Abstract
Objectives
Calumenin, a molecular chaperone, exerts a regulatory effect on the vitamin K-dependent γ-carboxylation redox cycle that inhibits transfer of the reduced vitamin K from VKORC1, the pharmacological target of warfarin, to the γ-carboxylase. Because of its polymorphic structure and central role in the warfarin metabolic pathway, a contributory role for calumenin to warfarin dose variability has been posited. The current study sought to validate modulation of therapeutic dosing requirements by a single nucleotide polymorphisms (SNP) occurring in the calumenin gene (CALU) reported in previous studies. The CALU SNP was further modeled to detect interaction with SNPs occurring in VKORC1, CYP2C9, and CYP4F2 genes and characterize any additional contribution to variability in therapeutic warfarin dose requirement.
Setting
The study was undertaken in an established, well-characterized cohort of subjects treated with warfarin in the Anticoagulation Clinic of Marshfield Clinic in Marshfield, Wisconsin.
Methods
Subjects (N=491) previously genotyped for SNPS known to contribute variability to therapeutic warfarin dose requirement were genotyped for CALU SNP rs1043550, using TaqMan assays. Contribution of CALU SNP rs1043550 was modeled relative to other genotypic and phenotypic characteristics including gender, diagnosis, age, body surface area, underlying indication for warfarin, comorbidities, and pharmacological exposures. Interaction between SNPs impacting on warfarin dose requirements and calumenin SNPs was also modeled.
Results
Small differences in warfarin dosing requirements detected among individuals encoding the mutant G allele in the calumenin SNP were not statistically or clinically significant relative to therapeutic warfarin dose requirement and did not independently contribute significantly to the warfarin dosing model. Interaction between calumenin and VKORC1 SNPs contributed only minor additional variability to that ascribed to the wild type VKORC1 genotype.
Conclusions
The impact of the CALU SNP on warfarin dose variability was minor and did not contribute significantly to therapeutic warfarin dose requirement in our study cohort. While no contribution was noted for the SNP examined in the present study, further examination of interaction between genetic elements contributing major impact on therapeutic warfarin dose requirements and genes exhibiting a lesser contribution is warranted.
Keywords: Calumenin, Warfarin, Vitamin K, Anticoagulation, VKORC1
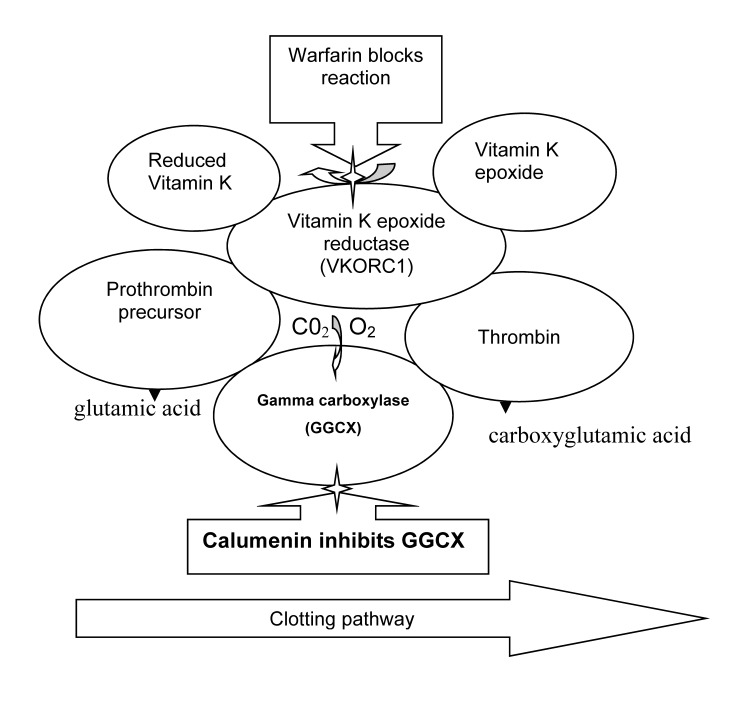
Warfarin effectively prevents thromboembolic events in patients, yet not without significant risk of adverse outcomes including serious bleeding events. The established mechanism of action for warfarin involves the drug’s inhibition of the enzyme vitamin K epoxide reductase complex, subunit 1 (VKORC1). VKORC1 catalyzes the redox reaction involved in regenerating vitamin K to its reduced form, which is required by the gamma carboxylase enzyme as a cofactor in converting prothrombin to thrombin (figure 1).1,2 Historically, approximately 20% of variability in warfarin dosing has been associated with the cumulative variability attributable to individual factors including age, presence of co-morbidities such as diabetes, cancer, renal or liver disease, diet, gender, smoking, and some concomitant medications. Genetic polymorphisms occurring in coding regions of the VKORC1, cytochrome P450 metabolic enzymes 2C9 (CYP2C9) and 4F2 (CYP4F2), collectively contribute an additional 35% of variability in therapeutic warfarin dose requirement.3 While dosing variability contributed by these factors has been validated in multiple populations, approximately 40% of variability remains unexplained.
Figure 1.
Diagram of the interactions between overlapping enzymatic pathways regulating clotting. Reduction in GGCX activity through in the presence of increased calumenin expression perturbs the vitamin K-dependent activity of VKORC1 whose enzymatic action regenerates vitamin K to its reduced state. Compensatory overexpression of VKORC1 results and is associated with increased resistance to warfarin.6
Recent research to explain residual variability in therapeutic warfarin dosing requirement has focused on the contribution of additional genetic polymorphisms in several proteins. Several studies have suggested a role for calumenin in coagulation, especially in the context of higher warfarin dose requirements. Calumenin, encoded by the CALU gene, is a 315 amino acid, 37 kDa, water-soluble, acidic protein that maps at human chromosome 7q32.4,5 The molecule belongs to a family of similar calcium binding proteins including reticulocalbin, Erc-55, and Cab-45, often referred to by the acronym CREC.4 CREC proteins share structural and functional similarities. Calumenin is a molecular chaperone localized in the endoplasmic reticulum and Golgi body and is the only CREC protein found throughout the secretory pathway.5 The CALU gene is polymorphic, with a total of 548 SNPs documented in NCBI (http://www.genecards.org/cgi-bin/carddisp.pl?gene=CALU&snp=548#snp).
A study by Wajih et al6 in a rat model demonstrated that calumenin exerted an inhibitory regulation on the gamma carboxylase (GGCX) enzyme in the vitamin K pathway that is central to its regeneration as a cofactor in the coagulation cascade (figure 1). By silencing the calumenin gene, a 5-fold increase in gamma carboxylase activity was noted.6 This study, that examined both normal rats and those with genetically encoded resistance to warfarin, demonstrated the central role of calumenin in the coagulation cascade and suggested a potential for increased warfarin resistance in association with SNPs that impact on the regulatory activity of calumenin.
Further, several studies have confirmed that, in humans, SNPs occurring in the CALU gene are associated with higher warfarin dose requirements in some populations. A study done in Israel by Vecsler et al7 enrolled ethnicities including Ashkenazi, Iraqi, and North Africans found that genetic profiles consistent for wild type VKORC1 and CYP2C9 but encoding variant allele G11A (R4Q) in the CALU gene (rs2290228) required highest warfarin doses. The highest frequency of patients encoding the variant ‘A’ allele at that site was found among the upper 10th percentile for warfarin dose (P <0.02), with a patient homozygous for the mutant allele requiring an exceptionally high daily warfarin dose of 20 mg.
Despite the association seen with rs2290228 in the Vecsler study,7 we chose to examine a different allelic variant, A29809G (rs1043550), occurring in the 3′UTR region, based on the following rationale: (1) the ethnicity of Vecsler7 study cohort that exhibited an increase in warfarin dose requirement as a factor of the rs2290228 allele did not align with that of the Northern European, Marshfield cohort whose heritage is predominantly German; (2) the minor allele frequency for rs1043550 is well represented in a Caucasian population; and (3) the study by Gonzalez-Conejero et al8 observed that CALU A29809G (rs1043550) enhanced the effect of VKORC1 1173C>T on acenocoumarol dose in a statistically significant manner (P = 0.001) in a Spanish Caucasian population. Therefore, it was of interest to see if this allele would impact warfarin dose in our Caucasian, northern European cohort, especially among patients requiring higher warfarin dosing.
Previous studies suggested that calumenin may contribute to variability in warfarin or acenocoumarol therapeutic dosing requirements, and observation of dosing outliers.8–10 The present study, undertaken in a previously modeled warfarin cohort at Marshfield Clinic, sought to validate putative lower therapeutic warfarin dosing requirements among individuals encoding the wild type allele of the CALU SNP rs1043550 (A29809G), as had previously been reported for CALU SNPs examined in other phenotypically and genotypically modeled Caucasian cohorts.11–13
Methods and Materials
Study Cohort, Setting, and Design
The study cohort was drawn from a population residing in the service area of Marshfield Clinic, which serves the population of northern and central Wisconsin. This population is predominantly Caucasian, of Northern European (largely German) descent. The cohort consisted of subjects on warfarin who were previously modeled for factors affecting therapeutic warfarin requirements. This cohort included 491 subjects enrolled in a study approved by the institutional review board for human subjects’ research for whom all other genetic and clinical data were available. Phenotypic characterization included gender, age, body surface area, underlying indication for warfarin, comorbidities, and pharmacological exposures. Subjects were previously genotyped for relevant SNPs of the CYP2C9, VKORC1, and CYP4F2 genes.12,13 Genotype determinations for SNP rs1043550 were not possible for two patients in the present study; therefore these patients were excluded from further analysis. Warfarin management of these subjects was accomplished by nurses in the Anticoagulation Clinic at Marshfield Clinic through use of algorithm-driven protocols for standardized patient anticoagulation assessment, warfarin dosing, laboratory coagulation testing, and documentation. The study cohort had excluded patients known to have underlying conditions that would alter drug metabolism, such as cancer, renal or hepatic insufficiency, or congestive heart failure. Patients under therapeutic management with long-acting barbiturates were also excluded.
Genotyping
Genomic DNA previously extracted from whole blood using the Autopure LS system (Gentra Systems Inc., Minneapolis Minnesota, USA) were used in the current study.11 Samples were encrypted with a study code before determination of genotype. Genotypes for CALU SNP (rs1043550) were determined using TaqMan assays (Applied Biosystems, Foster City, CA).14 Investigators performing the DNA analysis were blinded to phenotypic and genotypic data previously described.
Statistical Analysis
The candidate SNP was screened for association with both observed therapeutic dose and the prediction errors from the Marshfield pharmacogenetic dose prediction model,15 which includes recognized clinical (gender, age, body surface area) and genetic (CYP2C9, VKORC1, CYP4F2) factors as predictor variables. The data were reviewed graphically, and the Kruskal-Wallis test was used for statistical comparison.
Results
Gene Frequencies
The underlying indications warranting warfarin therapy and frequency distribution of genotype for CALU SNP rs1043550 are shown in table 1.
Table 1.
Frequency distribution of rs1043550 by indication for warfarin.
| AA | AG | GG | ||
|---|---|---|---|---|
| Indication | n (%) | n (%) | n (%) | Total |
| Atrial arrhythmias | 74 (28.3) | 124 (47.3) | 64 (24.4) | 262 |
| Prosthetic heart valves | 26 (27.1) | 40 (41.7) | 30 (31.2) | 96 |
| Thromboembolic disease | 15 (18.8) | 37 (46.2) | 28 (35.0) | 80 |
| Other (eg, cardiomyopathy, stroke) | 13 (24.5) | 21 (39.6) | 19 (35.9) | 53 |
| Total | 128 | 222 | 141 | 491 |
Genetic distribution of CALU SNPs found in the study cohort was as follows: AA (26%), AG (45%), and GG (29%). The allele frequency was A (49%) and G (51%). This differed from frequencies reported in the HapMap CEU population,10 (table 1, P <0.0001) but resembled other subpopulations of subjects from other studies with underlying cardiovascular disease conditions,16,17 although ethnicity differed in these studies compared to the study cohort. The study cohort showed deviation from Hardy-Weinberg equilibrium (P = 0.035), while the HapMap CEU does not (P = 0.49, see table 2).
Table 2.
Population comparison of genotype and allele frequencies for calumenin among a patient population of northern European ethnicity with an indication for warfarin compared to a population of Spanish patients with an indication for acenocoumarol and a second population with non-ST elevation acute coronary syndrome.*
| Allele Frequency | |||||
|---|---|---|---|---|---|
| Genotype Frequency | Major | Minor | |||
|
|
|||||
| Population | A/A | A/G | G/G | A | G |
| Marshfield warfarin-treated patients (N=491)* | 0.26 | 0.45 | 0.29 | 0.49 | 0.51 |
| Acenocoumarol-treated patients (Spanish population) N=20322 | 0.33 | 0.52 | 0.15 | 0.59 | 0.41 |
| nSTACS (Spanish population) N=37423 | 0.33 | 0.51 | 0.16 | 0.59 | 0.41 |
| HapMap CEU | 0.43 | 0.43 | 0.13 | 0.65 | 0.35 |
Distribution among these populations of patients with requirements for anticoagulation is compared to the HapMap CEU, a Northern European population in Utah more similar to the Marshfield population in ethnic makeup.10 Whereas the two Spanish cohorts of acenocoumarol-exposed subjects exhibit nearly identical genotype frequency distribution, distributions between the Marshfield warfarin cohort and HapMap CEU population are significantly different (P=0.035).
Genotype vs. Maintenance Dose
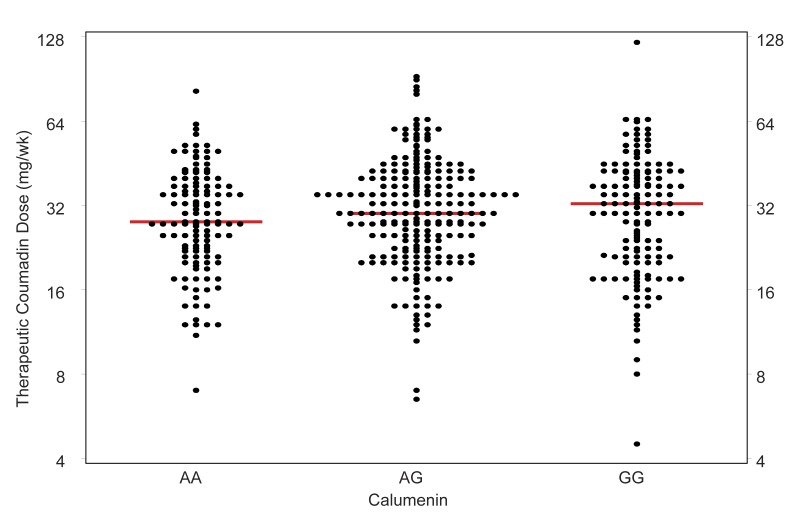
Median therapeutic warfarin dose (mg/week) varied slightly among the three calumenin genotypes: AA (28), AG (30), GG (32.5) as shown in figure 1. The data suggest that incremental periodicity of guanine composition may be marginally associated with therapeutic warfarin dosing. However, this potential association was not found to be statistically significant (P =0.376), and given the similarity of the estimates may be of limited clinical importance (table 3).
Table 3.
Warfarin dose and prediction errors by the calumenin variant.*
| AA | AG | GG | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| n | Mean | Median | n | Mean | Median | n | Mean | Median | P value | |
| Therapeutic warfarin dose (mg/wk) | 127 | 30.9 | 28.0 | 222 | 33.5 | 30 | 140 | 32.8 | 32.5 | 0.376 |
| Dose Prediction Error (mg/wk) | 127 | 0.4 | −0.9 | 222 | 1.3 | 0.4 | 140 | 1.6 | −0.1 | 0.638 |
Table based on 489 patients with complete clinical and genotypic data for the prediction model (2 patients excluded). Prediction error defined as the observed therapeutic dose minus the predicted dose.
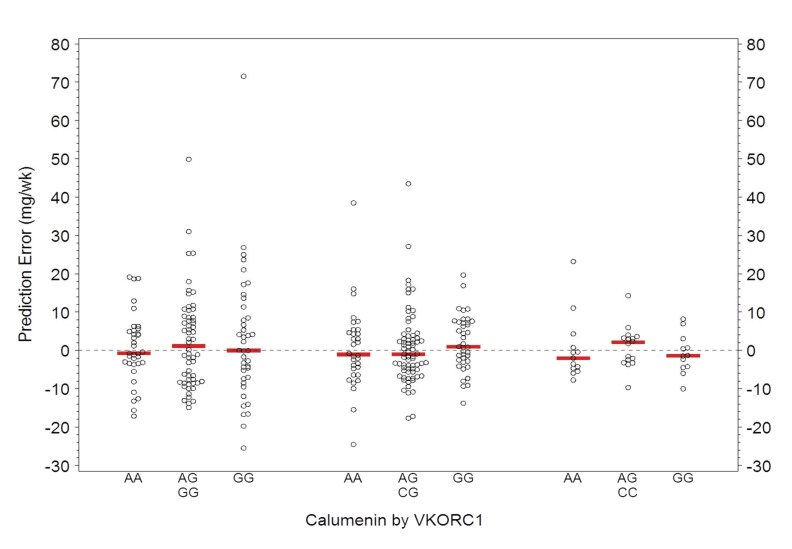
Calumenin SNP rs1045330 was not significantly associated with either therapeutic warfarin dose (figure 2) or with errors in predicted dose (figure 3) when modeled using the Marshfield pharmacogenetic dose prediction model.14 When the potential contribution of calumenin was examined with respect to VKORC1 genotype (for CYP2C9 *1/*1 only) to determine whether there was a modulatory effect, no association was observed (figure 2).
Figure 2.
Scatter plot showing therapeutic dose of warfarin for groups defined by calumenin variant (AA, AG, and GG). Each black dot corresponds to one patient. The red line indicates the median dosage of warfarin required for therapeutic anticoagulation.
Figure 3.
Scatter plot of prediction errors (actual therapeutic dose minus predicted dose) in the subset of patients with CYP2C9 *1/*1 for groups defined by calumenin (AA, AG, and GG) within VKORC1 (GG, CG, and CC). Red lines indicate the median error. Prediction errors by both calumenin and VKORC1 did not differ significantly by genotype.
Discussion
Wajih et al18 and Wallin et al19 first demonstrated that overexpression of calumenin in the liver contributed to genetic warfarin resistance. Calumenin inhibited the γ-carboxylase (GGCX), an enzyme found in the vitamin K pathway,18,19 thereby inhibiting the overall effect of warfarin (figure 1).19 This thesis was supported by studies demonstrating that siRNA silencing of calumenin enhanced functional factor IX (r-hFIX) production and overexpression of VKORC1, consequently increasing inhibition of the vitamin K-dependent cycle.6
In the current study, those patients encoding the calumenin SNP 1043550 exhibited nominal increases in mean therapeutic warfarin dosing requirements (figure 2), but these were not statistically significant, and the overall contribution to daily dosing requirement seen in this study was not deemed clinically significant. The subject numbers available provided substantial statistical power for overall tests of such an association, but power may have been limiting for detecting interactions with VKORC1. For example, using simulation,20 we estimate that the overall power would be 98% if we assume the true effect of CALU equals the moderate effect we demonstrated in our previous study for cytochrome P450 4F2 (CYP4F2).12 However, assuming that same effect size was limited to the subgroup with VKORC1=GG, the power within that subgroup would only be 46%. In addition, because CALU is associated with >500 SNPs, some of which are in linkage disequilibrium with CALU SNP 1043550, a role for calumenin cannot be discounted based on studies of other SNPs in other populations where frequency distributions for the various SNPs were seen to vary with race and ethnicity
A study in a German-Austrian population reported a statistically significant association between two CALU SNPs (rs2290228 C>T and rs2307040 G>A) in one individual who was homozygous T/T and G/G, respectively, and a substantially higher anticoagulation requirement (P = 0.0197).10 However, they were unable to confirm an interaction with VKORC1 1173C>T and A29809G (rs 1043550) in their population. These SNPS do not appear to be in linkage disequilibrium with the SNP rs1043550 examined in the current study.
Other studies have similarly observed a small contribution of CALU SNPs to warfarin dosing requirements. Applying a multiplex SNP approach and univariate analysis to examine genetic impact on therapeutic warfarin dose requirement, Wadelius et al21 identified a nominal association of the following genes or flanking regions: VKORC1; CYPs 2C9, 2C19, 2C18; CALU; GGXC; ORM1; ORM2; PROC; EPHX1; and SNPs that distinguish APOE E2, E3, and E4. However, following application of the Bonferroni Meff method to determine the significance cut point, and assuming independent contribution of each gene, only VKORC1 and the CYP SNPs remained significantly associated. G11A (R4Q) (rs2290228) in the CALU gene, that was associated with higher warfarin dose requirements in the study by Vecsler et al,7 showed no association in the largely Swedish Wadelius21 cohort; whereas 3′ untranslated region (UTR) SNP (rs11653), a non-synonymous SNP (rs2307040), and two intronic SNPs, rs339054 and rs1006023, showed marginal statistical significance. However, when all nominally associated genes were modeled and adjusted for variables with individual P values above 0.2 and low r2 values, CALU was not found to be a significant contributor. Although the Wadelius study21 is silent regarding association specifically of the SNP A29809G (rs1043550) examined in the present study and warfarin dose requirement, it is noteworthy that rs1043550 is in linkage disequilibrium with 3′UTR SNP rs11653,22 which initially exhibited marginal statistical significance prior to modeling in their largely Swedish cohort. Further, the non-synonymous SNP (rs 230740) was the same SNP identified in the study by Cadamuro,10 who reported on the patient homozygous for the G allele who exhibited the significantly higher dosing requirement to achieve anticoagulation.
Notably, based on allelic frequency data determined for the cohort in the present study, Hardy Weinberg equilibrium could not be demonstrated. Allelic distribution differed from that of the Spanish cohorts receiving anticoagulation therapy and differed significantly from the HapMap CEU population that was ethnically more similar to the Marshfield cohort. Notably, a study by Wang et al23 demonstrated that SNPs in linkage disequilibrium in a noncoding block at position +2255 locus of VKORC1 conferred twice the risk for arterial vascular diseases. We postulate that lack of demonstration of linkage disequilibrium for our cohort may be related to the fact that our cohort is highly selected for patients with underlying cardiovascular conditions requiring anticoagulation therapy, whereas the comparison population represented a general population of similar ethnic heritage. These data may suggest that allelic distribution of calumenin SNPs found in our study may similarly be highly associated with propensity for arterial vascular disease. However, this was not further examined in the present study. In addition, exclusion of patients known to have underlying conditions that would alter drug metabolism, such as cancer, renal or hepatic insufficiency, congestive heart failure, or those under therapeutic management with long-acting barbiturates may have potentially created a more skewed population, thereby contributing to limiting generalizability of conclusions.
Voora et al9 undertook resequencing of the CALU gene in a cohort of warfarin dosing outliers. The investigators reported that the intronic CALU SNP rs339097 was significantly associated with 14.5% higher warfarin dose requirements on average in African Americans (P = 0.03) and validated their results in two separate replication cohorts. This SNP was not significant in Caucasians, however, likely due to its low allele frequency (2%) among Caucasians. Notably, in their study, the SNP rs1043550 A>G, which was the focus of our study, was represented in the Caucasian subset of the discovery cohort in approximately equal distribution in both low dose and high dose outliers.9 Ethnicity representation among their Caucasian sub-cohorts was not defined. Interestingly, rs2290228 (G11A), which demonstrated some association with higher dose requirements in the Vecsler7 study that was focused in a non-Caucasian population, similarly was at a higher frequency among Caucasian high dose outliers in their discovery group, although the association did not achieve statistical significance when compared to its representation among low dose outliers (P = 0.09).
A recent study by Ramirez et al,16 that linked the electronic medical record to biorepositories storing genetic data, similarly validated the contribution of CALU SNP rs339097 and other CYP2C9 SNPs to therapeutic dose requirement prediction in African Americans. Inclusion of these SNPs in dosing models explained 41% of warfarin dose variation in African Americans, compared to only 29% explained in African Americans applying the algorithms of the International Warfarin Consortium, which was modeled in populations wherein the Black race was under-represented.16 These data validated an earlier study by Voora et al9 who also reported higher warfarin dose requirements in African American patients (P = 0.01) using two separate replication cohorts: one with subjects of mixed ethnicity and one consisting of African American subjects. However, while this SNP is rare in Caucasians of European descent, (allele frequency=0.002), it exhibits comparatively higher allele frequency among individuals of sub-saharan African and African American racial origin (allele frequency=0.14.) This SNP would, therefore, not be expected to contribute to variability in warfarin dosing requirements in the European, largely German, population in the present study.
Several genome wide association studies (GWAS) have focused on defining genetic determinants influencing warfarin dose requirements.17,24 The study by Takeuchi et al21 undertaken in a Swedish cohort (n=1053) had statistical power to detect genome wide significance at the level of P <1.5×10−7 for SNPs with moderate impact on warfarin dose requirements. This study identified a role only for VKORC1 and CYP2C9, with CYP4F2 contributing moderately to warfarin dose requirement. The GWAS study by Cooper et al,24 in a Caucasian population (n=181) and a replication cohort consisting of an additional 379 patients, was similarly powered (P <10−7) to detect SNPs contributing 20% variability in warfarin dose requirement and reported significant contributions by only VKORC1 and CYP2C9. CALU (rs3778760) was included among candidate genes influencing warfarin dose requirements, although it exhibited a genome wide association of 1.9×10−2, which was too low to meet the significance threshold of the study for a highly significant impact. This SNP demonstrated linkage disequilibrium with rs1043550, the SNP examined in the present study in a European population.24 Importantly, these studies did not examine whether there was interactive effect detectable among the genes that would enhance the impact on warfarin dose requirements relative to genes exhibiting significant but minor impacts on warfarin dose requirements.
In summary, data from our study and others support a minor contribution of CALU on warfarin dose requirement, but the extent of the effect varies strongly with race and ethnicity of the population under study and the minor allele frequencies of the SNPs. Further examination of gene interaction with respect to overall impact on warfarin dose requirements as demonstrated by Vecsler et al7 may be warranted.
Acknowledgements
We acknowledge editorial assistance from the Office of Scientific Writing and Publication at the Marshfield Clinic Research Foundation and assistance of Sharilyn Rennie for her contributions to early drafts of the manuscript.
Footnotes
Conflict of Interst Disclosure: JKB and RLB are co-inventors on a patent for the use of a CYP4F2 genetic test for warfarin dosing.
References
- 1.Suttie JW. Vitamin K-dependent carboxylase. Annu Rev Biochem 1985;54:459–477 [DOI] [PubMed] [Google Scholar]
- 2.Berkner KL. The vitamin K-dependent carboxylase. Annu Rev Nutr 2005;25:127–149 [DOI] [PubMed] [Google Scholar]
- 3.Glurich I, Burmester JK, Caldwell MD. Understanding the pharmacogenetic approach to warfarin dosing. Heart Fail Rev 2010;15:239–248 [DOI] [PubMed] [Google Scholar]
- 4.Yabe D, Taniwaki M, Nakamura T, Kanazawa N, Tashiro K, Tasuku H. Human calumenin gene (CALU): cDNA isolation and chromosomal mapping to 7q32. Genomics 1998; 49:331–333 [DOI] [PubMed] [Google Scholar]
- 5.Yabe D, Nakamura T, Kanazawa N, Tashiro K, Honjo T. Calumenin, a Ca 2+-binding protein retained in the endoplasmic reticulum with a novel carboxyl-terminal sequence, HDEF. J Biol Chem 1997;272:18232–18239 [DOI] [PubMed] [Google Scholar]
- 6.Wajih N, Hutson S, Wallin R. siRNA silencing of calumenin enhances functional factor IX production. Blood 2006;108:3757–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vecsler M, Loebstein R, Almog S, Kurnik D, Goldman B, Halkin H, Gak E. Combined genetic profiles of components and regulators of the vitamin K dependent γ-carboxylation system affect individual sensitivity to warfarin. Thromb Haemost 2006;95:205–211 [DOI] [PubMed] [Google Scholar]
- 8.González-Conejero R, Corral J, Roldán V, Ferrer F, Sánchez-Serrano I, Sánchez-Blanco JJ, Marín F, Vicente V. The genetic interaction between VKORC1 c1173t and calumenin a29809g modulates the anticoagulant response of acenocoumarol. J Thromb Haemost 2007;5:1701–1706 [DOI] [PubMed] [Google Scholar]
- 9.Voora D, Koboldt Dc, King CR, Lenzini PA, Eby CS, Porche-Sorbet R, Deych E, Crankshaw M, Milligan PE, McLeod HL, Patel SR, Cavallari LH, Ridker PM, Grice GR, Miller RD, Gage BF. A polymorphism in the VKORC1 regulator predicts higher warfarin dose requirements in African Americans. Clin Pharmacol Ther 2010;87:445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadamuro J, Dieplinger B, Felder T, Kedenko I, Mueller T, Haltmayer M, Patsch W, Oberkofler H. Genetic determinants of acenocoumarol and phenprocoumon maintenance dose requirements. Eur J Clin Pharmacol 2010;66:253–260 [DOI] [PubMed] [Google Scholar]
- 11.Hillman MA, Wilke RA, Caldwell MD, Berg RL, Glurich I, Burmester JK. Relative impact of covarieties in prescribing warfarin according to CYP2C9 genotype. Pharmacogenetics 2004;14:539–547 [DOI] [PubMed] [Google Scholar]
- 12.Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C, King CR, Brower A, Schmelzer JR, Glurich I, Vidaillet HJ, Yale SH, Qi Zhang K, Berg RL, Burmester JK. CYP4F2 genetic variant alters required warfarin dose. Blood 2008; 111:4106–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burmester JK, Berg RL, Yale SH, Rottscheit CM, Glurich IE, Schmelzer JR, Caldwell MD. A randomized controlled trial of genotype-based Coumadin initiation. Genet Med 2011;13:509–518 [DOI] [PubMed] [Google Scholar]
- 14.NCBI dbSNP Short Genetic Variations. Available at: http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?searchType=adhoc_search&type=rs&rs=rs1043550 Accessed October 4, 2012
- 15.Caldwell MD, Berg RL, Zhang KQ, Glurich I, Schmelzer JR, Yale SH, Vidaillet HJ, Burmester JK. Evaluation of genetic factors for warfarin dose prediction. Clin Med Res 2007; 5:8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez AH, Shi Y, Schildcrout JS, Delaney JT, Xu H, Oetjens MT, Zuvich RL, Basford MA, Bowton E, Jiang M, Speltz P, Zink R, Cowan J, Pulley JM, Ritchie MD, Masys DR, Roden DM, Crawford DC, Denny JC. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics 2012;13:407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet 2009;5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wajih N, Sane D, Hutson S, Wallin R. The inhibitory effect of calumenin on the vitamin k-dependent γ-carboxylation system. Characterization of the system in normal and warfarin-resistant rats. J Biol Chem 2004;279:25276–25283 [DOI] [PubMed] [Google Scholar]
- 19.Wallin R, Hutson SM, Cain D, Sweatt A, Sane DC. A molecular mechanism for genetic warfarin resistance in the rat. FASEB J 2001;15:2542–2544 [DOI] [PubMed] [Google Scholar]
- 20.Feiveson AH. Power by simulation. Stata J 2002;2:107–124 [Google Scholar]
- 21.Wadelius M, Chen LY, Ericsson N, Bumpstead S, Ghori J, Wadelius C, Bentley D, McGinnis R, Deloukas P. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet 2007;121:23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura R, Miyashita K, Kokubo Y, Akaiwa Y, Otsubo R, Nagatsuka K, Otsuki T, Okayama A, Minematsu K, Naritomi H, Honda S, Tomoike H, Miyata T. Genotypes of vitamin k epoxide reductase, γ-glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese Patients. Thromb Res 2007;120:181–186 [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Zhang W, Zhang Y, Yang Y, Sun L, Hu S, Chen J, Zhang C, Zheng Y, Zhen Y, Sun K, Fu C, Yang T, Wang J, Sun J, Wu H, Glasgow WC, Hui R. VKORC1 haplotypes are associated with arterial vascular diseases (stroke, coronary heart disease, and aortic dissection). Circulation 2006;113:1615–1621 [DOI] [PubMed] [Google Scholar]
- 24.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwartz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, Veenstra DL, Rettie AE, Rieder MJ. A genome wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 2008;112:1022–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]