Abstract
Cryoelectron microscopy of Mouse mammary tumor virus, a Betaretrovirus, provided information about glycoprotein structure and core formation. The virions showed the broad range of diameters typical of retroviruses. Betaretroviruses assemble cytoplasmically, so the broad size range cannot reflect the use of the plasma membrane as a platform for assembly.
Mouse mammary tumor virus (MMTV) can be endogenously transmitted through the germ line or exogenously transmitted via milk. The virus induces formation of mammary carcinomas. Its wide use as a model system has provided important insights into cellular signaling pathways, host-virus interactions, and the genetic basis of breast cancer (3, 12). The morphology of the virus and therefore the characteristic morphology of the genus have been described previously on the basis of negative-stain electron microscopy (11, 13). The Betaretrovirus spp. are distinguished from those of other genera by distinct surface projections and an eccentrically located core.
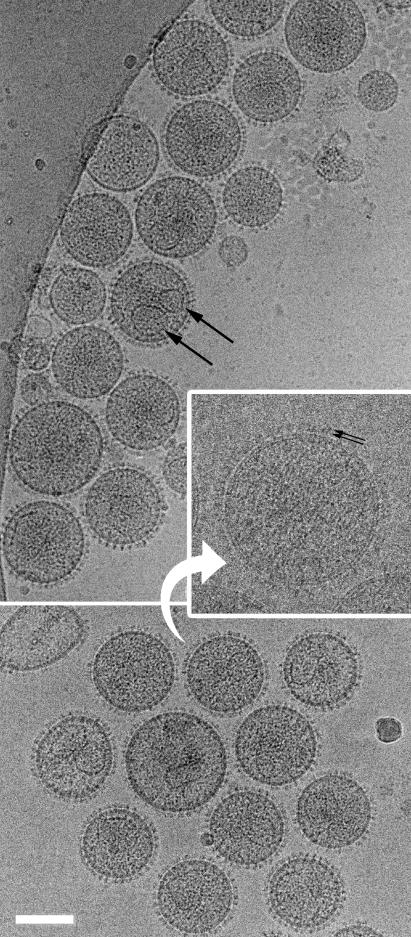
The virus was produced from GR cells, in which MMTV is endogenous. The cells were grown to confluence, and virus production was induced by overnight incubation in 0.1 μM dexamethasone. The cell supernatant was clarified by 45 min of centrifugation at 3,750 rpm and ultracentrifuged through a 20% sucrose cushion in a Beckman SW28 rotor for 2 h at 26,000 rpm. The pellet was resuspended overnight in TN buffer (25 mM Tris [pH 7.6], 100 mM NaCl) and stored at −20°C. Virus from four consecutive overnight inductions was thawed, pooled, pelleted by 1 h of ultracentrifugation, resuspended in 200 μl of TN buffer, and then separated into 18 fractions by ultracentrifugation into a 20 to 60% sucrose gradient in a Beckman SW41 rotor for 16 h at 40,000 rpm. A visible band indicated the virus-containing fractions. These fractions were pooled, pelleted by a further 4 h of ultracentrifugation, and resuspended in 100-μl TN buffer. Three independent preparations were made. Cryogrids were prepared and cryoelectron microscopy (cEM) was performed as described previously by using a Phillips CM200 FEG operated at 200 kV and a GATAN 626 Cryo-transfer holder (8, 16). cEM permits visualization of the virus in the unfixed, unstained, hydrated state, which eliminates the inherent problems with particle distortion when studying size and morphology by negative stain. The virions displayed classical MMTV morphology (Fig. 1). They are roughly spherical, with prominent glycoprotein knobs. The core is eccentrically located but in many virions is difficult to distinguish. Images were digitized on a UMAX 3000 scanner at 8-μm step size and manipulated by using SPIDER (7) and IMAGIC (14) image processing packages.
FIG. 1.
Two fields of MMTV virions. The range of particle size and core size is illustrated. Large black arrows indicate two cores within one virion. The inset shows a contrast transfer function corrected image taken closer to focus. Small black arrows indicate the two radially arranged densities in the glycoprotein. Scale bar, 200 nm.
No particles with double-shelled immature morphology (13) were observed. This finding contrasts with observations of other retroviruses, such as human immunodeficiency virus (HIV) (2), which consistently show a small fraction of immature particles.
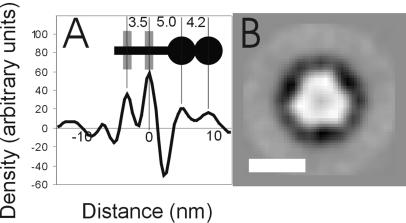
The virions exhibit prominent glycoprotein knobs on the surface. The spacing between the knobs is approximately 12 nm, which corresponds to about 400 knobs on an average virion or 1,200 copies of surface envelope protein. HIV, in contrast, is believed to have less than 50 copies of surface envelope protein (5, 10). The distance between spikes does not vary visibly with particle size. The knobs appear to have two radially distinct densities (Fig. 1). Radial density profiling was performed on contrast transfer function corrected images (6) by using the Fourier Bessel expansion method described previously (16). Three hundred sixty profiles were averaged for each particle used. Radial density profiles of eight virions were measured, aligned using the peak corresponding to the outer leaflet of the bilayer, and summed. The two densities of the knob are clearly visible at distances of 5.0 and 9.2 nm from the outer leaflet (Fig. 2A). The knob is similar in length to the 9.5 nm observed for HIV-1 (16) and shorter than the 14 nm observed for human foamy virus (15).
FIG. 2.
(A) The radial density profile of the membrane region of mature MMTV. Radial density profiles for eight MMTV virions were aligned and averaged. The two largest peaks correspond to the inner (left) and outer (right) leaflets of the bilayer. Outside the membrane, the glycoprotein consists of two distinct densities. The inset shows the schematic glycoprotein structure (black) with the lipid bilayer (grey), indicating the distances in nanometers between features. (B) A top view of the viral spike obtained by aligning, averaging, and applying threefold symmetry to images of 542 negatively stained spikes. Scale bar, 5 nm.
Five hundred forty two top views of viral glycoprotein knobs were collected from images of negatively stained virions. These views were translationally and rotationally aligned and averaged, suggesting threefold symmetry. This symmetry was applied, revealing a trimer with a central stain-filled depression (Fig. 2B) similar to that of earlier observations (13).
The viral core is eccentrically located. The core is irregularly sized and shaped and is often angular. It is not significantly more electron-dense than the rest of the virion, so interpretation is difficult. Figure 1 illustrates the range of core shapes seen. The dimensions of 117 single cores were measured. The observed ratio of long to short axes varied between 1 and 2.2, with dimensions varying between 45 nm along the short axis to 110 nm along the long axis. The data are inconsistent with a homogeneous population of cores being viewed from different angles and so reflect true variation in structure. The variation appears greater than that observed for HIV or murine leukemia virus (2, 17). Extensive attempts in our laboratory to repeat the isolation and purification of the viral core using the methods described by Cardiff and coworkers (4) were unsuccessful.
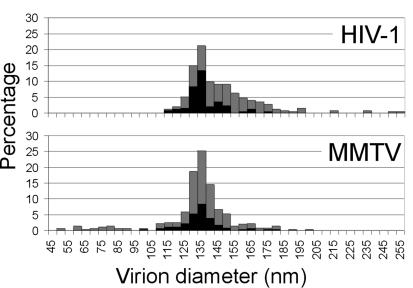
cEM permits reliable measurement of virus diameter, avoiding the problems associated with viral membrane collapse when using conventional techniques (11, 17). The diameters of viruses were measured to the outer edge of the lipid bilayer. The three preparations exhibited mean particle diameters of 130 ± 18 nm (n = 150), 129 ± 18 nm (n = 150), and 141 ± 14 nm (n = 300), with virus diameter ranging between 50 and 210 nm. The mean diameter is significantly smaller than that observed after negative staining (180 nm) and similar to that observed after freeze drying (125 nm) (11). Data from preparations 1 and 2 were combined (Fig. 3) and exhibit a size distribution of a breadth similar to that of HIV-1 (2) (Fig. 3). The large range of diameters observed is typical of retrovirus preparations (2, 9, 17). In most retroviruses, the Gag protein shell assembles either at the plasma membrane or on an internal membrane and the lipid bilayer plays a significant role in the assembly process (1). Assembly of Gag protein and nucleic acid in vitro in the absence of a lipid bilayer leads to the production of particles that are more homogeneous in size (16, 18). Betaretroviridae species, in contrast, assemble their Gag protein shells in the cytoplasm. The dramatic range in diameters seen within retrovirus preparations, therefore, does not result solely from the use of the lipid bilayer as an assembly platform.
FIG. 3.
The size distribution of mature HIV-1 (2) and MMTV virions. A histogram of the diameters to the outside of the lipid bilayer for all virions (grey bars) and for the subset of virions for which single cores were clearly distinguished (black bars) is shown.
The HIV-1 size distribution has a distinct shoulder towards higher radius formed by particles containing two cores (2) (Fig. 3). HIV-1 particles possessing a clearly interpretable single core are therefore smaller (mean value, 134 nm) than the overall distribution of virus diameters (mean value, 145 nm) (Fig. 3). The larger double-cored particles in HIV-1 are thought to arise from copackaging of two genomes (four RNA strands), which are then condensed into two separate cores. Copackaging may occur when domains of Gag protein assemble adjacent to one another on the plasma membrane and then bud into the same virion (2, 8). This process could not take place during cytoplasmic assembly. The lack of a shoulder in the MMTV size distribution is consistent with this hypothesis. The distribution of virions clearly containing a single core (mean diameters, 132 nm for preparations 1 and 2 and 141 nm for preparation 3) is similar to the overall size distribution of the virus (mean diameters, 130 and 141 nm) (Fig. 3). Some MMTV virions, however, do contain two cores (Fig. 1). In less than 3% of virions were two cores clearly distinguished, but 47% contain density that could be attributed to multiple cores.
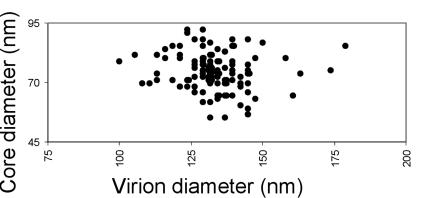
There is no significant correlation between the size of a virion and the apparent size of its core (Fig. 4). This finding implies that upon maturation core growth is not limited by the amount of protein available. Prior to maturation, the amount of Gag present is proportional to the surface area of the virion, meaning that the largest core-containing viruses, with a diameter 1.7 times that of the smallest core-containing viruses, will contain three times as much Gag protein yet produce similarly sized cores. Core growth must either be limited by a template such as the ribonucleoprotein or must proceed until the concentration of CA protein in the virus drops below a certain threshold. In MMTV, the absence of any correlation between number of cores, particle size, and core size suggests that cobudding of independent genome-containing Gag assemblies into the same particle is not the cause of size heterogeneity or of double-core formation.
FIG. 4.
The relationship between virus size and core size. The mean of the long and short axial lengths of the core were plotted against the virus diameter. There is no significant correlation.
MMTV exhibits the striking variability in virus diameter and in core size and shape that is characteristic of retroviruses. The presence of such variability in a betaretrovirus implies that it does not reflect the role of a lipid bilayer in the assembly process.
Acknowledgments
GR cells were a kind gift from Elena Buetti (Université de Lausanne, Lausanne, Switzerland). We acknowledge the contribution of Thomas Wilk (Vossius and Partner, Münich, Germany) who initiated work on MMTV in their laboratory. They thank Milan Nermut (National Institute for Biological Standards and Control, Potters Bar, Herts, United Kingdom) and Thomas Wilk for critical readings of the manuscript.
We acknowledge support from a Wellcome Trust Programme Grant (HH5FE) for this work. J.A.G.B. holds a Wellcome Trust Structural Biology Studentship. S.D.F. is a Wellcome Trust Principal Research Fellow.
REFERENCES
- 1.Briggs, J. A. G., T. Wilk, and S. D. Fuller. 2003. Do lipid rafts mediate virus assembly and pseudotyping? J. Gen. Virol. 84:757-768. [DOI] [PubMed] [Google Scholar]
- 2.Briggs, J. A. G., T. Wilk, R. Welker, H. G. Krausslich, and S. D. Fuller. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callahan, R., and G. H. Smith. 2000. MMTV-induced mammary tumorigenesis: gene discovery, progression to malignancy and cellular pathways. Oncogene 19:992-1001. [DOI] [PubMed] [Google Scholar]
- 4.Cardiff, R. D., M. J. Puentes, Y. A. Teramoto, and J. K. Lund. 1974. Structure of the mouse mammary tumor virus: characterization of bald particles. J. Virol. 14:1293-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chertova, E., J. W. Bess, B. J. Crise, R. C. Sowder, T. M. Schaden, J. M. Hilburn, J. A. Hoxie, R. E. Benveniste, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Haas, F., A. O. Paatero, L. Mindich, D. H. Bamford, and S. D. Fuller. 1999. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage phi 6. J. Mol. Biol. 294:357-372. [DOI] [PubMed] [Google Scholar]
- 7.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 8.Fuller, S. D., T. Wilk, B. E. Gowen, H. G. Krausslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 9.Kingston, R. L., N. H. Olson, and V. M. Vogt. 2001. The organization of mature Rous sarcoma virus as studied by cryoelectron microscopy. J. Struct. Biol. 136:67-80. [DOI] [PubMed] [Google Scholar]
- 10.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, S. R. Conley, J. P. Moore, J. L. Raina, H. Renz, H. R. Gelderblom, and P. L. Nara. 1992. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189:695-714. [DOI] [PubMed] [Google Scholar]
- 11.Nermut, M. V. 1973. Size, shape and surface structure of mouse mammary tumour virus (MTV). Brief report. Arch. Gesamte Virusforsch. 41:284-289. [DOI] [PubMed] [Google Scholar]
- 12.Ross, S. R. 2000. Using genetics to probe host-virus interactions; the mouse mammary tumor virus model. Microbes Infect. 2:1215-1223. [DOI] [PubMed] [Google Scholar]
- 13.Sarkar, N. H. 1987. Oncovirinae: type B oncovirus, p. 257-271. In M. V. Nermut and A. C. Stevens (ed.), Animal virus structure. Elsevier, Amsterdam, The Netherlands.
- 14.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 15.Wilk, T., F. de Haas, A. Wagner, T. Rutten, S. Fuller, R. M. Flugel, and M. Lochelt. 2000. The intact retroviral Env glycoprotein of human foamy virus is a trimer. J. Virol. 74:2885-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H. G. Krausslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]