Abstract
The potential transmission of porcine endogenous retroviruses (PERVs) has raised concern in the development of porcine xenotransplantation products. Our previous studies have resulted in the identification of animals within a research herd of inbred miniature swine that lack the capacity to transmit PERV to human cells in vitro. In contrast, other animals were capable of PERV transmission. The PERVs that were transmitted to human cells are recombinants between PERV-A and PERV-C in the post-VRA region of the envelope (B. A. Oldmixon, J. C. Wood, T. A. Ericsson, C. A. Wilson, M. E. White-Scharf, G. Andersson, J. L. Greenstein, H. J. Schuurman, and C. Patience, J. Virol. 76:3045-3048, 2002); these viruses we term PERV-A/C. This observation prompted us to determine whether these human-tropic replication-competent (HTRC) PERV-A/C recombinants were present in the genomic DNA of these miniature swine. Genomic DNA libraries were generated from one miniature swine that transmitted HTRC PERV as well as from one miniature swine that did not transmit HTRC PERV. HTRC PERV-A/C proviruses were not identified in the germ line DNAs of these pigs by using genomic mapping. Similarly, although PERV-A loci were identified in both libraries that possessed long env open reading frames, the Env proteins encoded by these loci were nonfunctional according to pseudotype assays. In the absence of a germ line source for HTRC PERV, further studies are warranted to assess the mechanisms by which HTRC PERV can be generated. Once identified, it may prove possible to generate animals with further reduced potential to produce HTRC PERV.
Xenotransplantation carries the concerns of cross-species transmission of infectious pathogens present in the donor species (7, 17, 18, 23). Studies indicate that many exogenous microorganisms can be eliminated from the donor herd by using various barrier methods and specialized qualified-pathogen-free rearing techniques (3, 25, 26). However, porcine endogenous retroviruses (PERVs) are an exception to this rule, as proviral copies are inherited through the germ line DNA. Three subgroups of PERV, PERV-A, -B, and -C, have been identified (24). Either PERV-A, PERV-B, or viruses derived from recombination events between PERV-A and PERV-C, are able to result in productive infection of human cells in vitro, such as the kidney epithelial 293 cell line (1, 10, 12-14, 16, 21, 24, 27). All three PERV subgroups can replicate in porcine cell lines (21, 24). Although in vitro transmission characteristics indicate a potential for in vivo transmission, infection by PERV of humans exposed to living porcine tissue has not been reported thus far (5, 6, 8, 19, 20, 22).
Genomic mapping studies have been used to investigate the distribution of the three subgroups of PERV in the genomic DNA of pigs. PERV-A and PERV-B loci have been identified in the germ line DNA of all pigs tested to date (11). Generally, PERV-A loci are present at a higher copy number than the PERV-B subgroup. In most pig breeds, the PERV-C subgroup either is absent or is represented by only a limited number of elements. In contrast, the copy number of PERV-C can be elevated in miniature swine (MS) (2, 21). Previous mapping data have shown that the majority of PERVs in the genome are defective due to mutations in their coding sequences (9, 15). Interestingly, those PERVs that are clearly replication competent have been isolated from the genomic DNAs of immortalized pig cell lines (10). In contrast, those clones isolated from primary pig cells have been of weak competence (15). However, because the transmission phenotype of the cells from which these mapping studies were produced is unknown, it is not possible to determine which, if any, loci are expressed and, moreover, whether these viruses can be transmitted.
In vitro transmission studies of an inbred herd of MS have identified animals that reproducibly do not transmit PERV to human cells (16). The identification of animals within the highly inbred MS herd that lack human-tropic replication-competent (HTRC) PERV (16) may circumvent the need for strategies such as specific breeding or gene targeting to remove HTRC PERV from the herd, which might be required in other outbred breeds of pig. On all occasions when HTRC PERV was transmitted to human cells, the viruses were recombinants between the PERV-A and PERV-C subgroups with recombination in the post-VRA region of the env gene (16), indicating that recombination may play a vital role in the generation of HTRC PERV. Accordingly, for analysis of the PERV burden of the genomic DNA of pigs to be complete, genomic mapping must identify (i) all HTRC PERVs in the genome that possess independent replication competence; (ii) long replication-defective PERV-A loci, i.e., those loci that contain an env open reading frame (ORF) beyond the VRA region, which might contribute toward the production of HTRC PERV via recombination; and (iii) transcriptionally silent but potentially replication-competent proviruses that might become activated and infect human cells. Genomic mapping of PERV loci can be used to achieve these goals.
In this study we investigated MS with known in vitro transmission phenotypes. Genomic libraries were constructed from peripheral blood mononuclear cells (PBMC) of two SLAd/d inbred MS (16); one of these (MS 13519) was capable of transmitting HTRC PERV-A/C recombinants to 293 cells in vitro, whereas MS 11852 lacked transmissible HTRC PERV and infected only the porcine cell line ST-IOWA. Study of these animals facilitates direct comparison of in vitro transmission data with genomic mapping data. We did not detect HTRC PERV in either of these libraries, which is indicative of the absence of HTRC PERV in the germ line DNA of MS. In addition, taken together with the data presented by Wood et al. (28), our data challenge the assumption that the most significant source of HTRC PERV is directly from intact germ line proviruses.
MATERIALS AND METHODS
Genomic library screening.
Lambda libraries were constructed from the PBMC of SLAd/d MS that had been classified as either a nontransmitter of HTRC PERV (MS 11852) (designated HTRC-NT) or a transmitter of HTRC PERV (MS 13519) (designated HTRC-T) (16). The library for MS 11852 was constructed from DNA partially digested with Sau3A1 and ligated into XhoI-digested lambda GEM-12 (Promega, Southampton, United Kingdom) that had been partially filled with Klenow and T and C deoxynucleotides to create an Sau3A1-compatible end. Titers of the library were determined in KW251 cells (Promega). For MS 13519, the library was prepared from DNA partially digested with MboI and ligated to BamHI-digested lambda DASH II vector (CMT Inc., Phillipsburg, N.J.), and titers were determined in P2 (MRA) cells (CMT Inc.). Neither library was amplified prior to use. For each library, a total of 1.5 × 106 PFU was plated on NZY agar plates. Duplicate filters were lifted by using Hybond NX (Amersham Pharmacia Biotech, Little Chalfont, Bucks, United Kingdom) and denatured and neutralized as described previously (9). Filters were UV-cross-linked and prehybridized for 30 min in Quickhyb solution (Stratagene, Amsterdam, The Netherlands). For gag screening, the probes and reagents used were as described by us previously (9). For env screening, a PERV-A-specific env probe was derived by using a nested set of primers specific to the variable region of the PERV-A envelope sequence. Primary PCR was performed on PK-15 cDNA by using conserved primers pCENV1 (5′-ACCAACGGCTGTGAAAGTCGAAG-3′) and pCENV2 (5′-AAGTACCATGATCTGGACTGCAC-3′). Secondary amplifications were carried out with specific primers PV4USU (5′-GGAGATGGAAAGATTGGCAACAG-3′) and PV4DSU (5′-CAGAGGTTGTATTGTAATCAGAG-3′). Products were purified, cloned into the vector pGEM-2T (Promega), and verified by restriction digestion and sequencing. An internal 334-bp insert was released by a SpeI-SacII digest and used as the PERV-A specific probe. Probes were labeled with [α-32P]dCTP by using Prime-it II (Stratagene). The library filters were hybridized and washed as described previously (9), exposed to X-ray film at −70°C for up to a week, and developed by using a Compact ×4 automatic film processor (X-ograph Imaging Systems, Gloucester, United Kingdom).
Clones that gave positive hybridizations on both the gag and env filters were then purified to homogeneity through multiple rounds of rescreening. Lambda DNA was prepared from liquid lysates of these clones by standard methodologies.
Analysis of proviral clones for ORFs.
To determine the lengths of the env genes of the lambda clones, genomic DNA was prepared from all gag- and env-positive lambda clones and analyzed by using PCR primer pairs external to the original env probe region. These primers (cenv-3 [5′-GGTTATAACAGGTGGTGGGCATG-3′] and cenv-4 [5′-CCAAGGAGACCTGTTGAACCGTC-3′]), which are conserved between all three subtypes of PERV, amplify a portion of the env gene and span the junction between the surface envelope (SU) and transmembrane (TM) protein-coding regions of env. PCRs were initiated by using the PCR core kit (Qiagen, Sussex, United Kingdom) with 50-μl mixtures containing 1.5 mM MgCl2, 1 U of Taq polymerase (Qiagen), and 200 ng of lambda DNA; reactions were cycled in an ABI9700 instrument (Perkin-Elmer Biosystems, Warrington, United Kingdom) at 94°C for 1 min 30 s followed by 35 cycles of 94°C for 15 s, 65°C for 30 s, and 72°C for 1 min and then followed by 72°C for 7 min, to yield a product of approximately 588 bp. Primers were used at a concentration of 10 pmol per reaction mixture.
Protein truncation test (PTT) PCR screening for gag and env ORFs was carried out as described previously on all clones that possessed long env genes (9). PCR products from clones that tested positive for full-length gag and env ORFs were cloned into the pCR2.1-Topo vector (Invitrogen, Paisley, United Kingdom). A minimum of two products for each clone were sequenced by ABI PRISM Dye terminator cycle sequencing on an ABI373 automated sequencer (Applied Biosystems, Warrington, United Kingdom). DNAs from all lambda clones that maintained full-length gag and env genes were digested with either SmaI or EcoRI in order to determine whether the clones isolated were unique.
Analysis of replication-defective PERV loci.
Lambda clones that tested negative by env PTT PCR but positive by gag PTT PCR were designated defective. To determine whether the negative env PCR was due to disrupted ORFs and not due to the presence of recombinant sequence not being amplified because of our primer selection, clones testing positive for env sequences according to PCR with the primers cenv3 and cenv4 but negative by env PTT PCR were amplified by long terminal repeat-long terminal repeat PCR as described previously (16), and the env region was sequenced.
Detection of PERV-A/C env recombinants by PCR.
Genomic DNAs were prepared from the PBMC of MS 11852 and MS 13519, and 293 cells were infected by a PERV-A/C recombinant and control plasmids containing PERV-A, -B, or -C env sequences. The DNAs were analyzed by using PCR primers specific to PERV-A/C env recombinant sequences. The primers A-VRA (forward) (5′-CCTACCAGTTATAATCAATTTAATTATGGC-3′) and C-TM (reverse) (5′-CTCAAACCACCCTTGAGTAGTTTCC-3′) were used to amplify sequences from 100 ng of genomic DNA in 50-μl reaction mixtures containing 25 pmol of primers in PCR master mix (Qiagen) with the following cycling parameters: 1 cycle of 94°C for 5 min; 35 cycles of 94°C for 10 s, 60°C for 30 s, and 72°C for 90 s; and 1 cycle of 72°C for 10 min.
Analysis of competence of PERV clones and Env proteins in vitro.
Transfection of lambda DNA was used to assess the replication competence of PERV loci, using techniques described by others (15). Briefly, 10 μg of DNA was prepared from clones by the liquid lysate method and transfected into the 293 human and the ST-IOWA porcine cell lines by using Lipofectamine Plus in accordance with the manufacturer's instructions (Invitrogen). As an internal control for transfection efficiency, each transfection also contained a 10-ng spike of the expression plasmid pEGFP-N1 (BD Biosciences Clontech, Palo Alto, Calif.), and transfected cells were examined after 48 h of culture by fluorescence microscopy. Cultures were maintained for up to 60 days and monitored for reverse transcriptase (RT) activity by using a sensitive enzyme-linked immunosorbent assay-based system (HS-Mn2+ kit; Cavidi Tech AB, Uppsala, Sweden) as described previously (15, 16). PERV-A60 DNA (1) was used as a positive control in the transfection assays.
Expression vectors were used to assess the competence of the Env proteins putatively encoded by the PERV-A proviral loci. Amplification of the env sequences from library clone DNA was carried out in 50-μl reaction mixtures containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, a 150 nM concentration of each primer, a 200 nM concentration of each deoxynucleoside triphosphate, 2.5 U of Amplitaq (Perkin-Elmer Biosystems), 200 ng of lambda DNA, and the primers env F (5′-GGATCCTAATACGACTCACTATAGGAACAGACCACCATGCATCCCACGTTAAGCCG-3′) and env R (5′-GCTCTAGACTAAGCGTAGTCTGGGACGTCGTATGGGTAGAACTGGGAAGGGTAGAGGTCAGT-3′) and were cycled as follows: 1 cycle of 95°C for 3 min; 30 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 2 min 10 s); and 10 min at 72°C. The primers used were previously described by us (9); the T7 RNA polymerase promoter sequence is incorporated in the sense primer, and the antisense primer is modified to contain an in-frame hemagglutinin epitope tag as utilized in the PTT assay (9). The PCR products were cloned into the pCR2.1-Topo cloning vector (Invitrogen) according to the manufacturer's instructions. The PERV-A envelope sequences were excised from the pCR2.1-Topo vector by digestion with SpeI and XbaI and subcloned into the expression vectors pCR3.1 (Invitrogen) and pCIneo at the XbaI site. The correct orientation of the env sequences was confirmed by digestion of the constructs with XhoI, and the constructs were sequenced on an ABI373 automated sequencer (Applied Biosystems). Plasmid DNAs were transfected into the retroviral packaging cell line TELCeB by using Lipofectamine (Invitrogen) according to the manufacturer's instructions. Stable clones were selected by the addition of 800 μg of G418 per ml 48 h after transfection. The infectious titers of the pseudotypes were determined by plating on 293 and ST-IOWA cells in the presence of 8 μg of Polybrene per ml and β-galactosidase staining as described previously (24).
Nucleotide sequence accession numbers.
Primers were derived from sequences taken from the GenBank database, for PERV-A, GenBank accession codes Y12238; for PERV B, Y17013/Y12239; and for PERV-C, AFO38600. Sequences for the clones Ntr1 to-4 are as follows: gag accession AY368583 to -6 and env genes accession numbers AY368587 to -9 and AY288779. For Tr1 to-4 env genes accession codes are AY368579 to -82; for Tr5, AY371067.
RESULTS AND DISCUSSION
To identify PERV clones that may be important in the formation of human-tropic viruses, we analyzed genomic DNA libraries made from MS PBMC genomic DNA. Each library contained an average insert size of approximately 18.2 kb and three to five genome equivalents. A primary screen to identify lambda clones that contained PERV-A proviral sequences was performed with (i) a PERV-A-specific env probe that is located within the VRA region and (ii) a gag probe, the sequence of which is conserved across the PERV subgroups (9). Double-positive clones were plaque purified prior to further analysis. A PCR-based secondary screen that analyzed the lengths of the gag and env genes of the clones was performed. In order for a clone to be included in third-round analysis, we required that the clone should produce near-full-length gag and env PCR products. Third-round analysis consisted of PTT and sequencing analysis of gag and env PCR products to detect loci that had full-length gag and env ORFs. In addition, those gag-positive clones that possessed env ORFs that extended beyond the VRA region were also included in final analysis, because these “long” defective PERV-A loci could be the genetic substrate for the formation of human-tropic PERV. Finally, restriction fragment length polymorphism analysis was performed on the qualifying clones to identify unique loci.
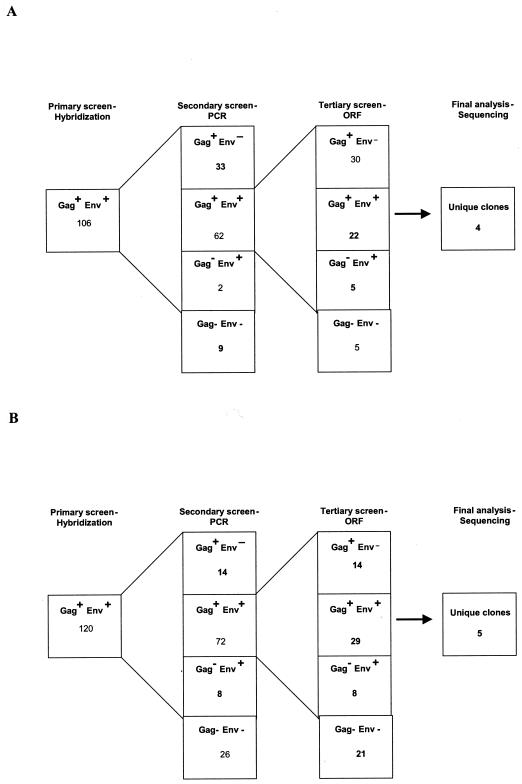
The first-round screen of the genomic library from MS 11852, an HTRC PERV-nontransmitting (HTRC-NT) pig, identified a total of 106 PERV-A-positive clones (Fig. 1A). PCR analysis of purified clones indicated that 62 of the clones possessed gag and long env genes. The remaining 44 clones were excluded due to truncations of either their env (33 of 44 cases) or gag (2 of 44 cases) genes or of both genes (9 of 44 cases). Within the 62 gag- and env-positive clones, 22 were found to be positive by both gag and env PTT PCR, indicating that these loci were potentially replication competent. The remaining 40 clones were excluded due to the absence of the env ORF (30 of 40 cases), the gag ORF (5 of 40 cases), or both ORFs (5 of 40 cases). RFLP analysis of the 22 double-positive loci indicated that 9 were unique, and of these, sequence analysis indicated that only 4 (NTr1 to -4) had full-length gag genes, i.e., genes of similar size to the gag genes of known replication-competent PERV. Analysis of the env genes of the four associated PERV-A sequences indicated that they possessed greater than 97% homology to each other and to the previously characterized germ line PERV-A env sequence (GenBank accession number AF435967) (data not shown) (11). Interestingly, none of these clones possessed full-length env ORFs due to two common point mutations that introduced stop codons at either nucleotide (nt) 1906 (CGA to TGA) or nt 1963 (CAA to TAA). The stop codon at nt 1963 terminates the putative Env proteins six amino acid residues before the termination site observed with PK15-derived PERV-A, which can grow to high titers in human cells in vitro (Fig. 2) (for complete sequence comparison, see http://www.vet.gla.ac.uk/retrovirus/PERV.html).
FIG. 1.
Genomic library screening results for transmitting and nontransmitting MS. (A) HTRC-NT MS11852; (B) HTRC-T MS13519. Lambda clones were identified by radioactive hybridization screening prior to PCR and sequencing.
FIG. 2.
Amino acid sequence comparison of the 3′ ends of the env regions of PERV-A clones isolated from both the HTRC-T 13519 (Tr1 to -5) and HTRC-NT 11852 (NTr1 to -4) libraries and PERV-A/C recombinants isolated from 293 cells infected with MS PBMC (t1.2c9 [accession number AF417229], t2a3 [AF417230], t2a5 [AF417231], and t6e5 [AF417232]). T142-derived PERV A/C recombinant sequences are those from the coculture of PBMC from animal 13519 (T142.1 [AY364234] and T142.8 [AY364236]). *, stop codon. Arrows highlight amino acid differences.
The genotypes of the PERV clones identified during analysis of the genomic DNA library made from MS 13519, an HTRC PERV-transmitting (HTRC-T) animal, are summarized in Fig. 1B. A total of 120 PERV-A-positive clones were identified during the primary screen of the library. PCR analysis of purified clones indicated that 72 of the clones initially possessed gag and env sequences. Of these 72 clones, 29 were positive for long gag and env genes as determined by PTT PCR. Of the remainder, 14 were defective in env, 8 were defective in gag, and the final 21 clones were negative for gag and env PTT PCR products. Within the 29 gag- and env positive clones, nucleotide sequencing revealed that five PERV-A clones (Tr1 to -5) possessed long gag and env genes. The remaining 24 clones were either defective PERV-A or PERV-B; the latter were probably identified due to cross-hybridization of the PERV-A probe during the lower-stringency hybridization employed in the primary screen.
Amino acid sequence analysis revealed no remarkable differences between the nucleotide sequences of the long env genes of the PERV-A loci present in the HTRC-T pig and HTRC-NT pigs (Fig. 2) (http://www.vet.gla.ac.uk/retrovirus/PERV.html), which is a reflection of the highly inbred nature of the MS herd (inbreeding coefficient, >0.9).
Studies of replication competence were performed with the PERV-A clones that may have possessed replication competence, i.e., the four isolated from the HTRC-NT library and two representative clones of the five isolated from HTRC-T library identified as possessing full-length gag ORFs as well as long env ORFs (i.e., truncated at nt 1906 or 1963). Purified lambda clone DNA was transfected into human (293) and pig (ST-IOWA) target cells, along with an enhanced green fluorescent protein reporter vector. Following transfection, approximately 10 to 30% of target cells expressed the enhanced green fluorescent protein reporter vector, indicating that the transfection was successful (data not shown). The cultures were monitored for supernatant RT activity for approximately 60 days. No production of RT activity was detected in either of the cell lines, indicating that the clones were not replication competent (Table 1). In contrast, RT activity was detectable with the control PERV-A clone (PERV-A60) after approximately 10 days in culture (data not shown).
TABLE 1.
Nucleotide sequence and replication competence analysis of PERV-A loci
| Clonea | gag ORF | env ORFb | Replication competencec in:
|
|
|---|---|---|---|---|
| 293 cells | ST-IOWA cells | |||
| NTr1 | Full length | Premature stop | Negative | Negative |
| NTr2 | Full length | Long env | Negative | Negative |
| NTr3 | Full length | Long env | Negative | Negative |
| NTr4 | Full length | Long env | Negative | Negative |
| Tr1 | Full length | Premature stop | Negative | Negative |
| Tr2 | Full length | Long env | Negative | Negative |
The NTr and Tr designations refer to loci identified from nontransmitting and transmitting MS, respectively.
Long env refers to env sequences terminated approximately 6 amino acids short of full length. Premature stop refers to a significantly truncated env ORF at nt 247 for NTr1 and nt 1183 for Tr1.
The PK15-derived control PERV transfection resulted in detectable replication within 10 days of transfection.
In order to further analyze the competence of the clones, representative env genes of long PERV-A loci were cloned into mammalian expression vectors and transfected into the murine leukemia virus-based pseudotype producer cell line TELCeB, which encodes β-galactosidase activity. As shown in Table 2, Env function was not detected for any of the clones when tested on 293 or ST-IOWA cells.
TABLE 2.
Infectivity of PERV pseudotypes for human and porcine cells
| PERV clonea | Infectivity (IU/ml) on the following target cell line:
|
|
|---|---|---|
| 293 | ST-IOWA | |
| PERV-A | 104 | 104 |
| PERV-B | 103 | 103 |
| PERV-C | <4 | 102 |
| NTr1 | <4 | <4 |
| NTr4 | <4 | <4 |
| Tr1 | <4 | <4 |
| Tr2 | <4 | <4 |
The NTr and Tr designations refer to loci identified from nontransmitting and transmitting MS, respectively.
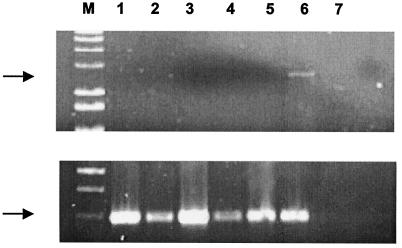
To date, it has been assumed that (i) the production of human-tropic PERV is governed by germ line proviral loci and (ii) the HTRC PERV-A/C recombinants detected by using in vitro transmission assays are tissue culture phenomena. However, the results reported by Wood et al. (28), indicating that PERV-A/C recombinants exist in vivo, raise the possibility that the viruses might not be present in the genomes of transmitting pigs. Therefore, we extended the analysis of the genomic DNAs and the genomic library DNAs (data not shown) from both animals in an attempt to identify the presence of recombinant viruses. By PCR, PERV-A/C recombinants were not detected in either the genomic DNA (Fig. 3) or the genomic library DNA from the transmitter animal. It is noteworthy that PERV-A/C recombinants were also not detected during the screening of the HTRC-NT and HTRC-T genomic libraries. These results are consistent with the hypothesis proposed by Wood et al. (28), indicating that these viruses are not present in the germ line DNA of MS and that recombination may be critical for the formation of human-tropic PERV.
FIG. 3.
Genomic DNAs isolated from transmitting and nontransmitting MS and analyzed by PCR for the presence of PERV-A/C recombinant env sequences (top) and pan-PERV env sequences (bottom), using conserved primers. Spiking experiments with recombinant PERV-A/C DNA confirmed the ability of the PCR to detect a single copy of PERV-A/C provirus (not shown). Lanes: 1, PERV-A control (PCR-A60); 2, PERV-B control (PCR-B17); 3, PERV-C env control (PoEV3); 4, MS13519; 5, MS11852; 6, 293 PERV-A/C genomic DNA; 7, 293 cell genomic DNA; M, size marker.
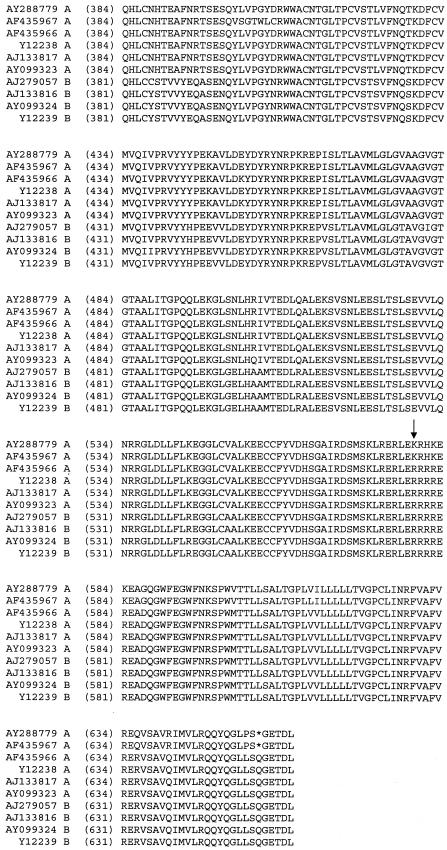
To investigate the potential importance of PERV recombination in more detail, we analyzed the functional env sequences that have been deposited in the GenBank database. We compared the env sequences of five functional PERV-A and four functional PERV-B GenBank sequences (isolated either from the PK-15 cell line [1, 4, 11] or from various pig breeds [4, 15], along with a representative PERV-A env sequence isolated from the MS genomic library. Alignment of these sequences indicated that all but one of the PERV-A env genes are recombinants with PERV-B within the TM region (Fig. 4), with the exception being an env sequence (accession number AF435967) which was derived from a weakly replication-competent PERV-A cloned from the genomic DNA of Large White pigs (15). Interestingly, in contrast to the apparent PERV-A/B recombinants identifiable in cell lines producing high titers of PERV, the proviral PERV-A sequences identified in the MS libraries appear to be nonrecombinant and most similar to the PERV-A sequence under accession number AF435967 (Fig. 4). The Env protein products encoded by the MS-derived sequences were nonfunctional in pseudotype assays (Table 2). Given the reproducible nature of the recombination with PERV-B, it is tempting to speculate that the generation of HTRC PERV-A env genes that can infect human cells at a significant titer, such as the PK-15 cell line-derived envelope described by Le Tissier et al. (11), may be the result of recombination between PERV-A and PERV-B elements in vitro.
FIG. 4.
Comparison of PERV-A and -B sequences from the GenBank database in the transmembrane region of the envelope. The GenBank accession number for each sequence is indicated at the start of the sequence. The nonrecombinant PERV-A sequence NTr3 (AY28879), which was isolated from MS11852, is included for comparison. Arrow, recombination site; *, stop codon.
In summary, in this study we were unable to identify PERV proviral loci derived from either an HTRC-T or an HTRC-NT MS library that possess replication competence in human 293 cells. Current data indicate that HTRC PERV possesses env genes that are recombinants between PERV-A and -C env sequences. Our PCR and genomic mapping studies indicate that these recombinant viruses are not present in the germ line DNA of MS, which, taken together with our observation that recombinant PERVs can be identified in the PBMC of MS (28), indicates that recombination is an important mechanism in the generation of PERVs that are able to infect human cells. The identification of human-tropic recombinant PERV in vitro but not in the germ line suggests that the recombination event may occur de novo in each pig. A number of mechanisms potentially underlie the generation of replication-competent PERV. First, because PERV loci were not identified in transmitting pigs that correlated with the potential to produce HTRC PERV, it is possible that these elements are being generated in pig cells from a number of defective germ line PERV elements. Thus, in a worst-case scenario, despite an organ or tissue being free of replication-competent retroviruses at the time of transplant, replication-competent retroviruses might be generated in the organ while it is in the xenograft recipient, thus exposing them to risk of infection. Second, the PERV-A/C recombinant may exist as an exogenous agent or may be generated as a result of infection of the pig with an infectious PERV. If this proves to be correct, it may be possible to dramatically reduce the rate of recombinant PERV formation via established specific-pathogen-free pig derivation procedures. Further studies are required to define the mechanism by which PERV-A/C recombinant viruses are formed so that appropriate strategies can be developed to remove these viruses from the herd and improve the safety of xenotransplantation.
Acknowledgments
This research was supported in part by Immerge BioTherapeutics Inc. and the Scottish Hospitals Endowment Research Trust RG6/02.
We gratefully acknowledge Y. Takeuchi (UCL, London, United Kingdom) for providing the PERV A-60 and PERV B-17 isolates and K. Patience for assistance in preparation of the manuscript.
REFERENCES
- 1.Bartosch, B., R. A. Weiss, and Y. Takeuchi. 2002. PCR-based cloning and immunocytological titration of infectious porcine endogenous retrovirus subgroup A and B. J. Gen. Virol. 83:2231-2240. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, S., C. Arnauld, and A. Jestin. 2000. Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J. Virol. 74:8575-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, D. A., J. F. Fryer, A. W. Tucker, P. D. McArdle, A. E. Hughes, V. C. Emery, and P. D. Griffiths. 2003. Porcine cytomegalovirus in pigs being bred for xenograft organs: progress towards control. Xenotransplantation 10:142-148. [DOI] [PubMed] [Google Scholar]
- 4.Czauderna, F., N. Fischer, K. Boller, R. Kurth, and R. R. Tonjes. 2000. Establishment and characterization of molecular clones of porcine endogenous retroviruses replicating on human cells. J. Virol. 74:4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinsmore, J. H., C. Manhart, R. Raineri, D. B. Jacoby, and A. Moore. 2000. No evidence for infection of human cells with porcine endogenous retrovirus (PERV) after exposure to porcine fetal neuronal cells. Transplantation 70:1382-1389. [DOI] [PubMed] [Google Scholar]
- 6.Elliott, R. B., L. Escobar, O. Garkavenko, M. C. Croxson, B. A. Schroeder, M. McGregor, G. Ferguson, N. Beckman, and S. Ferguson. 2000. No evidence of infection with porcine endogenous retrovirus in recipients of encapsulated porcine islet xenografts. Cell Transplant. 9:895-901. [DOI] [PubMed] [Google Scholar]
- 7.Fishman, J. A. 2000. Xenotransplantation from swine: making a list, checking it twice. Xenotransplantation 7:93-95. [DOI] [PubMed] [Google Scholar]
- 8.Heneine, W., A. Tibell, W. M. Switzer, P. Sandstrom, G. V. Rosales, A. Mathews, O. Korsgren, L. E. Chapman, T. M. Folks, and C. G. Groth. 1998. No evidence of infection with porcine endogenous retrovirus in recipients of porcine islet-cell xenografts. Lancet 352:695-699. [DOI] [PubMed] [Google Scholar]
- 9.Herring, C., G. Quinn, R. Bower, N. Parsons, N. A. Logan, A. Brawley, K. Elsome, A. Whittam, X. M. Fernandez-Suarez, D. Cunningham, D. Onions, G. Langford, and L. Scobie. 2001. Mapping full-length porcine endogenous retroviruses in a Large White pig. J. Virol. 75:12252-12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krach, U., N. Fischer, F. Czauderna, and R. R. Tonjes. 2001. Comparison of replication-competent molecular clones of porcine endogenous retrovirus class A and class B derived from pig and human cells. J. Virol. 75:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 12.Martin, U., V. Kiessig, J. H. Blusch, A. Haverich, K. von der Helm, T. Herden, and G. Steinhoff. 1998. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352:692-694. [DOI] [PubMed] [Google Scholar]
- 13.Martin, U., M. E. Winkler, M. Id, H. Radecke, L. Arseniev, R. Groteluschen, A. R. Simon, and G. Steinhoff. 2000. Transmission of pig endogenous retrovirus to primary human cells. Transplant. Proc. 32:1157. [DOI] [PubMed] [Google Scholar]
- 14.Martin, U., M. E. Winkler, M. Id, H. Radeke, L. Arseniev, Y. Takeuchi, A. R. Simon, C. Patience, A. Haverich, and G. Steinhoff. 2000. Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV). Xenotransplantation 7:138-142. [DOI] [PubMed] [Google Scholar]
- 15.Niebert, M., C. Rogel-Gaillard, P. Chardon, and R. R. Tonjes. 2002. Characterization of chromosomally assigned replication-competent gamma porcine endogenous retroviruses derived from a Large White pig and expression in human cells. J. Virol. 76:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oldmixon, B. A., J. C. Wood, T. A. Ericsson, C. A. Wilson, M. E. White-Scharf, G. Andersson, J. L. Greenstein, H. J. Schuurman, and C. Patience. 2002. Porcine endogenous retrovirus transmission characteristics of an inbred herd of miniature swine. J. Virol. 76:3045-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onions, D., D. K. Cooper, T. J. Alexander, C. Brown, E. Claassen, J. E. Foweraker, D. L. Harris, B. W. Mahy, P. D. Minor, A. D. Osterhaus, P. P. Pastoret, and K. Yamanouchi. 2000. An approach to the control of disease transmission in pig-to-human xenotransplantation. Xenotransplantation 7:143-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onions, D., D. Hart, C. Mahoney, D. Galbraith, and K. Smith. 1998. Endogenous retroviruses and the safety of porcine xenotransplantation. Trends Microbiol. 6:430-431. [DOI] [PubMed] [Google Scholar]
- 19.Paradis, K., G. Langford, Z. Long, W. Heneine, P. Sandstrom, W. M. Switzer, L. E. Chapman, C. Lockey, D. Onions, E. Otto, et al. 1999. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. Science 285:1236-1241. [DOI] [PubMed] [Google Scholar]
- 20.Patience, C., G. S. Patton, Y. Takeuchi, R. A. Weiss, M. O. McClure, L. Rydberg, and M. E. Breimer. 1998. No evidence of pig DNA or retroviral infection in patients with short-term extracorporeal connection to pig kidneys. Lancet 352:699-701. [DOI] [PubMed] [Google Scholar]
- 21.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 22.Pitkin, Z., and C. Mullon. 1999. Evidence of absence of porcine endogenous retrovirus (PERV) infection in patients treated with a bioartificial liver support system. Artif. Organs 23:829-833. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi, Y. 2000. Risk of zoonosis in xenotransplantation. Transplant. Proc. 32:2698-2700. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. Le Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tucker, A., C. Belcher, B. Moloo, J. Bell, T. Mazzulli, A. Humar, A. Hughes, P. McArdle, and A. Talbot. 2002. The production of transgenic pigs for potential use in clinical xenotransplantation: microbiological evaluation. Xenotransplantation 9:191-202. [DOI] [PubMed] [Google Scholar]
- 26.Tucker, A. W., F. McNeilly, B. Meehan, D. Galbraith, P. D. McArdle, G. Allan, and C. Patience. 2003. Methods for the exclusion of circoviruses and gammaherpesviruses from pigs. Xenotransplantation 10:343-348. [DOI] [PubMed] [Google Scholar]
- 27.Wilson, C. A., S. Wong, M. VanBrocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood, J. C., G. Quinn, K. M. Suling, B. A. Oldmixon, B A. Van Tine, R. Cina, S. Arn, C. A. Huang, L. Scobie, D. E. Onions, D. H. Sachs, H.-J. Schuurman, J. A. Fishman, and C. Patience. 2004. Identification of exogenous forms of human-tropic porcine endogenous retrovirus in miniature swine. J. Virol. 78:2494-2501. [DOI] [PMC free article] [PubMed]