other salmonid-derived mtDNA mutation rates (ND4, 9.7 × 10−9 (Wilson and Turner, 2009), and whole mtDNA genome, 1.537 × 10−8, (Jacobsen et al., 2012)) produced more recent divergence estimates; critically, however, all divergence times for the Spanish and French groups pre-date the last glacial maximum (Supplementary Information and Supplementary Figure 4).
Figure 5.
Estimate of divergence times from IMa2 using an ND1-derived divergence rate of 5.7 × 10−9 substitutions per site per year (Doiron et al., 2002).
Discussion
The high genetic diversity observed in Britain and Ireland, particularly in the SouthwestUK, suggests that Atlantic salmon did not colonise this region following the last glacial retreat from salmon expanding out of a single Iberian refuge in northern Spain (that is, H0 is rejected). Rather, the results are consistent with there being a second, cryptic western refuge for Atlantic salmon in northwest France (that is, H1A is accepted) with colonisation of Britain and Ireland from both refugia.
Genetic diversity and differentiation
The genetic diversity of 21 populations of Atlantic salmon in northwest Europe as revealed by microsatellite analysis (12 loci, 289 alleles) was found to be comparable with that reported for other regions (King et al., 2001; Tonteri et al., 2005; Säisä et al., 2005). Expected heterozygosity (He=0.808) was consistent with that reported for populations from Britain and Ireland by Säisä et al. (2005) (He=0.812) and higher than that reported for other regions (Baltic: He=0.73 (Säisä et al., 2005); northern Europe: He=0.56 (Tonteri et al., 2005); North America: He=0.60 (King et al., 2001)). The genetic divergence of salmon populations in northwest Europe, as measured using FST estimates, was lower in this study (Global FST=0.0436) than previously reported for populations in northwest Europe (FST=0.12 (Bourke et al., 1997); FST=0.114 (King et al., 2001)). However, the sampling range of this study was restricted compared with those of Bourke et al. (1997) and King et al. (2001). Within northwest Europe, genetic diversity was found to be highest in UKSouthwest (Ar=12.276, He=0.86) and lowest in UKSouth (Ar=7.396, He=0.767), although this latter estimate was still higher than previously reported for other regions (King et al., 2001; Säisä et al., 2005; Tonteri et al., 2005). Genetic divergence was highest between UKSouth and Spain (FST=0.114) and lowest between the UKSouthwest and UKNorthwest/Ireland (FST=0.026).
The genetic diversity of matrilineal Atlantic salmon populations in west/northwest Europe as revealed by PCR-RFLP of mtDNA and measured using average haplotype diversity estimates (Global H=0.509) was found to be comparable to that observed in previous studies (British Isles and Spain H=0.507 (Nilsson et al., 2001), higher than that reported for the Atlantic as a whole or the Baltic (H=0.478, H=0.217, respectively; Nilsson et al., 2001), but lower than that reported for northern Europe (H=0.543; Asplund et al., 2004)). In terms of PCR-RFLP variability within west/northwest Europe, genetic diversity was found to be highest in UKSouthwest (H=0.647) and lowest in Spain (H=0.166). Lower diversity in Spanish populations compared with those from the British Isles in PCR-RFLP mtDNA studies has been reported previously by Verspoor et al. (1999) and Consuegra et al. (2002).
All four composite mtDNA haplotypes observed in this study have been described previously (Nielsen et al., 1996) and are known to be widespread across Europe (Verspoor et al., 1999; Nilsson et al., 2001; Asplund et al., 2004). The consensus is that the evolution of these haplotypes predates the Pleistocene glaciations and hence all haplotypes were probably widespread before the last glacial maximum (Nilsson et al., 2001; Asplund et al., 2004; Säisä et al., 2005). Thus, the effects of the last glacial period would primarily be observed by lineage sorting through random drift, initially in the differing frequency distributions of these mtDNA haplotypes in different refugial populations, and subsequently in mixing of these haplotypes in postglacial processes, such as processes of secondary contact.
The dominance of the ‘Atlantic' (BBBB and BBBA) mtDNA haplotypes in Spain and high frequency of these haplotypes in Britain and Ireland has been noted previously (Verspoor et al., 1999; Consuegra et al., 2002; Ciborowski et al., 2007; Campos et al., 2008). However, the dominance of the ‘Baltic' (AAAA and AABA) haplotypes in northwest France has not been observed before, although none of the previous studies had sampled from this region.
Refugial locations for Atlantic salmon in northwest Europe
The British/Irish ice sheet extended to around 52°N at the time of the last glacial maximum, with permafrost extending over most of Britain and Ireland, except the far southwest corner of England (Murton and Lautridou, 2003). South of the ice sheet, the Iberian Peninsula (northern Spain and Portugal) has been widely accepted as a refuge for Atlantic salmon since bone relics dating back 40 000 years were uncovered in a cave in this region (Consuegra et al., 2002). The initial hypothesis set out in this study was to ascertain whether this was the sole source of contemporary Atlantic salmon populations in northwest Europe; results from this study suggest that this is highly unlikely, and there is evidence to propose that there was a distinct second refuge for Atlantic salmon in northwest France. Supporting evidence comes from both microsatellite and mtDNA data. Separation of the French populations from the Spanish populations is depicted in the clustering and phylogenetic analysis, where the French populations formed a distinct cluster in the 𝒮𝒯ℛ𝒰𝒞𝒯𝒰ℛℰ analysis and distinct groupings on both the microsatellite and mtDNA population trees. Furthermore, the allele size permutation test of microsatellite data indicated that French and Spanish populations were separated before the last glacial maximum, although some caution may be required in interpreting the results from the allele size permutation test, as recent work has shown that the FST estimator used in the calculations may be downwardly biased in cases of high heterozygosity (Meirmans and Hedrick, 2011), as was the case in this study. This result, however, was also supported by the analysis of divergence times with IMa2. This analysis indicated divergence times between populations in the two proposed refugia (Spain and France) to be considerably older (∼60 000 years) than were the divergence estimates of populations from Britain and Ireland with either of the continental refugial populations (Britain/Ireland–France ∼16 000 years; Britain/Ireland–Spain ∼20 000 years).
Hence, these regions (France and Spain) and the salmon within them appear to have been distinct during the last glacial period. The discontinuity of mtDNA haplotype frequencies also corroborates this theory. In line with previous studies (Verspoor et al., 1999; Consuegra et al., 2002; Ciborowski et al., 2007; Campos et al., 2008), modern Spanish populations are dominated by ‘Atlantic' haplotypes, although the high frequency of a distinctive HaeIII variant in Pleistocene salmon samples from northern Spain (Consuegra et al., 2002) appears more characteristic of contemporary Baltic populations, suggesting that Pleistocene salmon populations within the region may have differed somewhat from those found today. Contemporary French populations, however, are dominated by ‘Baltic' haplotypes. This indicates that modern French populations are unlikely to be related to contemporary Spanish populations, but may be a relic of forms distributed more widely in the west of Europe during the Pleistocene era. To our knowledge, northwest France has never been stocked with fish from the Baltic region, and due to the current migratory behaviour of Baltic salmon, whereby very few fish pass the Danish strait into the North Sea and Atlantic Ocean, it seems improbable that northern France would have been colonised naturally by Baltic salmon. Therefore, it seems likely that the high frequencies of these ‘Baltic' haplotypes exhibited by salmon populations in northwest France are due to lineage sorting through random drift. Thus, it appears that French populations were isolated from those in Spain during the last glacial period, over which time populations between the two regions differentiated genetically. Such a finding accords with the results from another biogeographic study within the region; Larmuseau et al. (2009) identified a similar distinct differentiation between a population of sand gobies from the Gironde river in western France and those from the Atlantic rivers of Portugal and Spain.
Thus, the data suggest that at least two refugia for Atlantic salmon were present in western Europe during the last glacial period, one in northern Spain (Iberian Peninsula) and one in northwest France, and there is increasing evidence that many species existed further north of the well-recognised southern refugia, in sheltered areas experiencing stable microclimates (Stewart and Lister, 2001). Northwest France may have been once such location and evidence emerging from a number of marine species over the past few years (Provan et al., 2005; Hoarau et al., 2007; Olsen et al., 2010; Panova et al., 2011) suggests that the Brittany coast and the area of Hurd Deep in the western English Channel may have been a refuge during the last glacial maximum. In a recent review, it was proposed that species whose current northerly range extends to 60°N or further would potentially have been able to persist in northerly refugia; this seemingly reflects the ecological and physiological adaptations these animals have to survive these conditions (Bhagwat and Willis, 2008). For example, the bullhead (Cottus gobio), a cold adapted freshwater fish, seems to have persisted in a refuge in the southern British Isles (Hanfling et al., 2002). In the case of Atlantic salmon, their current distribution extends to over 70°N in the Barents and Kara Seas (MacCrimmon and Gots, 1979) and they can exhibit high tolerance to extreme cold conditions; hence, it is plausible that Atlantic salmon could have persisted in the conditions in northern France during the late glacial period.
In earlier studies, the southern North Sea has been proposed as the most likely location of a second refuge for Atlantic salmon in northwest Europe (Verspoor et al., 1999; Asplund et al., 2004; Säisä et al., 2005). No indication of this was observed in this study, but our research focused on a more westerly region. An investigation centred on drainages of the North Sea would be necessary to explore this theory, although this would be difficult since many of the North Sea drainages are now extinct of salmon.
Colonisation of Britain and Ireland
A second objective for this study was to reconstruct the colonisation pathway of Atlantic salmon from these source populations into Britain and Ireland. Results are consistent with the hypothesis that Britain and Ireland are a secondary contact zone for Atlantic salmon expanding out from the Spanish and French refugial populations.
Evidence from the allele size permutation test, comparing populations within Britain and Ireland, indicated that stepwise mutations had not contributed to the differentiation of populations within the region, suggesting that these populations are likely to be descended from the same refugial population(s). Moreover, when populations from Britain and Ireland were compared with the potential source populations of France and Spain, the allele size permutation tests revealed a significant contribution of stepwise mutations to the observed differentiation. We interpret this as evidence that salmon from both France and Spain were successful in colonising Britain and Ireland. Analysis of divergence times in IMa2 suggested similar divergence times for populations from Britain and Ireland with France (∼16 000 years) and with Spain (∼20 000 years); these are broadly consistent with a pattern of recolonisation since the last glacial maximum,that is, approximately 20 000 years ago (particularly given the wide confidence intervals on such estimates; Supplementary Information and Supplementary Table 5).
Given our findings, it is possible to speculate on the colonisation route of Atlantic salmon out of these refugial populations through the exploration of matrilineal haplotype frequencies. The French populations were largely dominated by the ‘Baltic' haplotypes, whereas the Spanish populations were dominated by the ‘Atlantic' haplotypes. Within Britain, the highest frequency of ‘Baltic' haplotypes was in the region geographically most proximate to France, namely SouthUK. The proportion of ‘Baltic' haplotypes then decreased in SouthwestUK, where the frequency distribution of the ‘Baltic' and ‘Atlantic' haplotypes were most closely balanced, and decreased northwards in Britain and Ireland, where the ‘Atlantic' haplotypes began to dominate once more. This suggests that as the ice retreated, salmon from the French refuge first recolonized rivers in SouthUK, while salmon from the Spanish refuge moved into SouthwestUK and Ireland, where the two lineages came into secondary contact; ultimately, it appears that salmon from the Spanish refuge came to dominate more in the northern regions. Although speculative, perhaps similarities between refugial environments and recolonised regions, and potential associated local adaptations, can help explain the distribution of contemporary haplotypes.
Conclusions
The use of multiple classes of molecular markers has proved highly effective in resolving the phylogeographic history of Atlantic salmon in a region which to date has received little focused attention. In addition to the well-established refuge in the Iberian Peninsula, a cryptic western refuge in northwest France also seems to be likely, with subsequent colonisation of Britain and Ireland occurring from both refugia.
The clear distinction of the populations of northwest France from those in Spain has not been observed in previous studies. To protect effectively these two ancient lineages, it would be necessary to locate the boundary between the two refugia—an obvious avenue for future research. The ability to reliably identify such variation is important to safeguard the genetic diversity of the species, especially in these small populations existing in somewhat atypical warmer environments at the extreme southern edge of the species' European range.
Data archiving
All mtDNA ND1 sequences were deposited in EMBL with accession numbers HF586486–HF586505. Microsatellite data and mtDNA RFLP haplotype data were deposited in Dryad doi: 10.5061/dryad.0hj6g.
Acknowledgments
This project was funded by the Natural Environment Research Council (NERC), UK, through an Industrial CASE studentship (NER/S/C/2005/13466), with CASE partners, the Westcountry Rivers Trust. Additional funding was provided through the Atlantic Salmon Arc Project (ASAP), European Union INTERREG IIIB Project No. 040. We are grateful to Professor Paulo Prodohl (Queen's University, Belfast) for helpful suggestions on the analysis. We thank J-L Baglinière (INRA, Rennes) for access to french scale samples and partners in the ASAP project for additional samples.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Andersen BG, Borns WW. The Ice Age World. Scandinavian University Press: Oslo, Norway; 1994. [Google Scholar]
- Asplund T, Veselov A, Primmer CR, Bakhmet I, Potutkin A, Titov S, et al. Geographical structure and postglacial history of mtDNA haplotype variation in Atlantic salmon (Salmo salar L.) among rivers of the White and Barents Sea basins. Ann Zool Fenn. 2004;41:465–475. [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bernatchez L, Wilson LL. Comparative phylogeography of Nearctic and Palearctic fishes. Mol Ecol. 1998;7:431–452. [Google Scholar]
- Bhagwat SA, Willis KJ. Species persistence in northerly glacial refugia of Europe: a matter of chance or biogeographical traits. J Biogeogr. 2008;35:464–482. [Google Scholar]
- Bourke EA, Coughlan J, Jansson H, Galvin P, Cross TF. Allozyme variation in populations of Atlantic salmon located throughout Europe: diversity that could be compromised by introductions of reared fish. ICES J Mar Sci. 1997;54:974–985. [Google Scholar]
- Campos JL, Posada D, Morán P. Introgression and genetic structure in northern Spanish Atlantic salmon (Salmo salar L.) populations according to mtDNA data. Con Gen. 2008;9:157–169. [Google Scholar]
- Child AR, Burnell AM, Wilkins NP. The existence of two races of Atlantic salmon (Salmo salar L.) in the British Isles. J Fish Biol. 1976;8:35–43. [Google Scholar]
- Ciborowski KL, Consuegra S, Garcia de Leaniz C, Wang J, Beaumont M, Jordan WC. Stocking may increase mitochondrial DNA diversity but fails to halt the decline of endangered Atlantic salmon populations. Con Gen. 2007;8:1355–1367. [Google Scholar]
- Consuegra S, De Leaniz CG, Serdio A, Morales MG, Straus LG, Knox D, et al. Mitochondrial DNA variation in Pleistocene and modern Atlantic salmon from the Iberian glacial refugium. Mol Ecol. 2002;11:2037–2048. doi: 10.1046/j.1365-294x.2002.01592.x. [DOI] [PubMed] [Google Scholar]
- Crête-Lafrenière A, Weir LK, Bernatchez L. Framing the Salmonidae family phylogenetic portrait: a more complete picture from increased taxon sampling. PLoS One. 2012;7:e46662. doi: 10.1371/journal.pone.0046662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doiron S, Bernatchez L, Blier PU. A comparative mitogenomic analysis of the potential adaptive value of Arctic Charr mtDNA introgression in Brook Charr populations (Salvelinus fontinalis Mitchill) Mol Biol Evol. 2002;19:1902–1909. doi: 10.1093/oxfordjournals.molbev.a004014. [DOI] [PubMed] [Google Scholar]
- Estoup A, Largiader CR, Perrot E, Chourrout D. Rapid one-tube DNA extraction for reliable PCR detection of fish polymorphic markers and transgenes. Mol Mar Biol Biotechnol. 1996;5:295–298. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecoly Res. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J.1993. PHYLIP: Phylogeny inference package, Version 4.0 http://evolution.genetics.washington.edu/phylip.html . Accessed 5/3/2013..
- Finnegan AK, Stevens JR. Assessing the long-term genetic impact of historical stocking events on contemporary populations of Atlantic salmon (Salmo salar L.) Fish Manage Ecol. 2008;15:315–326. [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and backgroud selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- Griffiths A, Machado-Schiaffino G, Dillane E, Coughlan J, Horreo J, Bowkett A, et al. Genetic stock identification of Atlantic salmon (Salmo salar) populations in the southern part of the European range. BMC Genet. 2010;11:31. doi: 10.1186/1471-2156-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall HJ, Nawrocki LW. A rapid method for detecting mictochondrial DNA variation in the brown trout, Salmo trutta. J Fish Biol. 1995;46:360–364. [Google Scholar]
- Hanfling B, Hellemans B, Volckaert FAM, Carvalho GR. Late glacial history of the cold-adapted freshwater fish Cottus gobio, revealed by microsatellites. Mol Ecol. 2002;11:1717–1729. doi: 10.1046/j.1365-294x.2002.01563.x. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Charbonnel N, Freville H, Heuertz M. Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics. 2003;163:1467–1482. doi: 10.1093/genetics/163.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 1996;58:247–276. [Google Scholar]
- Hewitt GM. Post-glacial re-colonization of European biota. Biol J Linn Soc. 1999;68:87–112. [Google Scholar]
- Hey J. Isolation with migration models for more than two populations. Mol Biol Evol. 2010;27:905–920. doi: 10.1093/molbev/msp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SYW, Larson G. Molecular clocks: when times are a-changin'. Trends Genet. 2006;22:79–83. doi: 10.1016/j.tig.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Hoarau G, Coyer JA, Veldsink JH, Stam WT, Olsen JL. Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Mol Ecol. 2007;16:3606–3616. doi: 10.1111/j.1365-294X.2007.03408.x. [DOI] [PubMed] [Google Scholar]
- Jacobsen MW, Hansen MM, Orlando L, Bekkevold D, Bernatchez L, Willerslev E, et al. Mitogenome sequencing reveals shallow evolutionary histories and recent divergence time between morphologically and ecologically distinct European whitefish (Coregonus spp.) Mol Ecol. 2012;21:2727–2742. doi: 10.1111/j.1365-294X.2012.05561.x. [DOI] [PubMed] [Google Scholar]
- King TL, Eackles MS, Letcher BH. Microsatellite DNA markers for the study of Atlantic salmon (Salmo salar) kinship, population structure, and mixed-fishery analyses. Mol Ecol Notes. 2005;5:130–132. [Google Scholar]
- King TL, Kalinowski ST, Schill WB, Spidle AP, Lubinski BA. Population structure of Atlantic salmon (Salmo salar L.): a range-wide perspective from microsatellite DNA variation. Mol Ecol. 2001;10:807–821. doi: 10.1046/j.1365-294x.2001.01231.x. [DOI] [PubMed] [Google Scholar]
- Koskinen MT, Knizhin I, Primmer CR, Schlotterer C, Weiss S. Mitochondrial and nuclear DNA phylogeography of Thymallus spp. (grayling) provides evidence of ice-age mediated environmental perturbations in the world's oldest body of fresh water, Lake Baikal. Mol Ecol. 2002;11:2599–2611. doi: 10.1046/j.1365-294x.2002.01642.x. [DOI] [PubMed] [Google Scholar]
- Larmuseau MHD, Van Houdt JKJ, Guelinckx J, Hellemans B, Volckaert FAM. Distributional and demographic consequences of Pleistocene climate fluctuations for a marine demersal fish in the north-eastern Atlantic. J Biogeogr. 2009;36:1138–1151. [Google Scholar]
- Lericolais G, Auffret J-P, Bourillet J-F. The Pleistocene rivers of the English Channel region. J Quat Sci. 2003;18:245–260. [Google Scholar]
- Liu KJ, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- MacCrimmon HR, Gots BL. World distribution of Atlantic salmon, Salmo salar. J Fish Res Board Canada. 1979;36:422–457. [Google Scholar]
- Makhrov AA, Bolotov IN. Dispersal routes and species identification of freshwater animals in Northern Europe: a review of molecular evidence. Russ J Genet. 2006;42:1101–1115. [PubMed] [Google Scholar]
- McConnell SK, Oreilly P, Hamilton L, Wright JN, Bentzen P. Polymorphic microsatellite loci from Atlantic Salmon (Salmo salar)—genetic differentiation of North-American and European populations. Can J Fish Aquat Sci. 1995;52:1863–1872. [Google Scholar]
- McElroy D, Moran P, Bermingham E, Kornfield I. REAP—an integrated environment for the manipulation and phylogenetic analysis of restriction data. J Hered. 1992;83:157–158. doi: 10.1093/oxfordjournals.jhered.a111180. [DOI] [PubMed] [Google Scholar]
- Meirmans PG, Hedrick PW. Assessing population structure: FST and related measures. Mol Ecol Resour. 2011;11:5–18. doi: 10.1111/j.1755-0998.2010.02927.x. [DOI] [PubMed] [Google Scholar]
- Murton JB, Lautridou JP. Recent advances in the understanding of Quaternary periglacial features of the English Channel coastlands. J Quat Sci. 2003;18:301–307. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press: New York, NY, USA; 1987. [Google Scholar]
- Nielsen EE, Hansen MM, Loeschcke V. Genetic structure of European populations of Salmo salar L. (Atlantic salmon) inferred from mitochondrial DNA. Heredity. 1996;77:351–358. [Google Scholar]
- Nilsson J, Gross R, Asplund T, Dove O, Jansson H, Kelloniemi J, et al. Matrilinear phylogeography of Atlantic salmon (Salmo salar L.) in Europe and postglacial colonization of the Baltic Sea area. Mol Ecol. 2001;10:89–102. doi: 10.1046/j.1365-294x.2001.01168.x. [DOI] [PubMed] [Google Scholar]
- O'Reilly P, Hamilton L, McConnell SK, Wright JN. Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can J Fish Aquat Sci. 1996;53:2292–2298. [Google Scholar]
- Olsen JL, Zechman FW, Hoarau G, Coyer JA, Stam WT, Valero M, Aberg P. The phylogeographic architecture of the fucoid seaweed Ascophyllum nodosum: an intertidal ‘marine tree' and survivor of more than one glacial–interglacial cycle. J Biogeogr. 2010;37:842–856. [Google Scholar]
- Panova M, Blakeslee AMH, Miller AW, Mäkinen T, Ruiz GM, Johannesson K, et al. Glacial history of the North Atlantic marine snail, Littorina saxatilis, inferred from distribution of mitochondrial DNA lineages. PLoS One. 2011;6:e17511. doi: 10.1371/journal.pone.0017511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson S, Piertney SB, Knox D, Gilbey J, Verspoor E. Characterization and PCR multiplexing of novel highly variable tetranucleotide Atlantic salmon (Salmo salar L.) microsatellites. Mol Ecol Notes. 2004;4:160–162. [Google Scholar]
- Payne RH, Child AR, Forrest A. Geographical variation in the Atlantic salmon. Nature. 1971;231:250–252. doi: 10.1038/231250a0. [DOI] [PubMed] [Google Scholar]
- Peakell R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Resour. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Aguinagalde I, de Beaulieu J-L, Bittkau C, Brewer S, Cheddadi R, et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J, Wattier RA, Maggs CA. Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol Ecol. 2005;14:793–803. doi: 10.1111/j.1365-294X.2005.02447.x. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (Version-1.2)—Population-Genetics Software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Säisä M, Koljonen ML, Gross R, Nilsson J, Tähtinen J, Koskiniemi J, et al. Population genetic structure and postglacial colonization of Atlantic salmon (Salmo salar) in the Baltic Sea area based on microsatellite DNA variation. Canad J Fish Aquat Sci. 2005;62:1887–1904. [Google Scholar]
- Sarnthein M. Der Atlantic im Letzen Glazialen Maximum. Nova Acta Leopold. 2001;88:35–43. [Google Scholar]
- Slettan A, Olsaker I, Lie O. Atlantic salmon, Salmo salar, microsatellites at the SSOSL25, SSOSL85, SSOSL311, SSOSL412 loci. Anim Genet. 1996;26:281–282. doi: 10.1111/j.1365-2052.1995.tb03262.x. [DOI] [PubMed] [Google Scholar]
- Ståhl G.1987Genetic population structure of Atlantic salmonIn: Ryman N, Utter FM, (eds).Population Genetics and Fisheries Management University of Washington Press: Seattle, WA, USA; 121–140. [Google Scholar]
- Stewart JR, Lister AM. Cryptic northern refugia and the origins of the modern biota. Trends Ecol Evol. 2001;16:608–613. [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF. Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. CLUSTAL X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonteri A, Titov S, Veselov A, Zubchenko A, Koskinen MT, Lesbarreres D, et al. Phylogeography of anadromous and non-anadromous Atlantic salmon (Salmo salar) from northern Europe. Ann Zool Fenn. 2005;42:1–22. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Verspoor E.1986Identification of stocks in the Atlantic salmonIn: Stroud RH, (ed.).Proceedings of the Symposium on Future Atlantic Salmon Management Marine Recreational Fisheries Series, National Coalition for Marine Conservation: Savannah, GA, USA; 37–46. [Google Scholar]
- Verspoor E, Jordan WC. Genetic-variation at the Me-2 locus in the Atlantic salmon within and between Rivers—evidence for its selective maintenance. J Fish Biol. 1989;35:205–213. [Google Scholar]
- Verspoor E, McCarthy EM, Knox D. The phylogeography of European Atlantic salmon (Salmo salar L.) based on RFLP analysis of the ND1/16sRNA region of the mtDNA. Biol J Linn Soc. 1999;68:129–146. [Google Scholar]
- Wilson WD, Turner TF. Phylogenetic analysis of the Pacific cutthroat trout (Oncorhynchus clarki ssp.: Salmonidae) based on partial mtDNA ND4 sequences: a closer look at the highly fragmented inland species. Mol Phylogenet Evol. 2009;52:406–415. doi: 10.1016/j.ympev.2009.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.