Abstract
Cronobacter spp. is an emerging pathogen that causes meningitis, sepsis, bacteremia, and necrotizing enterocolitis in neonates and children. The present study developed an assay integrating real-time PCR and high resolution melting (HRM) analysis targeting the OmpA gene for the specific detection and rapid identification of Cronobacter spp. (formerly Enterobacter sakazakii) in powdered infant formula. Eleven Cronobacter field isolates and 25 reference strains were examined using one pair of primers, having the accuracy of 100% in reference to conventional methods. The assay was proved to be highly sensitive with a detection limit of 102 CFU/ml without pre-enrichment, and highly concordant (100%) when compared with ISO-IDF 22964 in 89 actual samples. The method performed for Cronobacter spp. detection was less than 24 h, drastically shortened, compared to several days using standard culturing method, it is probe-free and reduces a risk of PCR carryover. Moreover, all Cronobacter strains examined in this study were genotyped into two species according to their HRM profiles. The established method should provide a molecular tool for direct detection and simultaneous genotyping of Cronobacter spp. in powdered infant formula.
Introduction
Cronobacter spp., formerly named as Enterobacter sakazakii, is an emerging food-borne pathogen causing neonatal meningitis, sepsis and necrotizing enterocolitis and with 40–80% fatality rate to neonates, children and even in adults [1]–[4]. It has been ranked by The International Commission for Microbiological Specification for Foods as a bacterium having “severe hazard for restricted populations, life threatening or substantial chronic sequence long duration” [5]. Phylogenetic analysis of Enterobacter sakazakii has been studied by Lehner et al. [6] who revealed the presence of two phylogenetically distinct lineages based on the full length sequences of 16S rRNA, ie., most of the isolates and the type strain (ATCC 29544) forming one lineage, while type strain (ATCC 51329) formed the other lineage. Recently, E. sakazakii has been proposed to be reclassified as five genomospecies, namely: Cronobacter. muytjensii (type strain ATCC 51329), C. sakazakii (type strain ATCC 29544), C. turicensis, C. dublinensis and C. malonaticus [7], [8].
Powdered infant formulas (PIF) are common products intended for use as a food for infants due to their simulation and suitability as a complete or partial substitute for human breast milk [9]. However, increasing number of outbreaks among infants due to the consumption of PIF contaminated with harmful bacteria, first of all, Cronobacter spp., have been reported [1], [2], [4]. Non-sterile preparation and inappropriate handling of PIF can lead to exacerbation of Cronobacter infection in infants, causing illness with fatal consequence. Rapid, sensitive, simple and accurate techniques for early diagnosis of PIF contaminating pathogens, including Cronobacter spp., are urgently required [10], [11].
To date, various detection and diagnostic techniques for detection of Cronobacter spp. have been developed, including the traditional isolation and biochemical characterization directly from culture or after enrichment, or differential screening medium to identify samples potentially contaminated. However, the conventional method for the isolation and identification of Cronobacter spp. is time consuming and labor intensive, since it requires enrichment culture and then inoculation to selective agar followed by phenotypic identification, taking up to 7 days [12]–[14]. A number of alternative molecular methods for Cronobacter spp. have been investigated, including conventional PCR assay using 16S rRNA [6], OmpA or rpoB genes [15], [16], real time PCR using TaqMan and SYBR Green [3], [9], and the loop-mediated isothermal amplification [17]. However, real-time PCR requires expensive labelled probe, and LAMP may have high risk of amplicon contamination.
Many different genotyping methods have also been used to differentiate and genotype Cronobacter spp., which have been divided into four pulsotypes [1], and 16 fingerprint types [18], five species by PCR-RFLP [19], seven species by MLST [20], while the most authoritative classification was five species based on many biochemical and genetic studies [7], [8]. The major disadvantage of PFGE is its low stability, and the time taken (2 to 4 days). Morever, the high costs and time consuming protocols associated with sequencing for MLST have limited its use. PCR-RFLP has also been impeded by difficulties, such as the minor differences of band sizes between some species and the non-specific primers used for amplification.
Recently, high-resolution melting (HRM) analysis for fast, high-throughput post-PCR analysis of many pathogens has been developed. The HRM has been effectively used for species identification of five human hookworm species, variation scanning for differentiation, ie. between “cattle type” and “sheep type” of Mycobacterium avium [21] or genotyping of human hookworm [22], Pseudomonas savastanoi [23], noroviruses [24], Salmonella serovars [25], and the identification of recent and non-recent HIV infections [26]. To date, HRM has not yet been applied with real-time PCR for detection and genotyping of opportunistic foodborne pathogens Cronobacter spp. The real-time PCR platform with HRM supports is single-step closed tube, which can interrogate different classes of genetic polymorphisms. Moreover, it reduces turnaround time of the assay reported here to almost 1 h, eliminates the risk of contamination, and saves expense [21]–[26]. These features make it advantages for use in microbiology laboratories.
The out membrane protein A (OmpA), attached to host cell and persistence within macrophages, plays an important role in the brain damage [27]. It was previously reported that the OmpA region is suitable for the identification of Cronobacter spp. with higher specificity than ITS, 16S rRNA, zpx, gluA and gluB genes [28]. In the present study, we developed a real-time PCR assay coupled with HRM analysis, which was a more rapid, technically simpler detection and typing method for Cronobacter spp. in powdered infant formula, for the first time, targeting the OmpA gene.
Materials and Methods
Bacterial Strains
The reference strains used in this study were obtained from the American Type Culture Collection (ATCC). ATCC 51329 and ATCC 29544 were used as positive control in the detection assay and in all experimental procedures performed. Other 11 strains of Cronobacter spp. were isolated from the milk powder specimens in Zhongshan Entry Exit Inspection and Quarantine Bureau, Zhongshan, Guangdong Province, China during 2008 and 2010. All of the isolates were determined by Vitek 2 compact system (bioMerieux, Co., USA), and identified ultimately by the classical method ISO-IDF 22964 [12]. The 25 non-Cronobacter reference strains (Table 1) including Salmonella, Shigella and Escherichia coli were used to demonstrate the specificity of the detection assay. All bacterial cultures were maintained on Nutrient Agar plates (BD, USA).
Table 1. Bacterial species used in the real-time PCR and HRM analysis.
| Bacterial species | Source of strain | Result | |
| 1 | Cronobacter muytjensii | ATCC 51329 | + |
| 2 | Cronobacter sakazakii | ATCC 29544 | + |
| 3 | Escherichia coli | ATCC 11775 | – |
| 4 | Escherichia coli | ATCC 11229 | – |
| 5 | Escherchia coli | ATCC 25922 | – |
| 6 | Enterobacte aerogenes | ATCC 13048 | – |
| 7 | Proteus mirabilis | ATCC 12453 | – |
| 8 | Proteus vulgaris | ATCC 6380 | – |
| 9 | Listeria monocytogene | ATCC 19114 | – |
| 10 | Enterococcus faecalis | ATCC 29212 | – |
| 11 | Citrobacter freundii | ATCC 8090 | – |
| 12 | Staphylococcus aureus | ATCC 25923 | – |
| 13 | Klebsiella pneumoniae | ATCC 4352 | – |
| 14 | Salmonella cholerae suis | ATCC 10708 | – |
| 15 | Vibrio parahaemolyticus | ATCC 17802 | – |
| 16 | Salmonella Enteritidis | ATCC 13076 | – |
| 17 | Salmonella Typhimurium | ATCC 13311 | – |
| 18 | Yersinia enterocolitica | ATCC 27729 | – |
| 19 | Yersinia ruckeri | ATCC 29473 | – |
| 20 | Yersinia Kristensenii | ATCC 33639 | – |
| 21 | Shigella sonnei | ATCC 25931 | – |
| 22 | Campylobacter jejuni | ATCC 33291 | – |
| 23 | Pseudomonas aeruginosa | ATCC 27853 | – |
| 24 | Pseudomonas putida | ATCC 49128 | – |
| 25 | Clostridium perfringens | ATCC 13124 | – |
Genomic DNA Extraction
Single colony was picked up from agar plate and inoculated into 3 mL Nutrient broth (LuQiao, China) in a flask. The bacteria were cultured at 37°C for 18 h with shaking. One ml of each bacterial culture was centrifuged at 8000 g for 5 min, and the bacterial pastes were subjected to DNA extraction using a SK8192 DNA extraction Kit (Sangon Biotech, China). The bacterial genomic DNA samples (in TE buffer) were stored at −20°C until further use.
Primers, Real-time PCR and HRM Analysis
A pair of primers targeting the OmpA gene of Cronobacter spp. (ie., ESOMP5- F: 5′-GGTGAAGGATTTAACCGTGAACTT-3′ and ESOMP5-R: 5′- GCGCCTCGTTATCATCCAAA-3′), was synthesized by Takara Biotechnology (Dalian, China). Real-time PCR amplification of the OmpA target was performed in Lightcycler 480 (Roche, USA). The total reaction volume was 20 µL, which consisted of 1× Fast-Plus EvaGreen® qPCR Master Mix (Bio-rad, USA), reaction buffer containing dNTPs, MgCl2, fast-activating chemically-modified hotstart enzyme, Cheetah™ Taq, 0.3 µM of each primer and 1 µL genomic DNA as template.
The PCR was carried out with initiation at 95°C for 60 s, then 45 cycles of denaturation 95°C for 10 s, annealing at 60°C for 10 s and extension at 72°C for 20 s. When PCR amplification was completed, HRM analysis was performed by lowering the temperature to 60°C for 5 min, followed by increasing the temperature ramping from 60°C to 95°C at 0.11°C/s, 25 acquisitions/°C. In this process, the PCR amplicons were allowed to denature and re-anneal before the high resolution melting recording changes in fluorescence with changes in temperature (dF/dT) and plotting against changes in temperature. The HRM profile was then analyzed using HRM analysis software version 2.0.1 with fluorescence (melting curve) normalization by selecting the linear region before and after the melting transition, as reported previously [24]. The auto-group function of the software was applied to generate automatic genotype group by analyzing the normalized melting curves and clustering samples into groups with similar melting profiles. Hence the difference in the shape of the “normalized melting curve” will helped cluster samples into different subgroups, and the samples can be detected and grouped in the same time [21]–[26].
Specificity of the Detection Assay
To evaluate the specificity of the assay, 25 reference strains (Table 1) were cultured and maintained on Nutrient Agar (Oxoid, UK) plates. A single colony of pure culture was resuspended in 1 ml sterile, deionised powdered infant formula after which the genomic DNA was extracted as described above. Genomic DNA extract was used as template in the subsequent real-time PCR reactions. The specificity of the results was based upon the melting curve analysis and real time PCR amplification curve. Moreover, the identity of the amplicons was further confirmed by sequencing (Takara Biotech, China) and subsequent alignment of the obtained sequences with corresponding sequences available in GenBank.
Internal Control
A segment of the OmpA gene of Cronobacter spp., corresponding to nucleotides 113 to 182 (GenBank Accession No. GQ845410.1), was amplified from genomic DNA using the primers ESOMP5F/ESOMP5R to generate an amplicon. The 70 bp product was ligated into the pGEM-T Easy plasmid and used to transform competent E. coli DH5α cells. Plasmids with the target insert (OmpA-containing plasmids) were confirmed by sequencing, then quantified and used as internal control for real time PCR assay.
Detection Limit of the Assay
Artificially contaminated powdered infant formula which were tested negative for Cronobacter spp. by selective plating before use were prepared under double-blind conditions by directly spiking with Cronobacter spp. Briefly, 100 g of powdered infant formula were added to 900 mL BPW (Buffered Peptone Water) medium (Oxoid, UK). Then, two homogenates of each sample were prepared; one was confirmed to be negative for Cronobacter spp. according to ISO-IDF 22964 [12], and the other one was randomly inoculated with the described dilution under a double-blind condition.
The theoretical final concentration of Cronobacter spp. in each contaminated sample was determined using plating techniques. This was achieved by plating 200 µl of each serial dilution of Cronobacter spp. used on a nonselective media (Nutrient Agar, BD) in triplicate. Following overnight incubation at 36±1.0°C, the average count was determined for each dilution and the theoretical concentration of C. sakazakii per powdered infant formula sample was stated as CFU/ml. After artificial contamination (ranged from 10 to 107 CFU/mL) with diluted Cronobacter, aliquots of 1 mL of homogenates were collected, then genomic DNA was extracted and real-time PCR assay was performed in triplicate.
Results
Specificity of the Detection Assay and Confirmation of Amplicon Identity
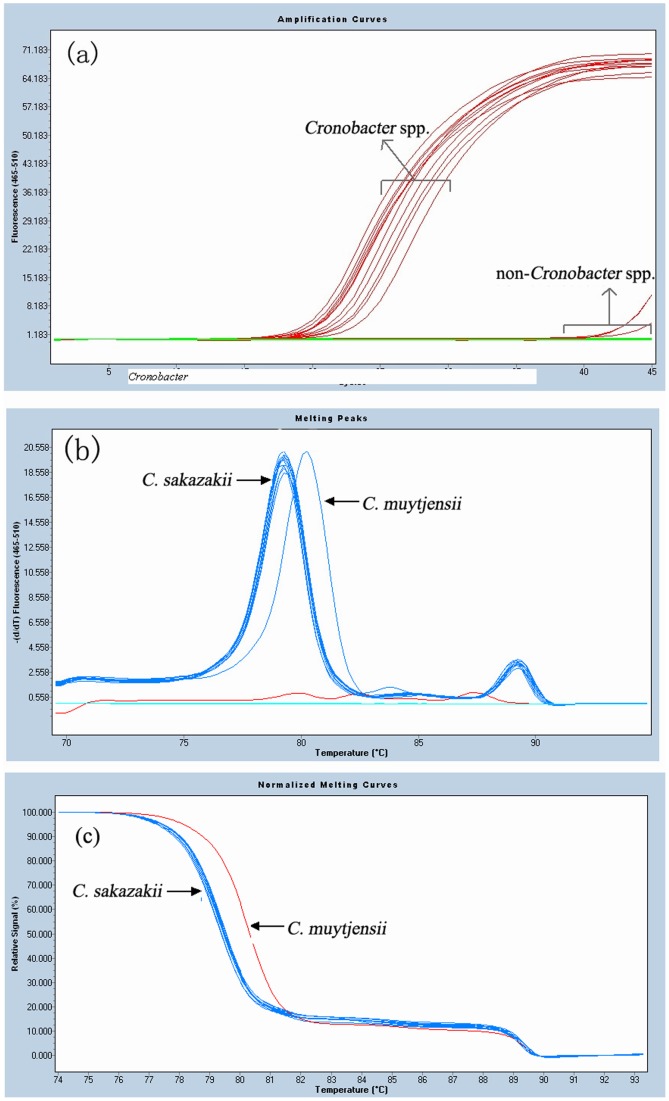
The specificity of the OmpA primers was evaluated using 11 Cronobacter isolates and 25 reference strains. Only strains of Cronobacter spp. produced positive signal (Figure 1a). All the non-Cronobacter strains and blank control had no amplification curve before 40 cycles. Agarose gel electrophoresis indicated that positive amplification products correlated to a size of 70 bp as expected, and nucleotide sequence analyses of the amplicons confirmed 100% identical to C. sakazakii (type strain ATCC 29544), while a difference of two bases was detected between C. sakazakii and C. muytjensii (type strain ATCC 51329) (BLASTn option, GenBank).
Figure 1. Real time PCR and HRM analysis for all bacterial strains.
(a) Real time PCR amplification for Cronobacter spp. and other reference strains. All the non-Cronobacter strains and blank control had no amplification curve before 40 cycles. (b) Melting peaks of Cronobacter spp. 1, C. sakazakii including type strain ATCC 29544 and all isolates, Tm, 79.23±0.05°C. 2, C. muytjensii including type strain ATCC 51329, Tm, 80.11°C. (c) Normalized melting curves for Cronobacter spp. strains.
Detection Limit and Reproducibility of the Assay
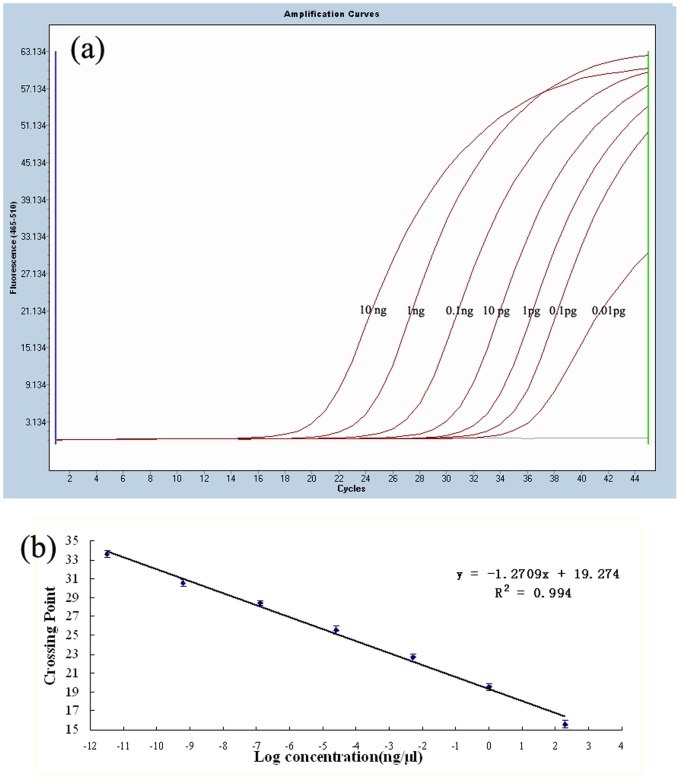
Using powdered infant formula samples artificially contaminated with Cronobacter tenfold serial dilutions were performed by dissolved into BPW at 10–107 CFU/mL to assay the analytical sensitivity. The results demonstrated that the lower limit of detection of samples without pre-enrichment was 102 CFU/mL. Separately, a series of 10-fold diluted genomic DNA (10 ng to 0.01 pg) was chosen to evaluate the sensitivity of the detection. The results showed that limit of the assay was 0.01 pg (Figure 2a), when serial 10 dilutions were tested along with a blank control. Besides, there a good linear correlation (R2 = 0.994) between the log concentrations of purified DNA (10 ng to 0.01 pg) and the crossing point (Cp) value (Figure 2b). The detection assay also displayed a high degree of reproducibility. Intra-run analysis successfully detected the presence of Cronobacter spp. in all 10 replicates with very little variation in Cp and melting-temperature (Tm) value. Successful detection of Cronobacter spp. in the same milk powder sample over a period of three consecutive days also indicated high inter-run reproducibility.
Figure 2. Real time PCR and linear correlation of 10-fold serial dilution ofCronobacter spp. genomic DNA.
(a) amplification curves of diluted genomic DNA of Cronobacter spp., 1–7: dilutions 10 ng to 0.01 pg, respectively. (b) a linear regression of the data providing a formula of y = −1.2709x+19.274(R2 = 0.994), dilutions 10 ng to 0.01 pg, respectively.
Comparison of the Method with ISO-IDF 22964 for DetectingCronobacter spp
The usefulness of the real-time PCR combined with HRM assay was also compared to the method ISO-IDF 22964 in actual samples. A total of 89 milk powder samples were investigated using the cultural method and real-time PCR with HRM in parallel, between May 2012 and March 2013. Twenty-five grams test portions were analysed in duplicate for each sample after BPW-mLST enrichments as recommended [12]. The results showed that 7 samples were positive,while 82 samples was negative, 100% concordance with the gold standard method (ISO-IDF 22964).
HRM Analysis forCronobacter spp
The sequence difference between the forward and reverse primers will bring different melting temperature (Tm) values and normalized melting curves. Constant HRM profiles with distinct Tm peaks were persistently obtained for all Cronobacter spp. strains. As shown in Figure 1b, there were two kinds of characteristic profiles for Cronobacter spp. The OmpA amplification product from ATCC 51329 had a Tm of 80.11°C, while the amplification product from ATCC 29544 and 11 isolates collected by us had an average 79.23°C ±0.05°C (Figure 1b).
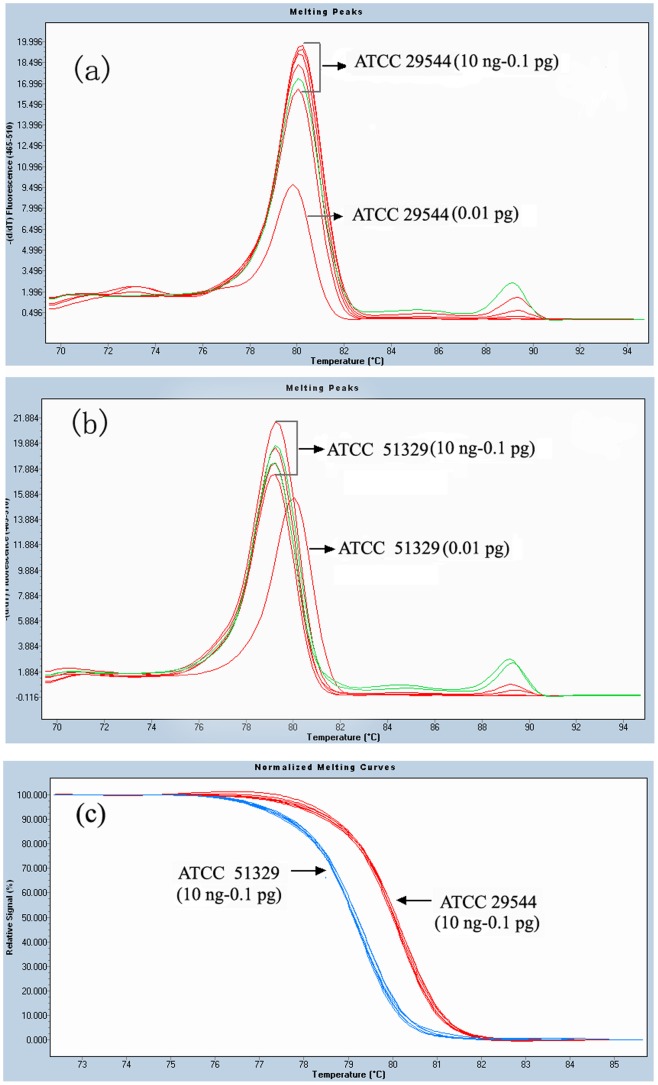
The HRM analysis with different concentrations (10 ng to 0.1 pg) of the template, appeared to be reliable. The melting peaks were obtained from each dilution (10 ng to 0.1 pg), it showed that Tm of ATCC 29544 was 79.20±0.04°C (Figure 3a), while Tm of ATCC 51329 was 80.06±0.08°C (Figure 3b). When the concentration of template was diluted to 0.01 pg, there was an obvious change, both in ATCC 51329 and ATCC 29544 (Figure 3a, 3b). Thus, the HRM profiles may be unreliable when the concentration of the template was too low (≤0.01 pg). As shown in Figure 3c, there were two obvious groups, one represented C. sakazakii (DNA ranged from 10 ng to 0.1 pg), and the other represented C. muytjensii ATCC 51329 (DNA ranged from 10 ng to 0.1 pg).
Figure 3. HRM analysis of 10-diluted genomic DNA (10 ng to 0.01 pg) from different type strains.
(a) Melting peaks of type strain (ATCC 29544), Tm, 79.21±0.04°C. (b) Melting peaks of type strain (ATCC 51329), Tm, ATCC 51329, 80.06±0.08°C. (c) Normalized melting curves of both type strains (10 ng to 0. 1 pg).
Discussion
The currently routine procedure for detection of Cronobacter spp. is laborious, taking up to 7 days for completion [12]–[14]. However, even with the additional enrichment step, the method developed in this study can detect Cronobacter spp. in about 24 h, and reduces the need to purchase expensive probes. Real time PCR combined with HRM analysis is a new genotyping technique with advantages over PFGE, PCR-RFLP and DNA sequencing. A differentiation has also been established for genomospecies within the Cronobacter spp. based on sequencing after routine PCR performance [19], [20], whereas, the single nucleotide polymorphism (SNP) can be detected by HRM instead of sequencing with much less cost and time.
Real time PCR targeting ITS sequences of Cronobacter spp. using SYBR Green has been developed [3], however, it can only detect gross differences between amplicons generated by real-time PCR, which commonly uses non-saturating dyes such as SYBR Green which will inhibit the PCR reaction. EvaGreen has been proven to be superior over SYBR Green in general, and high reaction efficiencies can be achieved [29]. The improved instrumentation for HRM analysis with a high rate of data acquisition, ideal optics, tight temperature control and adequate analysis software has enabled us to detect single-base difference by means of HRM analysis. In contrast to traditional melting curve analysis, the information in HRM analysis is contained in the shape of the melting curve, rather than just the calculated Tm, so HRM analysis may be considered a form of spectroscopy and the accuracy of the dissociation vs temperature (i.e. melting) curve is as sensitive as 0.01°C [30].
We failed to find appropriate primers targeting 16S rRNA and ITS for detection and subtyping of Cronobacter spp. simultaneously, while acquired a good performance by using OmpA gene as genetic marker. Moreover, the length of PCR product is important for HRM analysis. Analyzing short amplicons can assist genotypic discrimination in HRM analysis. Whenever possible, analyzing amplicons smaller than 100 bp is preferable, especially when sites with a known polymorphism are investigated. Reducing amplicon size increases the difference in signal. Although it is highly recommended to standardize amount of DNA templates to minimize reaction-to-reaction variability in HRM assays, however, it is not found to be extraordinarily critical for distinguishing sequence difference in our study.
It is interesting that phenotypic characteristics of all 11 Cronobacter isolates were consistent with that of C. sakazakii described previously [7]. Moreover, the OmpA sequences of these isolates were identical to that of C. sakazakii. As shown in HRM profiles in Figure 1, all 13 Cronobacter strains clustered into two obvious groups, one represented C. sakazakii (including ATCC 29544 and 11 isolates), and the other represented C. muytjensii (ATCC 51329). These results exhibited a clear correlation between phylogenetic analysis and HRM profiles. Currently five genomospecies are commonly recognized within the genus Cronobacter spp. [7], [8], but previous studies have shown that C. sakazakii is the predominant species, especially in PIF [19], [31]–[33]. OmpA sequences of three Cronobacter species were available in GenBank. As shown in Figure 4, there were ≥1 different bases in the segment between the forward and reverse primers in each species, which resulted in differences in melting temperature (Tm) values and HRM profiles. Therefore, the method we developed in this study has potential to differentiate all five recognized Cronobacter species, and this will be tested when strains representing other Cronobacter species become available.
Figure 4. Alignment of theOmpA gene from Cronobacter spp. available in GenBank.
1: Cronobacter muytjensii (accession number: DQ000206.1); 2: Cronobacter sakazakii (accession number: GQ845410.1); 3: Cronobacter turicensis z3032 (accession number: FN543093.2).
In conclusion, the present study developed an assay combining real-time PCR and HRM analysis for rapid and specific detection and simultaneous genotyping of Cronobacter spp. in powdered infant formula. This method has potential for multi-genotying of all recognized Cronobacter species, and can be used for epidemiological studies of Cronobacter spp.
Funding Statement
This work was supported by The Science & Technology Program of Zhongshan (Grant No. 20123A299) to XQC, The Science Fund for Creative Research Groups of Gansu Province (Grant No. 1210RJIA006) to XQZ, and the Science & Technology Program of The General Administration of Quality Supervision, Inspection and Quarantine of the People's Republic of China (Grant No. 2008IK007) to HQY. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Caubilla-Barron J, Hurrell E, Townsend S, Cheetham P, Loc-Carrillo C, et al. (2007) Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J Clin Microbiol 45: 3979–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hunter CJ, Petrosyan M, Ford HR, Prasadarao NV (2008) Enterobacter sakazakii: an emerging pathogen in infants and neonates. Surg Infect (Larchmt) 9: 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Y, Cai X, Zhang X, Gao Q, Yang X, et al. (2006) Real time PCR using TaqMan and SYBR Green for detection of Enterobacter sakazakii in infant formula. J Microbiol Meth 65: 21–31. [DOI] [PubMed] [Google Scholar]
- 4. O'Brien S, Healy B, Negredo C, Anderson W, Fanning S, et al. (2009) Prevalence of Cronobacter species (Enterobacter sakazakii) in follow-on infant formulae and infant drinks. Lett Appl Microbiol 48: 536–541. [DOI] [PubMed] [Google Scholar]
- 5. Hoque A, Ahmed T, Shahidullah M, Hossain A, Mannan A, et al. (2010) Isolation and molecular identification of Cronobacter spp. from powdered infant formula (PIF) in Bangladesh. Int J Food Microbiol 142: 375–378. [DOI] [PubMed] [Google Scholar]
- 6. Lehner A, Tasara T, Stephan R (2004) 16S rRNA gene based analysis of Enterobacter sakazakii strains from different sources and development of a PCR assay for identification. BMC Microbiol 4: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iversen C, Lehner A, Mullane N, Bidlas E, Cleenwerck I, et al. (2007) The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol Biol. 7: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, et al. (2008) Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol 58: 1442–1447. [DOI] [PubMed] [Google Scholar]
- 9. Seo KH, Brackett RE (2005) Rapid, specific detection of Enterobacter sakazakii in infant formula using a real-time PCR assay. J Food Prot 68: 59–63. [DOI] [PubMed] [Google Scholar]
- 10.WHO (2007) Guidelines for the safe preparation, storage and handling of powdered infant formula. WHO website. Available: http://www.who.int/foodsafety/publications/micro/pif2007/en/. Accessed 2012 Aug 8.
- 11. Kandhai MC, Heuvelink AE, Reij MW, Beumer RR, Dijk R, et al. (2010) A study into the occurrence of Cronobacter spp. in The Netherlands between 2001 and 2005. Food Control 21: 1127–1136. [Google Scholar]
- 12.International Organization for Standardization (2006) ISO-IDF 22964|IDF/RM 210: 2006. Specifies a method for the detection of Enterobacter sakazakii in milk and milk products.
- 13. Druggan P, Iversen C (2009) Culture media for the isolation of Cronobacter spp. Int J Food Microbiol 136: 169–178. [DOI] [PubMed] [Google Scholar]
- 14. Turcovský I, Kuniková K, Drahovská H (2011) Biochemical and molecular characterization of Cronobacter spp. (formerly Enterobacter sakazakii) isolated from foods. Antonie Van Leeuwenhoek 99: 257–269. [DOI] [PubMed] [Google Scholar]
- 15. Mohan NMK, Venkitanarayanan KS (2006) Cloning and sequencing of the OmpA gene of Enterobacter sakazakii and development of an OmpA-targeted PCR for rapid detection of Enterobacter sakazakii in infant formula. Appl Environ Microbiol 72: 2539–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stoop B, Lehner A, Iversen C, Fanning S, Stephan R (2009) Development and evaluation of rpoB based PCR systems to differentiate the six proposed species within the genus Cronobacter . Int J Food Microbiol 136: 165–168. [DOI] [PubMed] [Google Scholar]
- 17. Liu X, Fang J, Zhang M, Wang X, Wang W, et al. (2012) Development of a loop-mediated isothermal amplification assay for detection of Cronobacter spp. (Enterobacter sakazakii). World J Microbiol Biotechnol 28: 1013–1020. [DOI] [PubMed] [Google Scholar]
- 18. Ye Y, Wu Q, Xu X, Yang X, Dong X, et al. (2010) The phenotypic and genotypic characterization of Enterobacter sakazakii strains from infant formula milk. J Dairy Sci 93: 2315–2320. [DOI] [PubMed] [Google Scholar]
- 19. Strydom A, Cameron M, Corli Witthuhn R (2011) PCR-RFLP analysis of the rpoB gene to distinguish the five species of Cronobacter . Food Microbiol 28: 1472–1477. [DOI] [PubMed] [Google Scholar]
- 20. Joseph S, Sonbol H, Hariri S, Desai P, McClelland M, et al. (2012) Diversity of the Cronobacter genus as revealed by multi-locus sequence typing. J Clin Microbiol 50: 3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Douarre PE, Cashman W, Buckley J, Coffey A, O'Mahony JM (2012) High resolution melting PCR to differentiate Mycobacterium avium subsp. Paratuberculosis “cattle type” and “sheep type”. J Microbiol Meth 88: 172–174. [DOI] [PubMed] [Google Scholar]
- 22. Ngui R, Lim YA, Chua KH (2012) Rapid detection and identification of human hookworm infections through high resolution melting (HRM) analysis PLoS One. 7: e41996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gori A, Cerboneschi M, Tegli S (2012) High-resolution melting analysis as a powerful tool to discriminate and genotype Pseudomonas savastanoi pathovars and strains. PLoS One 7: e30199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tajiri-Utagawa E, Hara M, Takahashi K, Watanabe M, Wakita T (2008) Development of a rapid high-throughput method for high-resolution melting analysis for routine detection and genotyping of noroviruses. J Clin Microbiol 47: 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zeinzinger J, Pietzka AT, Stöger A, Kornschober C, Kunert R, et al. (2012) One-step triplex high-resolution melting analysis for rapid identification and simultaneous subtyping of frequently isolated Salmonella serovars. Appl Environ Microbiol 78: 3352–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cousins MM, Swan D, Magaret CA, Hoover DR, Eshleman SH (2012) Analysis of HIV using a High Resolution Melting (HRM) diversity assay: automation of HRM data analysis enhances the utility of the assay for analysis of HIV incidence. PLoS One 7: e51359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mittal R, Wang Y, Hunter CJ, Gonzalez I, Prasadarao NV (2009) Brain damage in newborn rat model of meningitis by Enterobacter sakazakii: a role for outer membrane protein A. Lab Invest. 89: 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jaradat ZW, Ababneh QO, Saadoun IM, Samara NA, Rashdan AM (2009) Isolation of Cronobacter spp. (formerly Enterobacter sakazakii) from infant food, herbs and environmental samples and the subsequent identification and confirmation of the isolates using biochemical, chromogenic assays, PCR and 16S rRNA sequencing. BMC Microbiol 9: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eischeid AC (2011) SYTO dyes and EvaGreen outperform SYBR Green in real-time PCR. BMC Res Notes 4: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin D, Luo Y, Zhang Z, Fang W, Ye J, et al. (2012) Rapid molecular identification of Listeria species by use of real-time PCR and high-resolution melting analysis. FEMS Microbiol Lett 330: 72–80. [DOI] [PubMed] [Google Scholar]
- 31. Miled-Bennour R, Ells TC, Pagotto FJ, Farber JM, Kérouanton A, et al. (2010) Genotypic and phenotypic characterisation of a collection of Cronobacter (Enterobacter sakazakii) isolates. Int J Food Microbiol 139: 116–125. [DOI] [PubMed] [Google Scholar]
- 32. Terragno R, Salve A, Pichel M, Epszteyn S, Brengi S, et al. (2009) Characterization and subtyping of Cronobacter spp. from imported powdered infant formulae in Argentina. Int J Food Microbiol 136: 193–197. [DOI] [PubMed] [Google Scholar]
- 33. Strydom A, Cameron M, Witthuhn RC (2012) Phylogenetic analysis of Cronobacter isolates based on the rpoA and 16S rRNA genes. Curr Microbiol 64: 251–258. [DOI] [PubMed] [Google Scholar]