Abstract
Understanding the biochemistry of DNA replication of the plant DNA viruses is important for the development of antiviral strategies. Since DNA replication is little studied in plants, a genetically tractable, easily culturable, eukaryotic model system is required to pursue such studies in a facile manner. Here we report the development of a yeast model system that supports DNA replication of a chosen geminivirus strain, Indian mung bean yellow mosaic virus. The replication of plasmid DNA in the model system relies specifically on the virus-derived elements and factors. Usage of this model system revealed the role of at least one hitherto unknown viral factor for viral DNA replication. The episomal characteristic of single-strandedness of replicated plasmid DNA was shown, and the expression of viral genes was also confirmed. This model system is expected to shed light on the machinery and mechanism involved in geminiviral DNA replication in plants.
The complexities of host features might hinder the addressing of specific issues related to host-virus interactions, including biosynthesis of viral nucleic acids. The factors required for DNA replication of the plant DNA viruses are difficult to discover in a systematic manner by employing the current approaches of plant biology. The variability associated with DNA transfections of plant protoplasts, the problems related to plant transformation, and other issues limit the progress of studies of DNA replication of viruses such as geminiviruses, which are potent phytopathogens. Moreover, the intermediates of replicated products are difficult to isolate from infected plant tissues. The lack of genetic mutants and of sequence information for the host genomes also compounds the problem. In order to circumvent these difficulties, we looked for a model eukaryotic host that supports DNA replication of the geminiviruses.
Geminiviruses represent a family of viruses which are characterized by twin icosahedral particles, each of which encapsidates a small (∼2.7 kb) single-stranded circular DNA. Though no crop is immune to the geminiviruses, each strain of geminivirus can infect crop plants within a narrow window of host specificity (19). Indian mung bean yellow mosaic virus (IMYMV), a member of the Geminiviridae family, possesses bipartite genomes, namely DNA-A and DNA-B, and is a menace in legume cultivation (18). The 2,745-bp DNA-A component encodes the information for viral DNA replication, transcription, and encapsidation, whereas the 2,616-bp DNA-B encodes two movement proteins responsible for virus translocation.
Studies with viruses related to IMYMV have suggested that the viral DNA frequently, but not exclusively, replicates in a rolling circle mode (RCR) (10, 16, 21) The replication origins of all geminiviruses include an invariant 9-mer sequence (TAATATT↓AC) which is site-specifically nicked (at the position of the arrow) by a virus-encoded replication initiator protein (Rep; also called AC1 or AL1) to initiate RCR (28). For efficient replication, Rep is assisted by a replication enhancer (REn; also called AC3 or AL3) (27). The roles of other virus-encoded factors and the multitude of host factors used for the viral replication process have remained obscure until now, despite the implication of a few host factors (1, 5, 12, 14, 30, 31).
The putative zone of initiation of RCR has been located in a common region (CR) of both the DNA-A and DNA-B components of IMYMV by use of in silico analyses. A zone of about 200 bp, called CR-A (in DNA-A), is organized in a stem-loop structure, and the invariant 9-mer sequence is located in the predicted loop region. The IMYMV DNA-A component codes for Rep, REn, and other proteins, such as AC2, AC4, and AC5, whose direction of transcription is similar to that of AC1 (Fig. 1A). The roles of AC4 and AC5 in determining viral growth or pathogenesis are currently unknown. The general role of Rep in the initiation of RCR has been suggested in view of its high-affinity binding and nicking-closing activities at the CR sequences (13). The similar biochemical activities of a 43-kDa recombinant, 6×His-Rep, of IMYMV have also been demonstrated (18).
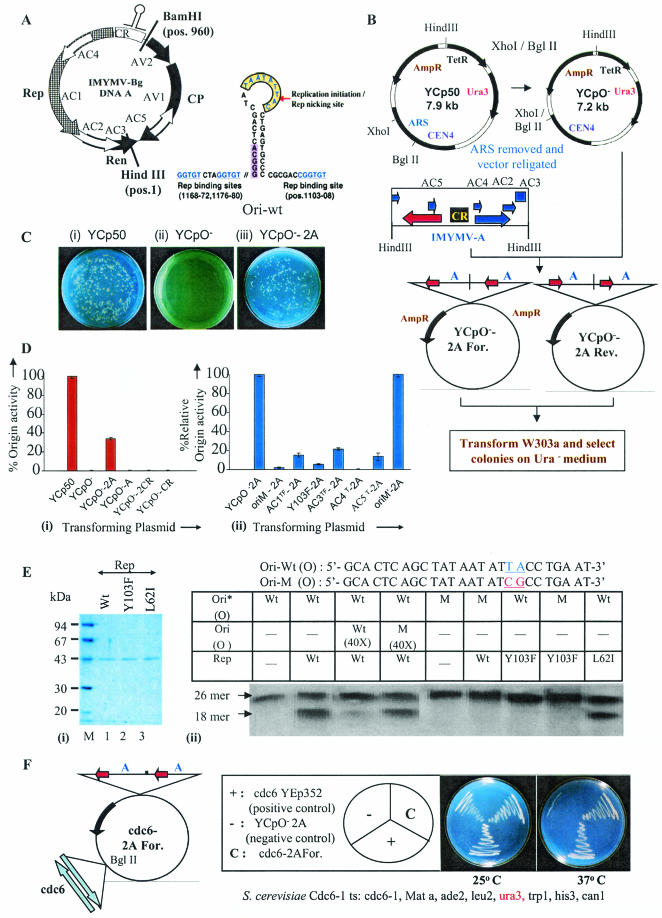
FIG.1.
Genetic studies on geminiviral replication in yeast. (A) Genome organization of IMYMV Black-gram isolate (IMYMV-Bg) DNA-A. An enlargement of the characteristic hairpin loop region containing the conserved nonamer sequence TAA TAT TAC where Rep initiates RCR (Rep cutting site [arrow]) is shown to the right. The various ORFs with their directions of transcription are shown. The putative Rep-binding sites are underlined. The unique sites of BamHI and HindIII are marked. (B) Strategy used to make the replication-competent virus recombinant plasmid YCpO−-2A. The nomenclature YCpO−-2A-For or -Rev was on the basis of orientation of the AV1 ORF with respect to the AmpR gene in YCpO−. (C) Representative plates comparing colonies obtained of the transformed yeast W303a strain (MATa leu2 ura3 his trp1 ade2) with plasmid YCp50 (i), YCpO− (ii), or YCpO−-2A (iii). (D) Panel i, % origin activity of the various constructs relative to YCp50, which was given a value of 100; panel ii, mutation of different ORFs in the DNA-A (in the YCpO−-2A background), namely, AC1, AC3, AC4, and AC5, led to a decrease in origin function (% relative origin activity) compared to plasmid YCpO−-2A, which was given a value of 100 in this case. The superscripts T and TF stand for termination and termination followed by frameshift mutations, respectively. (E) Panel i, SDS-polyacrylamide gel electrophoresis profile showing the apparent homogeneity in preparations of recombinant Rep and its mutant proteins. AC4T and L62I-Rep represent the same mutant. Panel ii, in vitro site-specific cutting activity of various recombinant versions of Rep with either a wild-type (Ori Wt) or mutant (OriM) oligonucleotide (sequences are indicated at the top). The 43-kDa Rep Wt was able to efficiently cut the 5′-labeled (*) Ori-Wt oligonucleotide (lane 2), and this site-specific nicking was competed out by 40× unlabeled Ori Wt (lane 3), but not by OriM (lane 4). 1× oligonucleotide represents about 1 ng of the 26-mer oligonucleotide. (F) cdc6 complementation. cdc6-YEp352 and YCpO−-2A plasmids were used as a positive and negative control, respectively. The plasmids used for transformation are shown in the sector diagram. All colonies were scored on Ura dropout plates.
Since the processes of viral DNA replication are difficult to study with infected plant tissues, we examined whether Saccharomyces cerevisiae cells could support replication of IMYMV viral DNA. If budding yeast cells can act as a proper host for viral DNA replication, prior knowledge of yeast genetics and the biochemistry of DNA replication can be applied toward understanding the mechanism of IMYMV DNA replication. Toward this goal, the DNA-A component was cloned into an engineered yeast shuttle plasmid, which by itself was deficient in autonomous replicating sequence (ARS)-related activity. The DNA-A component conferred replication activity on this recombinant plasmid, and this activity was monitored by counting the CFU of the plasmid-transformed auxotrophs of yeast in the selective medium. Here we report for the first time that the DNA-A of IMYMV can efficiently replicate as a recombinant plasmid within S. cerevisiae cells and suggest that the mechanism of replication of this small viral DNA genome could be investigated better by using this model.
MATERIALS AND METHODS
Cloning strategy.
The ARS was removed from the shuttle plasmid YCp50 by XhoI and BglII digestion to generate the plasmid YCpO− after end filling and religation. A HindIII cassette of the IMYMV DNA-A, obtained by digestion of its pUC clone, was recloned as either a monomer or a dimer into YCpO− as shown in Fig. 1B.
Mutagenesis.
Different open reading frames (ORFs) of the viral A genome were subjected to site-directed mutagenesis by using a QuikChange site-directed mutagenesis kit from Stratagene according to the manufacturer's protocol. The mutagenized IMYMV genome was introduced as a single copy or tandem dimer into YCpO−.
Yeast transformation.
The transformation of yeast cells with the plasmids was performed essentially by the high-efficiency transformation method, using lithium acetate, single-stranded DNA, and polyethylene glycol, of Agatep et al. (2). Single-stranded DNA was used as the carrier DNA to enhance the efficiency of the plasmid-mediated transformation. The Ura auxotroph W303a was transformed with a plasmid containing the Ura3 marker, and the colonies were scored on synthetic defined (SD) Ura− plates. The original activity of each plasmid was defined as the number of colonies scored on a Ura dropout plate when the Ura auxotroph of yeast, i.e., the W303a strain, was transformed with 6 μg of plasmid DNA (for YCpO−-2A) or 715 fM (for others) plasmid DNA.
PCR amplification.
The 5.6-kb DNA insert present at the HindIII cloning region of YCpO−2A was PCR amplified with Taq DNA polymerase and Pfu at a ratio of 9:1 in the presence of 5% dimethyl sulfoxide, using primers flanking the cloning site viz, namely YCpFwd (5′-CAC ATT TCC CCG AAA AGT GC-3′) and YCpRev (5′-GTT AGA TTT CAT ACA CGG TG-3′). The 230-bp CR-A region was amplified in the usual manner by using the CR5 (5′-GGG GAA TTC CCC TTG GCA TAT TTG AAG TC-3′) and CR3 (5′-ATC GGA TCC GAT TGA ACG ACT AAA GAT AAG-3′) primers specific for the CR (combination X). The DNAs from the RCR origin to the 5′ and 3′ termini were also amplified by using the Z (Ori WtR and CR5) and Y (Ori Wt and CR3) primer combinations, respectively, as shown in Fig. 2D, panel ii. The sequence of Ori Wt is shown in Fig. 1E, panel ii, and Ori WtR is complementary to Ori Wt.
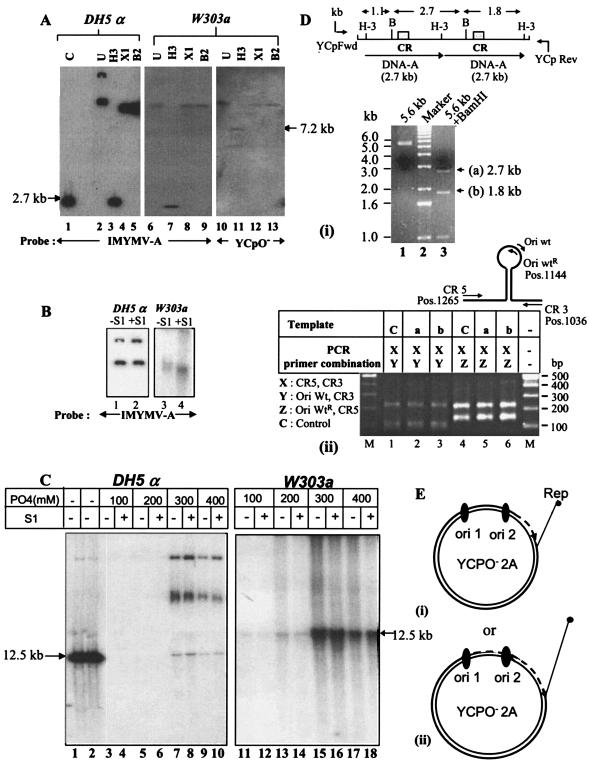
FIG. 2.
Southern blot and PCR analyses of plasmid products replicated in the W303a yeast strain. (A) Southern blotting of replicated plasmids. Plasmid DNA was isolated from transformed yeast by using a standard protocol (11) or from E. coli DH5α and was treated with a unique cutter, such as HindIII, XhoI, or BglII (H-3, X1, or B2). The digested DNA was resolved by electrophoresis in a 0.7% agarose gel, Southern blotted, and probed as shown. A 2.7-kb IMYMV DNA-A fragment obtained by HindIII digestion of plasmid pUC18-A was used as a length standard (lane 1). (B) Comparison of S1 nuclease sensitivities of plasmid DNAs derived from bacterial and yeast sources. S1 nuclease was used at a concentration of 2 U/μg of DNA at 37°C for 30 min. (C) Characterization of single-strandedness of plasmids replicated in yeast. For further confirmation of the presence of single-stranded DNA, 5 μg of YCpO−-2A plasmid DNA isolated from either E. coli DH5α or S. cerevisiae W303a was allowed to bind separately to mini-HAP columns and step eluted with phosphate buffer (pH 6.8) as indicated. M13 single-stranded DNA and double-stranded RF markers were used as standards during HAP chromatography and were eluted with 75 to 150 mM and 220 to 350 mM phosphates, respectively. The different fractions of eluted plasmids were either mock treated or treated with S1 nuclease. The DNAs of various fractions were sufficiently resolved by agarose gel electrophoresis, Southern blotted, and autoradiographed. The slower migrating bands represent multimeric forms of the 12.5-kb YCpO−-2A. The quantitation of DNA was carried out by measuring the intensities of the 12.5-kb DNA bands in the appropriate lanes with Kodak ID2.0 software. The single-stranded DNA-containing fractions were derived from lanes 11 and 13 (without S1) and 12 and 14 (with S1). Similarly, only the double-stranded DNA fractions from the yeast source were recovered from lanes 15 and 17 (without S1) and 16 and 18 (with S1). The positions of the 12.5-kb DNAs are marked by arrows. (D) Integrity of double copies of DNA-A during replication in yeast. Panel i, the DNA present at the HindIII site of the yeast-derived plasmids was amplified by using primers YCpFwd and YCpRev. The purified 5.6-kb amplified product (lane 1) was digested with BamHI (position 960) to obtain three fragments, of 2.7 kb (a), 1.8 kb (b), and 1.1 kb, respectively (lane 3). The top diagram shows the locations of CRs in the BamHI fragments. Panel ii, fragments a and b containing one origin (CR) were each purified and used as templates for amplification with duplex PCR primer combinations as indicated in the table. For each amplification, two bands of the expected sizes were obtained. The relative positions of the primers employed are shown at the top. The IMYMV DNA-A was used as control template C. (E) Model indicating possible modes of replication of YCpO−-2A plasmid in yeast. We hypothesize that only one of the origins, either ori1 (i) or ori2 (ii), is able to initiate RCR generating full-length YCpO−-2A.
Purification of recombinant Rep protein.
The DNA containing the AC1 (Rep) ORF was amplified from the wild-type and mutagenized templates by using gene-specific primers with BamHI and HindIII sites at the N and C termini, respectively, and was cloned into vector pGEMT (Promega). AC1 was then excised as a BamHI-HindIII cassette and recloned into pET28a. Escherichia coli strain BL-21 (DE3) harboring the recombinant plasmid was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), resuspended in buffer A (20 mM Tris-HCl [pH 8.0], 300 mM NaCl, 0.1% Triton X-100, 1 mM dithiothreitol [DTT], 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml, and 1 mg of benzamidine/ml), and lysed by sonication. The soluble fraction of the sonic lysate was subjected to chromatography through a Ni-nitrilotriacetic acid column. The bound proteins were eluted with buffer A containing 250 mM imidazole and were dialyzed against buffer B (25 mM Tris-HCl [pH. 8.0], 75 mM NaCl, 2.5 mM EDTA, 5.0 mM MgCl2, 2.5 mM DTT, 25% glycerol). The dialysate was stored at a concentration of 500 ng/μl at −20°C until use.
In vitro assay for cleavage function of Rep.
A 26-mer Ori-Wt oligonucleotide (5′-CGACTCAGCTATAATATTACCTGAGT-3′) encompassing the hairpin region of IMYMV DNA-A was 5′ end labeled by using T4 polynucleotide kinase. Approximately 1 ng of the primer was incubated with 5 μg of purified Rep protein in 20 μl of cleavage buffer (25 mM Tris-HCl [pH 8.0], 75 mM NaCl, 2.5 mM EDTA, 5.0 mM MgCl2, 2.5 mM DTT) at 37°C for 30 min. The reaction was terminated by the addition of 6 μl of loading buffer (1% sodium dodecyl sulfate [SDS], 25 mM EDTA, 10% glycerol) and heating to 90°C for 2 min. The products were resolved in a 15% acrylamide-urea gel and analyzed by autoradiography. One unit of activity of the site-specific cleavage was defined as the activity required to cleave 50% of 1 ng of 5′-labeled 26-mer oligonucleotide at 37°C in 30 min. A 5′-labeled 18-mer oligonucleotide is released after the cleavage reaction.
Indirect immunofluorescence staining for confocal imaging.
Transformed yeast cells grown to mid-log phase were fixed and permeabilized according to standard protocols. The cell suspension was layered onto poly-lysine-coated coverslips, blocked with 3% bovine serum albumin in phosphate-buffered saline, and immunostained with an Indian cassava mosaic virus coat protein-specific primary antibody and an Alexa fluor 488 (Molecular Probes)-labeled goat anti-rabbit immunoglobulin G secondary antibody. The cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) (0.2 μg/ml) for 20 min. Confocal laser scanning (Radiance 2100; Bio-Rad) was performed with a Nikon microscope (×60/1.4 oil objective; Plane Apo, Tokyo, Japan). The excitation wavelength for Alexa fluorescence was 488 nm (argon laser), and fluorescence was detected through emission filter HQ515/30 (high-quality band pass, centered at 515 nm, with a 30-nm bandwidth). DAPI fluorescence was excited by a blue diode (405 nm) and detected through emission filter HQ442/45.
RESULTS
Two tandem copies of the viral genome can replicate in yeast.
A 715-bp XhoI-BglII fragment containing the ARS function was removed from the yeast shuttle vector YCp50, and the resultant plasmid, YCpO−, was able to replicate only in E. coli and not in yeast (Fig. 1B and C). YCpO− served as the base vector wherein various forms of IMYMV DNA-A were cloned at the HindIII site. When one copy of the A component (Fig. 1A and B) was cloned, the recombinant plasmid (YCpO−-A) did not replicate in yeast, as evidenced by the appearance of only a few very tiny yeast colonies, which did not grow at all in Ura− culture medium (Fig. 1D, panel i). A closer look at the A genome revealed that the HindIII cloning site truncated the C terminus of ORF AC3 by four amino acids (Fig. 1A). These missing amino acids were restored in place when two copies of the HindIII-digested A genomes were cloned in tandem, resulting in plasmid YCpO−-2A For or YCpO−-2A Rev (Fig. 1B). With both of these plasmids, the origin function was recovered to about 34% that obtained with the YCp50 plasmid (Fig. 1C and D). Significantly, neither one nor two tandem copies of only the replication origin, i.e., CR-A, could confer a DNA replication function on the YCpO− plasmid (Fig. 1D, panel i). Hence, the CR-A segment alone does not have any ARS-related activity in yeast cells.
Virus-specific elements and factors determine the replication function of the plasmid YCpO−-2A.
For determination of the virus specificity of replication of the recombinant plasmid YCpO−-2A, a battery of site-specific mutants of the DNA-A component was constructed, and the origin activities of the plasmids bearing two tandem copies of the mutant A component were assayed. The seventh and eighth nucleotides of the invariant 9-mer sequence were changed from TA to CG to generate the mutated origin OriM. The plasmid YCpO−-OriM-2A had about 30 times less colony-forming ability in yeasts than the YCpO−-2A construct (Fig. 1D, panel ii). As shown in Fig. 1E, panel ii, a 5′-labeled 26-mer oligonucleotide containing the OriM mutation, i.e., Ori*M (O), could not be site-specifically cleaved even by excessive amounts of the recombinant IMYMV Rep protein (lane 6). Hence, it is possible that RCR initiation was extremely inefficient with the OriM plasmid. Another origin mutant, i.e., OriM′, with an A→T mutation 32 nucleotides upstream of the 5′ end of the 9-mer sequence, was constructed (YCpO−-OriM′-2A) in which neither the DNA binding nor the specific cutting site of the recombinant IMYMV Rep protein was affected (data not shown). A similar mutation in tomato golden mosaic virus DNA did not alter the replication origin activity of tomato golden mosaic virus (17). As expected, the replicative efficiency of YCpO−-OriM′-2A was the same as that of plasmid YCpO−-2A (Fig. 1D, panel ii).
IMYMV Rep contains many motifs that are characteristic of nicking-closing or type 1 topoisomerase-like enzymes, and a bioinformatic analysis revealed that a tyrosine residue (Y103) at conserved motif III could be responsible for the site-specific nicking-closing action (15). Accordingly, a mutation, Y103F, was introduced by an A→T change at nucleotide 1553 of the DNA-A genome, and the resulting plasmid construct was named YCpO−-Y103F-2A. The colony-forming ability with this plasmid was also about 20 times lower than that with the YCPO−-2A plasmid (Fig. 1D, panel ii). The site-specific nicking activity of the recombinant Y103F-Rep was also lost when the assay was performed with the Ori-Wt oligonucleotide (Fig. 1E, panel ii, lane 7). Thus, the failure of replication of the mutant YCpO−-2A plasmid (YCpO−-Y103F-2A) correlated with the loss of site-specific nicking at the origin. To address this point further, a termination (amber; T) coupled with a frameshift (F) mutation was engineered after the second codon of Rep by inserting a T nucleotide at position 1253 of DNA-A, and the corresponding plasmid construct was called YCpO−-AC1TF-2A. Though we expected no growth of yeast with this plasmid, transformants could still be observed. However, the efficiency of transformation was about 12-fold less than that with plasmid YCpO−-2A (Fig. 1D, panel ii). It remains to be seen if some leaky expression of Rep was responsible for this residual growth.
Since the AC3 ORF codes for REn (27), an amber termination coupled with a frameshift mutation (TF) was introduced after the 19th amino acid of the AC3 ORF by the addition of an “A” nucleotide at position 2438 of DNA-A, and the corresponding mutant plasmid was called YCpO−-AC3TF-2A. The transformation efficiency with the mutant plasmid was about fivefold less than that with plasmid YCpO−-2A (Fig. 1D, panel ii). Thus, this result corroborates the earlier finding that the AC3 protein functions as an accessory factor for replication (27, 29).
Usage of the model system.
Using the yeast transformation assay, we explored the roles of the AC4 and AC5 proteins. The reading frames of Rep and AC4 are different but overlapping, as shown in Fig. 1A. An opal codon (T) at the 9th residue, a cysteine (C9Op), of the AC4 ORF was introduced by a C→A change at position 1429 of DNA-A, and the resulting recombinant plasmid was called YCPO−-AC4T-2A. The colony-forming ability was about 30-fold less with this mutant plasmid (Fig. 1D, panel ii). Although this mutation in AC4 resulted in an L62I change in Rep, recombinant His-tagged L62I Rep was as efficient as Wt-Rep in site-specific cutting activity (Fig. 1E, panel ii, compare lanes 2 and 9). The measured specific activities of Wt-Rep, Y103F Rep, and L62I Rep were 380, 5, and 320 U/μg, respectively. Moreover, this mutation did not affect the DNA binding or ATPase activity of Rep (data not shown). These results indicate that Rep-mediated replication initiation might not be a problem for plasmid YCpO−-AC4T-2A. However, it remains to be determined whether the transcription control of Rep or/and other essential functions of Rep, such as the interaction pattern with other yeast replicative factors, are affected by the AC4 mutation. The next ORF examined was AC5, which partly overlaps with the ORF encoding CP but is transcribed in an opposite direction. An opal codon (T) was introduced at the 7th codon of AC5 (i.e., C7Op) by a C→A change at the 159th nucleotide position, and the corresponding plasmid was called YCpO−-AC5T-2A. The transformation efficiency of this plasmid was reduced about sevenfold compared to that of plasmid YCpO−-2A (Fig. 1D, panel ii). Thus, studies with the yeast model predict that the hitherto uncharacterized AC5 protein contributes significantly to geminiviral DNA replication and also suggest that the AC4 protein might be linked directly or indirectly to the viral replication process.
Plasmid cdc6-YCpO−-2A complements the host cdc6 defect.
The growth-related genetic defect of the yeast host can be complemented by a replicating plasmid incorporating the appropriate host gene. A validation of this principle was illustrated with the plasmid YCpO−-2A and the yeast cdc6 gene. The yeast CDC6 is required for prereplicative complex formation at the yeast replication origins (7), and hence S. cerevisiae cdc6(ts) fails to grow at 37°C. However, the mutant yeast harboring a known, episomally replicating plasmid, cdc6-YEp352 (YEp352 plasmid with cloned cdc6 gene), grows normally (20) at the nonpermissive temperature (Fig. 1F). A 3.8-kb EcoRI fragment containing the cdc6 gene along with its promoter was derived from the cdc6-YEp352 plasmid, its ends were engineered, and finally it was cloned into the BglII site of plasmid YCpO−-2A. The transformation efficiencies of the resultant plasmid, cdc6-2A-For, and the control plasmid, cdc6-YEp352, were similar (341 versus 502) when the colonies were scored on Ura dropout plates. As seen in Fig. 1F, the cdc6 defect of S. cerevisiae at the nonpermissive temperature was complemented quite efficiently by plasmid cdc6-YCpO−-2A. These observations confirm that plasmid YCpO−-2A can act as an independently replicating vector in yeast.
The characteristics of episomal RCR products of YCpO−-2A.
The similarity in the efficiencies of yeast transformation mediated by plasmid YCpO−-2A and a known, extrachromosomally replicating group of plasmids, namely YCp50 and cdc6-YEp352, strongly suggested the episomal nature of plasmid YCpO−-2A in yeast. To reveal some of these episomal characteristics, we biochemically compared YCpO−-2A plasmid DNAs isolated from E. coli (Cairn's mode of replication) and yeast (RCR mode) sources, since the modes of plasmid YCpO−-2A replication in these two hosts were supposedly different. The copy number (11) of plasmid YCpO−-2A in yeast was lower (≤20-fold) than that in E. coli, and this difference perhaps occurred due to the presence of the CEN4 region, which was functional only in yeast (6). When they were linearized with either XhoI or BglII, the E. coli-derived products migrated differently from unrestricted DNA, whereas such changes were not obvious with the yeast-derived products. Both forms generated a 2.7-kb viral DNA-A fragment as well as the YCpO− backbone of 7.2 kb when digested with the HindIII enzyme (Fig. 2A). As shown in Fig. 2B, the yeast-derived products were more sensitive to limited S1 nuclease treatment than the ones derived from E. coli. These observations suggested that the overall sizes of the circular portions of monomeric DNA derived from both sources was perhaps similar but that the yeast forms had more single-stranded DNA. The chromosomal DNA markers cdc6, orc2, and gal4 did not hybridize at the banding position of YCpO−-2A DNA, thus ruling out the integration of YCpO−-2A with the yeast chromosomal DNA (data not shown). Since the presence of a single strand in the replicated DNA is the hallmark of RCR, the aspects of single-strandedness were examined further. The isolated DNA products were subjected to chromatography through a hydroxyapatite (HAP) column and step eluted by using defined concentrations of phosphates. Figure 2C shows that the E. coli-derived products were double stranded and S1 resistant, whereas about 10 to 15% (occasionally even 30%) of the yeast-derived products were mostly single stranded and were partially digestible by S1 nuclease treatment. The ratio of the 12.5-kb DNA recovered from the single-strand-containing fractions (Fig. 2C, lanes 11 and 13) to the amount obtained from the fractions containing only double-stranded forms of DNA (Fig. 2C, lanes 15 and 17) gave a measure of the fractional yield of single-stranded DNA.
We expected a release of 2.7 kb of viral DNA after RCR of plasmid YCpO−-2A from both origins (24, 26). However, no such release or deletion was detected in Southern blotting experiments (Fig. 2A, lane 6, and Fig. 2C, lanes 15 and 16). This observation forced us to look carefully at the size of the viral DNA insert present in the HindIII cloning region of the plasmid that had replicated in yeast. This region was PCR amplified by using primers flanking the cloning site. The amplified product of 5.6 kb was purified and digested with BamHI, a unique cutter of the viral DNA-A component. The pattern of digestion (shown in Fig. 2D, panel i) revealed that two copies of DNA-A were present and that there was no major deletion of viral sequences in the replicated DNA. The expected CRs of the 2.7- and 1.8-kb BamHI fragments (Fig. 2D, panel i, top) were amplified separately by using the X primer combination, and no significant deletion was detected in the estimated 230-bp CR sequences (Fig. 2D, panel ii, compare the upper bands of lanes 2 and 3 with that of lane 1 or similar bands of lanes 5 and 6 with that of lane 4). DNAs from the RCR origin (i.e., the nicking point of the Rep protein) to the 5′ and 3′ termini were also amplified by using the Z and Y primer combinations, respectively. As shown in Fig. 2D, panel ii, no deletions were detected in those fragments (about 120 and 110 bp) (lower bands of lanes 4 to 6 and lanes 1 to 3, respectively). Subsequently, the amplified CR DNAs, isolated from each of the lanes 1 to 3 shown in Fig. 2D, panel ii, were sequenced, and the nucleotide sequences of all of them were found to be exactly the same, i.e., without even a single base change in the 229-bp DNA. Similar PCR-based assays were also performed with three other regions of the YCpO− backbone, and in none of the cases was any deletion observed (not shown), indicating that no rearrangement occurred during RCR. The maintenance of integrity of the viral DNA sequences led us to propose the model shown in Fig. 2E. Of the two origins of YCpO−-2A, only one might initiate at a time, and the replication synthesis perhaps terminates at the site from where it begins. With this model, therefore, we expect no release of viral DNA after RCR.
Expression of viral genes from plasmid YCpO−-2A.
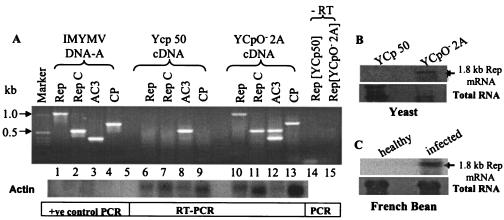
Active viral factors must have been produced in yeast so that plasmid YCpO−-2A DNA could replicate. For visualization of the presence and levels of viral transcripts and proteins, a few candidate viral targets, namely, the AC1, AC3, and CP genes, were chosen (Fig. 3 and 4). Reverse transcription (RT)-PCR data, obtained with the total RNA (4) from yeasts harboring the appropriate plasmids, revealed the presence of viral transcripts of expected sizes (Fig. 3A, lanes 10 to 13), and such transcripts were not found in the absence of the viral genome (Fig. 3A, lanes 6 to 9). The isolated RNA samples were also examined by Northern blot analysis using radiolabeled Rep DNA. Figure 3B shows the presence of a 1.8-kb Rep mRNA in yeast harboring the YCpO−-2A plasmid. Rep-specific mRNA of the same size was also detected in the leaves of IMYMV-infected French bean plants (Fig. 3C). Hence, it seems that the viral transcripts produced in yeast and plants were equivalent.
FIG. 3.
Viral transcripts in S. cerevisiae bearing plasmid YCpO−-2A and in infected French bean plants. (A) RT-PCR from RNAs of yeast samples transformed with various plasmids. The PCR products of cDNAs derived from yeast samples (4) bearing YCpO−-2A plasmids are shown for the pairs of primers specific for the Rep (lanes 10 and 11), AC3 (lane 12), and CP (lane 13) genes of IMYMV. RepC is the C-terminal part of Rep. The expected pattern of amplification is shown in lanes 2 to 5 of the positive (+ve) control PCR panel that was obtained from direct amplification from a template of viral DNA-A. The cDNA prepared from the total RNA extracted from YCp50 plasmid-bearing yeast was used as a negative control (lanes 6 to 9). A negative (−RT) control is shown in lanes 14 and 15 indicating that the PCR products are not the artifacts of DNA contamination in the processed RNA samples. A nonspecific band at around 550 bp was observed for both YCp50 control and YCpO−-2A samples in the case of AC3 primers (lanes 8 and 12). Southern blotting with an actin probe was carried out to show uniformity in the loading of cDNA templates (bottom panel). (B) Northern blot analysis of total RNA extracted from the yeast strain W303a bearing the YCpO−-2A plasmid. The presence of a 1.8-kb Rep transcript was revealed by probing with radiolabeled AC1 DNA. Such a transcript was absent from the RNA samples of yeast transformed with the YCp50 control plasmid, as expected. This result is consistent with the observation made with IMYMV-infected French bean plants (C). The total RNAs isolated from the appropriate sources are shown at the bottom for panels B and C.
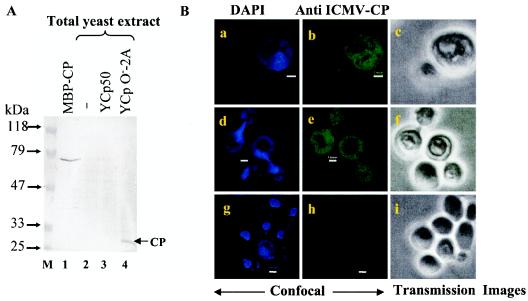
FIG. 4.
Expression of the viral CP in S. cerevisiae transformed with YCpO−-2A. (A) Western blot analysis of IMYMV CP. The total protein was prepared from untransformed and transformed yeast by resuspension of the cell pellet from 1 ml of culture, obtained at the mid-log phase, in 1× sample buffer (0.06 M Tris-HCl [pH 6.8] 10% [vol/vol] glycerol, 2% [wt/vol] SDS, 5% [vol/vol] 2-mercaptoethanol, 0.0025% [wt/vol] bromophenol blue) and boiling for 10 min. The plasmids used for transformation are indicated at the top of each lane (lanes 2 to 4). Fifty micrograms of each protein sample was separated in an SDS-10% polyacrylamide gel and immunoblotted with a heterologous Indian cassava mosaic virus CP antibody. Three hundred nanograms of purified maltose binding protein fusion of CP (MBP-CP) was used as a positive control. (B) Confocal imaging of yeast cells bearing the YCpO−-2A plasmid. The CP was expressed throughout the yeast cells (b and e), and the recognition specificity of the CP antibody was highlighted by the absence of CP staining in the control cells harboring plasmid Ycp50 (h). Panels a, d, and g show nuclei stained with DAPI, and panels c, f, and i show transmission images in white light. Bars, 1 μm. The same sets of cells are shown in rows, and cells at different growth stages are shown in columns.
Fig. 4A shows that the CP, with an expected size of 28 kDa, was expressed in cells bearing the YCpO−-2A plasmid (lane 4). Since the measured intensity of the recombinant IMYMV CP (i.e., maltose binding protein fusion of CP) was about two times that of yeast-expressed CP (compare lanes 1 and 4), the estimated expression of CP was about 0.3% of the total soluble proteins of yeast. It was reported earlier that the CP of a member of the Begomovirus family, namely Squash leaf curl virus, localized in the nucleus of the Xanthi protoplast (22). However, our confocal imaging studies revealed that IMYMV CP was present in the yeast nucleus as well as the cytosol, but not in the large vacuolar region. The CP was partitioned evenly in the small buds (Fig. 4B, panels b and e).
DISCUSSION
Taken together, the data presented above establish that budding yeast supports geminiviral DNA replication and that the plasmid YCpO−-2A replicates by using the viral replication origin and virus-encoded factors. The viral factors were perhaps required for essential replication initiation, whereas the yeast factors provided the replication elongation and maturation machinery. A similar partitioning of function might be required within the host plant itself for the replication of geminiviral genomes (9). Given that plants and yeast (S. cerevisiae) are divergent in evolution, it is indeed remarkable to observe the conservation in the mechanism and machinery of replication in these systems. However, the reported model system may not exactly mimic the plant host with regard to the nature and copy numbers of the replicated products, their subcellular distributions, the activities of the viral proteins, etc. Despite these shortcomings, the development of the yeast model has many advantages, as mentioned below.
The model system has already identified the AC5 viral factor as an important contributor to IMYMV DNA replication. The direct or indirect effect of the ORF AC4 in viral replication remains to be elucidated in detail. By using the genome-wide mutation screen applied to both viral and yeast genomes, it will be possible to identify factors responsible for YCpO−-2A plasmid DNA replication. A large repertoire of yeast mutants is already available. Hence, the rational usage of these mutants might pinpoint the host factors required for viral DNA replication. A yeast genome-wide two-hybrid analysis using viral replication factors as baits may select most of the yeast factors that are necessary for all facets of IMYMV DNA replication. The equivalent plant factors can subsequently be identified on the basis of structural and functional homologies. The availability of yeast cell extract (25) might pave the way for in vitro reconstitution of the replication of viral DNA. In other words, the system described here opens up avenues of exploration that were hitherto not possible with currently available protocols in plant virology. In addition, the yeast model can also serve as a screening system for the rapid identification of inhibitors of viral replication. The fact that yeast can be used as a model system for such studies was highlighted first by studies on RNA viruses such as flock house virus and brome mosaic virus (8, 23). Our studies raise the possibility that the yeast system could also be used as a host for other DNA viruses that are currently difficult to study in their natural hosts. This possibility may not be very remote, since papillomavirus DNA replication has already been established in S. cerevisiae (3, 32).
Acknowledgments
We are grateful to all of the colleagues and readers, especially D. Chattoraj and K. V. S. Rao, whose valuable comments have enriched the manuscript. Gratitude is expressed to Nilanjan Roy, Pratima Sinha, and Justin Donato for their gifts of plasmids and strains. The help of C. Tanwar with confocal microscopy is duly acknowledged.
The partial financial assistance of DBT, New Delhi, India, is acknowledged. Vineetha Raghavan and Punjab S. Malik were supported by Senior Research Fellowships from the Council of Scientific and Industrial Research, New Delhi, India, and University Grants Commission, New Delhi, India, respectively.
REFERENCES
- 1.Ach, R. A., T. Durfee, A. B. Miller, P. Taranto, L. Hanley-Bowdoin, P. C. Zamryski, and W. Gruissem. 1997. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 17:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agatep, R., R. D. Kirkpatrick, D. L. Parchaliuk, R. A. Woods, and R. D. Gietz. 1998. Transformation of Saccharomyces cerevisiae by lithium acetate/single stranded DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol. Technical Tips Online [Online.] http://tto.trends.com.
- 3.Angeletti, P. C., K. Kim, F. J. Fernandes, and P. F. Lambert. 2002. Stable replication of papillomavirus genomes in Saccharomyces cerevisiae. J. Virol. 76:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., London, United Kingdom.
- 5.Castillo, A. G., D. Collinet, S. Deret, A. Kashoggi, and E. R. Bejarrano. 2003. Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 312:381-394. [DOI] [PubMed] [Google Scholar]
- 6.Clark, D. D., and B. R. Peterson. 2003. Analysis of protein tyrosine kinase inhibitors in recombinant yeast lacking the ERG6 gene. Chembiochem 4:101-107. [DOI] [PubMed] [Google Scholar]
- 7.Cocker, J. H., S. Piatti, C. Santacanale, K. Nasmyth, and J. F. X. Diffley. 1996. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379:180-182. [DOI] [PubMed] [Google Scholar]
- 8.Duane Price, B., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez, C. 2000. DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J. 19:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeske, H., M. Lutgemeier, and W. Preiss. 2001. DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic virus. EMBO J. 20:6158-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohzaki, H., Y. Ito, and Y. Murakami. 1999. Context-dependent modulation of replication activity of Saccharomyces cerevisiae autonomously replicating sequences by transcription factors. Mol. Cell. Biol. 19:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong, L. J., and L. Hanley-Bowdoin. 2002. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14:1817-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, B., S. A. A. Behjatnia, I. B. Dry, J. W. Randles, and M. A. Rezaian. 2003. High-affinity Rep-binding is not required for the replication of a geminivirus DNA and its satellite. Virology 305:353-363. [DOI] [PubMed] [Google Scholar]
- 14.Luque, A., A. P. Sanz-Burgos, E. Ramirez-Parra, M. M. Castellano, and C. Gutierrez. 2002. Interaction of geminivirus Rep protein with replication factor C and its potential role during geminivirus DNA replication. Virology 302:83-94. [DOI] [PubMed] [Google Scholar]
- 15.Orozco, B. M., A. B. Miller, S. B. Settlage, and L. Hanley-Bowdoin. 1997. Functional domains of a geminivirus replication protein. J. Biol. Chem. 272:9840-9846. [DOI] [PubMed] [Google Scholar]
- 16.Orozco, B. M., and L. Hanley-Bowdoin. 1996. A DNA structure is required for geminivirus replication origin function. J. Virol. 70:148-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orozco, B. M., H. J. Gladfelter, S. B. Settlage, P. A. Eagle, R. N. Gentry, and L. Hanley-Bowdoin. 1998. Multiple cis elements contribute to geminivirus origin function. Virology 242:346-356. [DOI] [PubMed] [Google Scholar]
- 18.Pant, V., D. Gupta, N. R. Choudhury, V. G. Malathi, A. Varma, and S. K. Mukherjee. 2001. Molecular characterization of the Rep protein of the blackgram isolate of Indian mung bean yellow mosaic virus. J. Gen. Virol. 82:2559-2567. [DOI] [PubMed] [Google Scholar]
- 19.Paximadis, M., A. M. Idris, I. Torres-Jerez, A. Villarreal, M. E. Rey, and J. K. Brown. 1999. Characterization of tobacco geminiviruses in the Old and New World. Arch. Virol. 144:703-717. [DOI] [PubMed] [Google Scholar]
- 20.Piatti, S., C. Langauer, and K. Nasmyth. 1995. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a “reductional” anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14:3788-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preiss, W., and H. Jeske. 2003. Multitasking in replication is common among geminiviruses. J. Virol. 77:2972-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin, S., B. M. Ward, and S. Lazarowitz. 1998. The bipartite geminivirus coat protein aids BR1 function in viral movement by affecting the accumulation of viral single-stranded DNA. J. Virol. 72:9247-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires co-expression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. USA 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rigden, J. E., I. B. Dry, L. R. Krake, and M. A. Rezaian. 1996. Plant virus DNA replication processes in Agrobacterium: insight into the origins of geminiviruses? Proc. Natl. Acad. Sci. USA 93:10280-10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seki, T., and J. F. X. Diffley. 2000. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl. Acad. Sci. USA 97:14115-14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selth, L. A., J. W. Randles, and M. A. Rezaian. 2002. Agrobacterium tumefaciens supports DNA replication of diverse geminivirus types. FEBS Lett. 516:179-182. [DOI] [PubMed] [Google Scholar]
- 27.Settlage, S. B., A. B. Miller, W. Gruissem, and L. Hanley-Bowdoin. 2001. Dual interaction of a geminivirus replication accessory factor with a viral replication protein and a plant cell cycle regulator. Virology 279:570-576. [DOI] [PubMed] [Google Scholar]
- 28.Stanley, J. 1995. Analysis of African cassava mosaic virus recombinants suggests strand nicking occurs within the conserved nonanucleotide motif during the initiation of rolling circle DNA replication. Virology 206:707-712. [DOI] [PubMed] [Google Scholar]
- 29.Sunter, G., M. D. Hartitz, S. G. Hormuzdi, C. L. Brough, and D. M. Bisaro. 1990. Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology 179:69-77. [DOI] [PubMed] [Google Scholar]
- 30.Xie, Q., A. P. Sanz-Burgos, H. Guo, J. A. Garcia, and C. Gutierrez. 1999. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39:647-656. [DOI] [PubMed] [Google Scholar]
- 31.Xie, Q., P. Suarez-Lopez, and C. Gutierrez. 1995. Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. EMBO J. 14:4073-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao, K. N., and I. H. Frazer. 2002. Replication of bovine papillomavirus type 1 (BPV-1) DNA in Saccharomyces cerevisiae following infection with BPV-1 virions. J. Virol. 76:3359-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]