Abstract
Despite increasing characterization of pestivirus-encoded proteins, functions for nonstructural (NS) proteins NS2, NS2-3, NS4B, and NS5A have not yet been reported. Here we investigated the function of bovine viral diarrhea virus (BVDV) uncleaved NS2-3. To test whether NS2-3 has a discrete function, the uncleaved protein was specifically abolished in two ways: first by inserting a ubiquitin monomer between NS2 and NS3, and second by placing an internal ribosome entry site between the two proteins (a bicistronic genome). In both cases, complete processing of NS2-3 prevented infectious virion formation without affecting RNA replication. We tested the hypothesis that uncleaved NS2-3 was involved in morphogenesis by creating a bicistronic genome in which NS2-3 was restored in the second cistron. With this genome, both uncleaved NS2-3 expression and particle production returned. We then investigated the minimal regions of the polyprotein that could rescue an NS2-3 defect by developing a trans-complementation assay. We determined that the expression of NS4A in cis with NS2-3 markedly increased its activity, while p7 could be supplied in trans. Based on these data, we propose a model for NS2-3 action in virion morphogenesis.
The genus Pestivirus includes the important animal pathogens Classical swine fever virus, Border disease virus, and Bovine viral diarrhea virus (BVDV). BVDV infection of cattle can result in asymptomatic infection and seroconversion or a variety of pathologies including fatal mucosal disease (reviewed in reference 29). The genome organization and translation initiation of the pestiviruses closely resemble those of hepatitis C virus. Hepatitis C virus is the sole member of the Hepacivirus genus, which, along with the flaviviruses and pestiviruses, makes up the family Flaviviridae. Because of its close relation to hepatitis C virus, BVDV is increasingly being studied as a surrogate for this important human pathogen (reviewed in reference 23).
BVDV is a single-stranded RNA virus of positive polarity with a nonsegmented genome of approximately 12.3 kb. The genome is not capped, and on entry into the cell, translation is initiated at an internal ribosome entry site (IRES) formed by the highly conserved 5′ nontranslated region (NTR). A single polyprotein is produced and is co- and posttranslationally processed by viral and cellular enzymes. With the exception of the nonvirion protein Npro, the structural proteins are found at the amino terminus of the polyprotein, while the nonstructural (NS) proteins make up the carboxy-terminal portion. The viral proteins are sequentially designated Npro-C-Erns-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B. Npro is an autoprotease that catalyzes the cleavage after its own carboxy terminus. C is the highly basic nucleocapsid protein and is followed by the viral envelope proteins Erns, E1, and E2. The NS proteins are thought to be components of the membrane-associated viral replicase complex. NS3 has protease, helicase, and NTPase activities and is responsible for the cleavage of the NS proteins downstream from and including its own carboxy terminus. NS4A is a cofactor for the NS3 protease activity. NS5B is the viral RNA-dependent RNA polymerase, which catalyzes the asymmetric accumulation of genomic plus-strand over minus-strand RNA (reviewed in reference 23).
The mechanisms of pestivirus virion assembly are not well understood. Electron microscopy of infected cells has suggested that particles bud into the lumen of modified endoplasmic reticulum structures (15). This hypothesis is also supported by the absence of viral envelope protein expression on the cell surface (16, 17, 41). In pestivirus virions, the envelope proteins form E2 homodimers and E1-E2 heterodimers (40). The interaction of E1 and E2 with the endoplasmic reticulum chaperone calnexin has been shown to be important for the assembly of the heterodimers (6). Recently, Harada et al. have shown that p7 is essential for virion assembly (18). p7 is a small, mostly hydrophobic protein that is cleaved inefficiently from the carboxy terminus of E2 by signal peptidase. Neither p7 nor uncleaved E2-p7 is thought to be incorporated into viral particles (11).
p7 forms a leader sequence to properly orient NS2 in the membrane. NS2 is a 54-kDa hydrophobic protein of unknown function. Cleavage of the polyprotein between NS2 and NS3 has been found to correlate with the ability of BVDV to kill cells in tissue culture. Based on this phenomenon, BVDV is divided into two biotypes: cytopathic and noncytopathic. All autonomous cytopathic and noncytopathic strains reported to date express uncleaved NS2-3; cytopathic BVDV also produces discrete NS3. Both cytopathic and noncytopathic viruses can be found in animals suffering from mucosal disease. Since these viruses are antigenically related and are nearly identical in sequence, cytopathic BVDV is thought to develop from the noncytopathic virus within the infected animal (reviewed in reference 29). Cytopathic BVDV genomes can arise in a variety of ways, all of which affect processing at the NS2/NS3 junction. Cellular or viral insertions, deletions, and even point mutations have been found to give rise to NS3 expression and resulting cytopathogenicity (3, 5, 21, 26, 37). NADL, the American prototype cytopathic BVDV, contains a 270-nucleotide insertion in NS2 that potentiates the partial cleavage of NS2-3 (26). This insertion has recently been identified as coding for a 90-amino-acid portion of a cellular J domain protein (Jiv 90) (33). Removal of this insert results in NADL Jiv 90− (formerly cIns−), a noncytopathic BVDV strain in which NS2-3 remains uncleaved.
Large in-frame deletions of the structural proteins have also been shown to result in cytopathic viruses. These genomes are capable of autonomous RNA replication and can be incorporated into particles in the presence of a helper virus expressing the structural proteins (39). These genomes are termed defective interfering RNAs, as they reduce the ability of the helper virus to propagate (39). In the case of one such genome, DI9, Npro is fused directly to NS3, eliminating the first two NS proteins (p7 and NS2). The ability of DI9 to replicate autonomously excludes the possibility of an essential function for p7 and NS2 in RNA replication (5). Defective interfering-like genomes created in the laboratory that have not been confirmed to have the defective interfering phenotype are termed subgenomic (sg) RNAs.
In this study we investigated the function of uncleaved NS2-3. Uncleaved NS2-3 expression was abolished with either ubiquitin or an internal ribosome entry site (IRES) to genetically dissociate the structural genes (C, Erns, E1, and E2), p7, and NS2 from the replicase genes (NS3 to NS5B). Surprisingly, despite efficient RNA replication of these genomes, separation of NS2 from NS3 prevented the production of infectious virus. These results demonstrate that uncleaved NS2-3 is required for infectious virion production.
MATERIALS AND METHODS
Cell culture.
Madin-Darby bovine kidney (MDBK) cells were propagated in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing sodium pyruvate and 10% horse serum.
Plasmid constructs.
Plasmids were constructed by standard methods. Constructs were checked by restriction enzyme digestion, and PCR products were verified by sequencing. Descriptions of the constructs and cloning strategies are presented below; exact sequences are available upon request. The backbone for the following constructs is the bacterial artificial chromosome vector BeloBAC11. This single-copy vector is used to maintain large and unstable DNA fragments.
NADL Jiv 90−/Ubi.
The NADL Jiv 90−/Ubi genome contains the entire NADL Jiv 90− genome with a ubiquitin (Ubi) monomer inserted between NS2 and NS3. The sequence contains a G to S mutation at position 76 of ubiquitin. The BglII/BglII fragment of pACNR/NADL (nucleotides 4584 to 5644) (26), encompassing Jiv 90 in NS2 and the NS2/NS3 junction region, was cloned into BglII-digested pRS2 (pUC19 with a modified polylinker). The Jiv 90 sequence was then deleted by recombinant PCR, resulting in pRS2/NADL del (26). For cloning convenience, the pRS2/NADL del polycloning sites from PstI to NsiI were eliminated by digestion with PstI and NsiI, which produce compatible cohesive ends, and religation.
The ubiquitin monomer sequence was amplified from pTM3-HCV-Ubi-NS5B (C. Lin and C. M. Rice, unpublished) by PCR with the following modifications: the ubiquitin amino terminus was tagged with the BVDV NS2 carboxy-terminal sequence; the ubiquitin carboxy terminus was connected to the NS3 amino terminus containing an engineered PstI site; the BglII site in the ubiquitin amino terminus was eliminated by a silent mutation; and the ubiquitin cleavage consensus site was mutated from RGG to RGS (GGA to AGT), as found in the cytopathic Osloss BVDV strain (9). Next, part of the NS3 sequence (nucleotides 5153 to 5835) was amplified by PCR from pACNR/NADL. A PstI site was engineered into the amino terminus of NS3. Finally, the BamHI/PstI fragment of ubiquitin and the PstI/SalI fragment of NS3 were cloned into BamHI- and SalI-digested pRS2/NADL del, creating pRS2/NADL Jiv 90−-Ubi-NS3. The BglII/BglII fragment from pRS2/NADL Jiv 90−-Ubi-NS3 was then used to replace the corresponding portion of pACNR/NADL, resulting in NADL Jiv 90−/Ubi.
Npro-E2/sg.
The Npro-E2/sg genome contains the 5′ NTR to E2 region of the NADL genome, followed by the encephalomyocarditis virus (EMCV) IRES, Npro, NS3, and the remainder of the NADL sequence. Nucleotides 3386 to 3583 of pACNR/NADL were amplified by PCR. A stop codon was introduced after G1066 (end of E2), and unique MluI and NsiI sites were engineered downstream of the stop. The PCR product was digested with NspI and NsiI and ligated, with the 587-nt RsrII/NspI fragment of pACNR/NADL into Npro-PAC/NS2-sg (see below) digested with RsrII and NsiI. The RsrII/SacI fragment from this subclone (p11/RsrII-NspI), along with the 2,849-nt XbaI/RsrII fragment of pACNR/NADL, was cloned into pNpro-NS3 (T. M. Myers, E. Mendez, I. Frolov, C. L. Murray, E.V. Agapov, and C. M. Rice, unpublished data) digested with XbaI and SacI.
Npro-NS2/sg.
The Npro-NS2/sg genome contains the 5′ NTR to NS2 region of the NADL Jiv 90− genome, followed by the EMCV IRES, Npro, NS3, and the remainder of the NADL sequence. A strategy similar to that used for Npro-E2/sg was used except that nucleotides 4973 to 5152 of pACNR/NADL were amplified and digested with Bsp120I and NsiI. The stop codon is at R1589 (nucleotide 5152, at the end of NS2).
pACNR-NADL-pol−.
The pACNR-NADL-pol− genome is the full-length NADL genome with an inactivated NS5B polymerase. pBVDV NS5B GDD-AGG (nucleotides changed from GGG GAT GAT to GCG GCC GGC; gift of Marc Collett) was digested with SmaI and partially digested with EagI to isolate a 5,374-nucleotide fragment. This fragment was cloned into pACNR/NADL digested with the same enzymes.
Npro-PAC/NS2-sg.
The Npro-PAC/NS2-sg genome contains the first 39 amino acids of Npro followed by ubiquitin and puromycin acetyltransferase (PAC) in the upstream cistron. The downstream cistron contains a number of internal deletions in the structural region to preserve proper polyprotein topology and processing. Npro is followed by the 10 amino-terminal amino acids of C, a leucine codon (CTT), the 18 carboxy-terminal amino acids of capsid, the 9 amino-terminal amino acids of Erns, and the 48 carboxy-terminal amino acids of E2. Nucleotides 247 to 502 of pACNR/NADL were amplified and cloned into pRS2 with SphI and an engineered EcoRI site. An MluI site was engineered in frame with capsid, upstream of EcoRI (FR1). A 224-nucleotide fragment containing ubiquitin was PCR amplified from pTM3/Ubi-NS2 (22). BamHI and MluI sites were engineered upstream of ubiquitin, and this fragment was also cloned into pRS2 with the engineered BamHI and natural SacII site at the end of ubiquitin (FR2).
The puromycin acetyltransferase gene was isolated from pTSG/PAC (12). The CCG CGG TGG C sequence, representing the carboxy terminus of ubiquitin and a SacII site, was engineered upstream of the puromycin acetyltransferase AUG. This fragment was cloned into pRS2 with SacII and BssHII (FR3). The SphI/MluI (FR1), MluI/SacII (FR2), and SacII/BssHII (FR3) fragments were then assembled into SphI- and BssHII-digested pRS2 (pA). The BssHII/Ecl136II fragment of pTSG/PAC (containing the puromycin acetyltransferase carboxy terminus) was then ligated, along with the AclI/SacI fragment of pEMCV/BVDV (13), into pRS2 digested with BssHII and SacI (pB). AclI was blunt ended to allow ligation to Ecl136II. The SphI/BssHII fragment of pA and the BssHII/SacI fragment of pB were then ligated together into SphI- and SacI-digested pRS2 (pSph-Npro-Ubi-PAC-EMCV IRES). The 2,025-nucleotide SphI/SacI fragment of pSph-Npro-Ubi-PAC-EMCV IRES was then cloned into pNS2/sg or pNS2 Jiv 90−/sg (see below).
pNS2/sg and pNS2 Jiv 90−/sg contain the BVDV IRES and Npro followed by the first 10 amino acids of C, a leucine residue (CTT), the last 18 amino acids of C, the first 9 amino acids of Erns, the last 48 amino acids of E2, p7, and the remainder of the NADL or NADL Jiv 90− NS proteins. Nucleotides 695 to 916 were amplified from pACNR/NADL. The PCR product was cloned into pRS2 with SacI and an engineered HindIII (FR4). Nucleotides 1142 to 1225 were then amplified from pACNR/NADL and also cloned into pRS2 with HindIII and PstI sites engineered into the PCR (FR5). Finally, nucleotides 3440 to 4049 of pACNR/NADL were amplified and cloned into pRS2 with engineered HindIII and PstI sites (FR6). The fragments SacI/HindIII (FR4), HindIII/PstI (FR5), and PstI/BsmI (FR6) were then cloned into pACNR/NADL (pNS2/sg) or pACNR/NADL Jiv 90− (pNS2 Jiv 90−/sg) cut with SacI and BsmI.
Npro-E2/NS2-sg.
The Npro-E2/NS2-sg genome consists of the first cistron of Npro-E2/sg and the second cistron of pNS2/sg. The SacI/AatII fragment of pNS2/sg was replaced with the corresponding fragment from Npro-E2/sg.
Npro-E2/NS2-Ubi-sg.
The Npro-E2/NS2-Ubi-sg genome consists of the first cistron of Npro-E2/sg and a second cistron identical to that of pNS2/sg except with a ubiquitin monomer, with the G76S change, inserted at the NS2/NS3 junction. The MfeI/NsiI fragment from Npro-E2/NS2-sg was replaced with the corresponding fragment from NADL Jiv 90−/Ubi.
Npro-NS2/NS2-Ubi-sg.
The Npro-NS2/NS2-Ubi-sg genome consists of the first cistron of Npro-NS2/sg and the second cistron of Npro-E2/NS2-Ubi-sg. The RsrII/MluI fragment of Npro-E2/NS2-Ubi-sg was replaced with the corresponding fragment from Npro-NS2/sg.
p7-NS2-3-4A/sg.
The p7-NS2-3-4A/sg genome expresses Npro and p7-NS2-3-NS4A from the BVDV IRES. The cloning strategy left 27 amino acids of C, 7 amino acids of Erns, 48 amino acids of E2 between Npro and p7, and 37 amino acids of NS4B after NS4A. The downstream cistron, driven by the EMCV IRES, is identical to that of Npro-E2/sg. The XbaI/StuI fragment of Npro-E2/sg was replaced with the XbaI/DraI fragment of pNS2 Jiv 90−/sg.
Ubi-NS2-3-4A/sg.
The Ubi-NS2-3-4A/sg genome contains the BVDV IRES and 39 amino acids of Npro, followed by Ubi-NS2-3-NS4A. The cloning strategy left 37 amino acids of NS4B in the first cistron. The downstream cistron, driven by the EMCV IRES, is identical to that of Npro-E2/sg. Nucleotides 1147 to 5120 of p7-NS2-3-4A/sg were amplified by PCR. A SacII site and ubiquitin cleavage consensus sequence (RGG) were engineered immediately upstream of NS2. The PCR product was digested with SacII and MluI and ligated to the SphI/MluI fragment of p7-NS2-3-4A/sg and the SphI/SacII (ubiquitin) fragment of pSph-Npro-Ubi-PAC-EMCV IRES.
p7-NS2-3/sg.
The p7-NS2-3/sg genome expresses Npro and p7-NS2-3 from the BVDV IRES. The cloning strategy left 27 amino acids of C, 7 amino acids of Erns, and 48 amino acids of E2 between Npro and p7. Eight amino acids of NS4A remained in the first cistron, followed by five random amino acids and a stop codon. The downstream cistron, driven by the EMCV IRES, is identical to that of Npro-E2/sg. SpeI and MluI of p7-NS2-3-4A/sg were blunt ended and ligated.
p7-NS2-3(S1842A)-NS4A/sg.
The p7-NS2-3(S1842A)-NS4A/sg genome is identical to p7-NS2-3-4A/sg except for the S1842A change in NS3 in the first cistron. The BsrGI/DrdI fragment of p7-NS2-3-4A/sg was replaced with the corresponding fragment derived from pTM3/BVDV 1598-3035 S1842A (43).
p7-NS2-3(S1842A)/sg.
The p7-NS2-3(S1842A)/sg genome is identical to p7-NS2-3(S1842A)-NS4A/sg except that the first cistron contains a stop codon immediately after NS3. Nucleotides 241 to 2668 and nucleotides 2220 to 4765 of p7-NS2-3(S1842A)-NS4A/sg were amplified by PCR. The 2220 to 4765 fragment had a stop codon and MluI site engineered into the reverse primer. The 241 to 2668 product digested with SphI and PflMI and the 2220 to 4765 fragment digested with PflMI and MluI were then ligated into p7-NS2-3(S1842A)-NS4A/sg digested with SphI and MluI.
RNA transcription and transfection of MDBK cells.
Plasmids were linearized with SbfI and blunt ended by addition of 2 U of Klenow and 1.2 U of T4 DNA polymerase followed by 0.8 μM deoxynucleoside triphosphate mix. Reactions were extracted once with phenol and twice with chloroform and precipitated with ethanol and 3 M sodium acetate. One microgram of template was transcribed in a 20-μl reaction with the T7-Megascript kit (Ambion). Reactions were incubated for 2 h at 37°C, and the RNA was cleaned up with the RNeasy kit (Qiagen). RNA was quantified by radiolabel incorporation or by absorbance at 260 nm.
For electroporation, MDBK cells (70% confluent) were trypsinized, washed three times with ice-cold RNase-free phosphate-buffered saline (AccuGene PBS; BioWhittaker, Rockland, Maine), and resuspended at 2 × 107 cells/ml in PBS; 5 μg of each RNA to be electroporated was mixed with 0.4 ml of the cell suspension and immediately pulsed with a Bio-Rad Gene Pulsar (1.1 kV, 25 μF, infinite resistance, two pulses) or with a BTX ElectroSquare Porator ECM 830 (900 V, 99 μs, five pulses). Electroporated cells were diluted in 10 ml of DMEM-10% horse serum and plated on tissue culture dishes.
Analysis of virus-specific RNAs.
MDBK cells were labeled with [3H]uridine (60 μCi/ml) or H332PO4 (200 μCi/ml) in the presence of actinomycin D (1 μg/ml) for 8 to 9 h. Total cellular RNA was isolated with TRIzol (Gibco-BRL), denatured by treatment with glyoxal and dimethyl sulfoxide, and separated by electrophoresis with a 1% agarose gel containing 10 mM phosphate buffer. For [3H]uridine, the gels were fixed with methanol, impregnated with 3% 2,5-diphenyloxazole (PPO) in methanol, and equilibrated in water. All gels were dried and exposed to X-ray film at −70°C.
Immunoprecipitation.
MDBK cells were labeled in methionine-deficient DMEM (Life Technologies) containing 2% fetal calf serum and Expre35S protein labeling mix (100 μCi/ml; New England Nuclear) for 6 h. After labeling, the MDBK cells were washed with PBS and lysed with 0.5% sodium dodecyl sulfate (SDS) in TNE (50 mM Tris-Cl, pH 7.5, 1 mM EDTA, 200 mM NaCl, 20 μg of phenylmethylsulfonyl fluoride, 0.3 ml per 35-mm well and 5 × 105 cells). SDS lysates were sheared through a 26-gauge needle, heated to 70°C for 10 min, and clarified by centrifugation at 12, 000 × g for 10 min. Clarified lysates were diluted 1:5 with TNE containing 0.5% Triton X-100 and bovine serum albumin (BSA, 1 mg/ml); 2.5 μl of antiserum (G40) was added and incubated overnight at 4°C with rocking. G40 is a rabbit polyclonal antiserum specific for NS3 that has been described elsewhere (7, 8). Staphylococcus aureus (Pansorbin; Cabiochem, San Diego, Calif.) was washed three times with TNAS (TNE containing 0.5% Triton X-100, BSA [1 mg/ml], and 0.1% SDS); 25 μl of the washed cells was added to the lysates, and incubation was continued for 1 h at 4°C. Immunoprecipitates were washed three times with TNAS and once with TNE. Washed precipitates were resuspended in Laemmli sample buffer, heated at 75°C for 10 min, and centrifuged at 12,000 × g for 1 min. Immunoprecipitated proteins were separated by SDS-10% or 12% polyacrylamide gel electrophoresis (PAGE) and visualized by autoradiography.
Western blots.
Cells were lysed with passive lysis buffer (Promega, Madison, Wis.) 24 h postelectroporation and clarified by centrifugation at 18, 000 × g for 30 s. Lysates were separated on a SDS-10% PAGE and transferred to a nitrocellulose membrane. Blots were blocked for 1 h with 5% milk-TBST (0.02 M Tris, pH 7.4, 0.19 M sodium chloride, 0.1% Tween 20). For NS3 detection, polyclonal antiserum (G40) was diluted 1:500 in 3% BSA and incubated for 1 h. After washing with TBST, the secondary antibody (goat anti-rabbit immunoglobulin-horseradish peroxidase, Pierce) was added at 1:10,000 dilution in 5% milk-TBST and incubated for 30 min at room temperature. For β-actin, the primary antibody (mouse anti-β-actin, Sigma) was diluted 1:5,000 in 5% milk-TBST and the secondary antibody (rabbit anti-mouse immunoglobulin-horseradish peroxidase, Pierce) was diluted 1:2,500 in 5% milk-TBST. After removal of the secondary antibody by washing with TBST, blots were developed with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.).
BVDV plaque assay and focus-forming assay.
The BVDV plaque and focus-forming assays were conducted as previously described (26). Briefly, MDBK cells (70 to 80% confluent) were infected with 10-fold dilutions of virus. Following 1 h of adsorption at 37°C, cells were washed once with DMEM-10% horse serum, overlaid with 0.5 or 0.8% agarose in DMEM-5% horse serum, and incubated at 37°C. After 3 days, cells were fixed with 7% formaldehyde, the agarose plugs were removed, and the monolayers were stained with crystal violet or used for focus-forming assays. For the focus-forming assay, fixation was followed by washing with PBS. The cells were permeabilized with PBS-0.25% Triton X-100 for 15 min, washed with PBS, and incubated for 90 min with a 1:1,000 dilution of polyclonal antiserum anti-49 (30) in PBS-2% BSA. After washing with PBS, cells were incubated for 90 min with a 1:1,000 dilution of rabbit anti-bovine IgG-horseradish peroxidase (Sigma) in PBS-2% BSA. Cells were then washed with PBS and incubated with peroxidase substrate (Vector Laboratories) following the manufacturer's instructions.
Infectious center assay.
The infectious center assay was used to estimate the amount of infectious RNA electroporated into cells. A sample of the electroporated cells was diluted 1:10 and 2, 20, and 200 μl was added to separate preseeded monolayers of MDBK cells. Following 3 h of incubation, cells were washed once with DMEM-10% horse serum, overlaid with 0.5% agarose in DMEM-5% horse serum, and incubated at 37°C. After 3 days the cells were fixed and stained with crystal violet (for cytopathic virus) or used for the focus-forming assay (noncytopathic virus) as above.
Immunofluorescence.
Cells plated on culture slides (Becton Dickinson, Franklin Lakes, N.J.) were washed with Dulbecco's phosphate-buffered saline (DPBS; Gibco) and fixed for 20 min with 50% methanol-50% acetone. After washing twice, anti-NS3 antibody (monoclonal antibody 184) (10) was added at a 1:2,000 dilution in DPBS-5% goat serum. Cells were incubated for 1 h at room temperature and then washed twice with DPBS-5% goat serum. The secondary antibody (goat anti-mouse IgG-Alexa 594; Molecular Probes, Eugene, Oreg.) was added at a 1:2,000 dilution in DPBS-5% goat serum and cells were incubated for 20 min at 4°C. Hoechst 33342 (1 μg/ml) was added, and cells were incubated an additional 10 min at 4°C. Cells were washed three times with DPBS-5% goat serum and coverslips were mounted. Slides were viewed with a Nikon Eclipse TE 300 microscope.
Flow cytometry.
The trans-complementation assay produced very low numbers of viral particles, which were incapable of cell-to-cell spread. Flow cytometry was used to count the cells infected with these particles and thus determine the titer. Infected cells in a 35-mm2 dish were trypsinized, washed twice with DPBS, and fixed with 0.5% paraformaldehyde for 20 min at room temperature. Cells were blocked and permeabilized with PBS-5% goat serum-0.1% sodium azide-0.1% saponin for 20 min at room temperature. Blocking solution was removed and primary antibody (monoclonal antibody 184) was added at 1:2,000 dilution in SB (PBS-0.5% BSA-0.1% sodium azide-0.1% saponin). Cells were incubated for 1 h at room temperature and then washed twice with SB. Secondary antibody (goat anti-mouse IgG-Alexa 488; Molecular Probes, Eugene, Oreg.) was added at 1:1,000 dilution in SB, and cells were incubated for 30 min at room temperature. Cells were washed twice with SB and analyzed by flow cytometry on a FACSCalibur (Becton Dickinson), counting 104 cells for the replication assay and 105 cells for the supernatant assay.
Titers of supernatants were calculated from the average percentage of NS3-positive cells. Infected wells were estimated to contain 106 cells; this number was calculated by seeding cells under the same conditions as the assay and counting 24 wells in two independent platings. Since the viruses are incapable of cell-to-cell spread, flow cytometry counts were expected to reflect the number of cells infected by the initial inoculum. Inspection of the immunofluorescence assay fields, however, suggested that many cells were dividing during the incubation time, artificially increasing the titers. From the immunofluorescence fields of several experiments, it was determined that approximately 25% of cells had not divided, 60% had divided in two, and 15% occurred in clusters of three. The numbers of infected cells obtained by flow cytometry were therefore adjusted by multiplying by 0.6, discounting NS3-positive cells that had arisen from division and not de novo infection.
RESULTS
Genomes that lack uncleaved NS2-3 replicate but do not produce infectious virus.
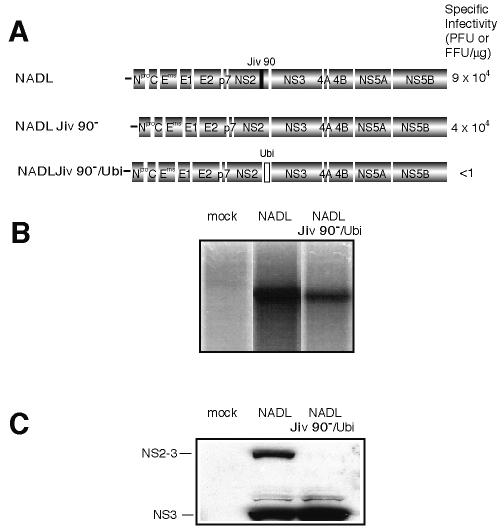
As an initial investigation of uncleaved NS2-3 function, we wished to regulate the extent of processing between the two proteins. To this end, we engineered an NADL Jiv 90− genome with a ubiquitin monomer (Ubi) inserted between NS2 and NS3 (NADL Jiv 90−/Ubi, Fig. 1A). A single amino acid change was engineered in the ubiquitin cleavage consensus site (RGG to RGS) with the intention of partially abolishing processing. This mutation is thought to contribute to attenuated cleavage in a naturally occurring BVDV strain, Osloss (9, 27). In vitro-transcribed NADL Jiv 90−/Ubi RNA was electroporated into cells in parallel with wild-type NADL RNA. Replication of NADL Jiv 90−/Ubi was visualized by labeling cells with H332PO4 in the presence of actinomycin D to inhibit cellular RNA synthesis. Total RNA was isolated, denatured, and analyzed by agarose gel electrophoresis (Fig. 1B).
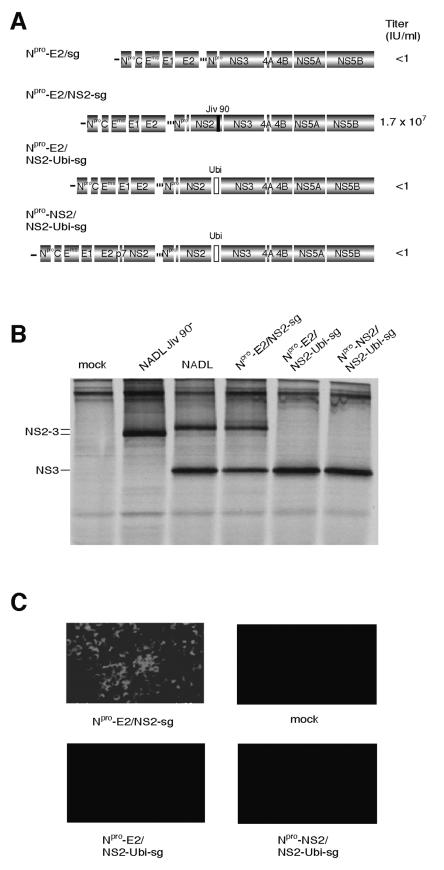
FIG. 1.
Insertion of ubiquitin between NS2 and NS3 in NADL Jiv 90− abolishes infectious particle production. (A) Schematic representations of the genomes of NADL, NADL Jiv 90−, and NADL Jiv 90−/Ubi. Specific infectivities (PFU or FFU/μg) were determined by infectious center assay and are the averages of two independent experiments. (B) RNA replication. MDBK cells were electroporated with the indicated RNA and labeled with H332PO4 in the presence of actinomycin D for 8 h starting at 16 h postelectroporation. Total cellular RNA was isolated with TRIzol, denatured, and separated on a 1% agarose gel. (C) Protein expression. Cells electroporated with the indicated RNA were lysed 10 h postelectroporation, separated by SDS-10% PAGE, Western blotted, and probed with NS3-specific polyclonal antiserum G40.
Analysis of protein expression by Western blot showed that NADL Jiv 90−/Ubi produced NS3 in similar amounts to wild-type NADL (Fig. 1C). Surprisingly, unlike NADL and Osloss, NADL Jiv 90−/Ubi did not express detectable uncleaved NS2-3 or NS2-Ubi-NS3. The lack of uncleaved NS2-3 suggested that the G to S mutation had no inhibitory effect on cleavage at the Ubi/NS3 junction in the NADL Jiv 90− background. We then determined the infectivity of the NADL and NADL Jiv 90−/Ubi RNAs by infectious center assay. While NADL Jiv 90− and its cytopathic parent, NADL, gave almost 105 PFU or focus-forming units (FFU) per μg of electroporated RNA, no infectious centers were detected for NADL Jiv 90−/Ubi (Fig. 1A). These results suggested that while NADL Jiv 90−/Ubi was able to replicate and produce NS3 in transfected cells, it was not capable of spread to produce a plaque or focus.
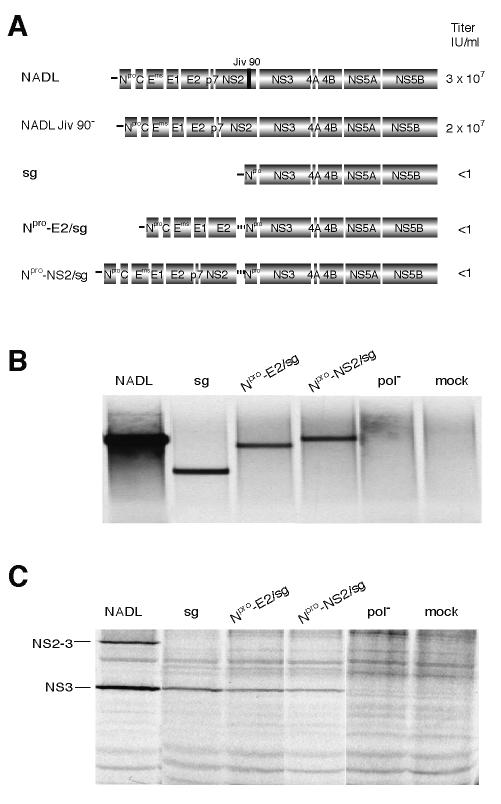
We speculated that abolition of uncleaved NS2-3 production might be detrimental to infectious particle production; however, it was also possible that ubiquitin expression in the NADL Jiv 90−/Ubi polyprotein interfered with an essential function of NS2. Bicistronic genomes were therefore created as an alternative method of abolishing uncleaved NS2-3 expression. In the bicistronic genomes, an internal ribosome entry site (IRES) separated the structural genes, p7, and NS2 from the replicase genes (Fig. 2A). The first cistron encoded Npro and the structural proteins (C, Erns, E1, and E2) and was terminated after E2 (Gly 1066, Npro-E2/sg) or NS2 (Arg 1589, Npro-NS2/sg). The second cistron (/sg) consisted of a subgenome with Npro fused to NS3, driven by the encephalomyocarditis virus (EMCV) IRES. Adaptive mutations in Npro and NS3 of this cistron were required to allow high levels of replication (T. M. Myers, E. Mendez, I. Frolov, C. L. Murray, E. V. Agapov, and C. M. Rice, unpublished data). A direct effect of the NS3 mutation on particle production was ruled out by engineering the change into NADL Jiv 90−. The mutation had no effect on viral titers (T. M. Myers, E. Mendez, I. Frolov, C. L. Murray, E. V. Agapov, and C. M. Rice, unpublished data).
FIG. 2.
Insertion of an IRES between NS2 and NS3 in NADL Jiv 90− abolishes infectious particle production. (A) Schematic representation of the genomes of NADL, NADL Jiv 90−, subgenome (sg), Npro-E2/sg, and Npro-NS2/sg. Titers were determined by plaque or focus-forming assay. The two open reading frames of the bicistronic constructs are driven by the BVDV IRES (black line) and the EMCV IRES (dotted line). (B) RNA replication. MDBK cells were electroporated with the indicated RNAs and labeled with [3H]uridine in the presence of actinomycin D for 12 h starting at 5 h postelectroporation. Total cellular RNA was isolated with TRIzol, denatured, and separated on a 1% agarose gel. (C) Protein expression. MDBK cells electroporated with the indicated RNAs were metabolically labeled with [35S]methionine for 6 h beginning at 12 h postelectroporation. Expression of NS3 was analyzed by immunoprecipitation with NS3-specific polyclonal antiserum G40 and separation by SDS-10% PAGE.
Replication of Npro-E2/sg and Npro-NS2/sg was confirmed by electroporation of in vitro transcribed RNA into MDBK cells. The cells were labeled with [3H]uridine in the presence of actinomycin D (Fig. 2B). Both the bicistronic genomes were able to replicate, compared to a negative control construct with a mutated polymerase. Protein expression from the electroporated bicistronic genomes was monitored in parallel by immunoprecipitation of [35S]methionine-labeled cell lysates with an NS3-specific antiserum (Fig. 2C).
Media harvested from electroporated cells were analyzed for infectious virus by plaque assay, focus-forming assay, and immunofluorescence of inoculated naïve cell monolayers. These assays revealed that no infectious virus was produced by replication of the bicistronic RNAs. In the case of Npro-NS2/sg, which encodes all of the BVDV proteins except for uncleaved NS2-3, this finding supported the NADL Jiv 90−/Ubi observations. In the case of Npro-E2/sg, this result confirmed that p7 or both p7 and NS2 are required for infectious particle production (18).
Bicistronic genomes can be packaged into virions.
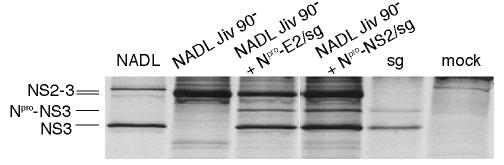
Given the artificial nature of the bicistronic genomes and the possibility that the EMCV IRES might interfere with RNA packaging, we next determined whether the bicistronic RNAs could be incorporated into virions. NADL Jiv90− is known to be able to package defective interfering genomes (39), and we therefore tested whether NADL Jiv90− was able to package the bicistronic RNAs. Since NADL Jiv90− produces only NS2-3 and the bicistronic genomes produce only cleaved NS3, the expression of these proteins provided markers for the packaged RNAs upon infection of naïve cells.
Cells were infected with NADL Jiv90− 48 h before electroporation with Npro-E2/sg or Npro-NS2/sg. The culture supernatants were harvested 24 h later and used to infect naïve cells. If the bicistronic RNAs could be packaged, cells supporting the replication of NADL Jiv90− and a bicistronic construct should produce two types of viral particles, each containing one of the genomes. [35S]methionine labeling of cells infected with the culture supernatants and immunoprecipitation with an anti-NS3 antiserum were used to determine which genomes had been packaged. Cells infected with NADL alone expressed NS2-3 and NS3, whereas those infected with NADL Jiv90− alone expressed only NS2-3 (Fig. 3). Lysate from subgenome-transfected cells was run as a size marker for NS3 and uncleaved Npro-NS3. Supernatants from cells electroporated with Npro-E2/sg or Npro-NS2/sg after infection with NADL Jiv90− transduced expression of NS3 and minor amounts of uncleaved Npro-NS3 in addition to uncleaved NS2-3 (Fig. 3). The presence of NS3 in the newly infected cells shows that the bicistronic genomes could be incorporated into infectious particles.
FIG. 3.
Bicistronic genomes can be packaged by NADL Jiv90−. MDBK cells were infected with NADL Jiv 90− at a multiplicity of infection (MOI) of 1 48 h before electroporation with bicistronic RNAs. Media were harvested at 48 h postelectroporation and used to infect naïve MDBK cells. The newly infected cells were labeled with [35S]methionine for 6 h beginning at 12 h postelectroporation, and expression of NS3 was analyzed by immunoprecipitation with NS3-specific antiserum G40) and separation by SDS-12% PAGE. Lysates from NADL-, NADL Jiv 90−-, subgenome-, and mock-infected cells were used as controls for native protein sizes. The positions of NS3, NS2-3, and uncleaved Npro-NS3 from the subgenomic cistron are indicated.
Structural proteins encoded by bicistronic genomes are functional.
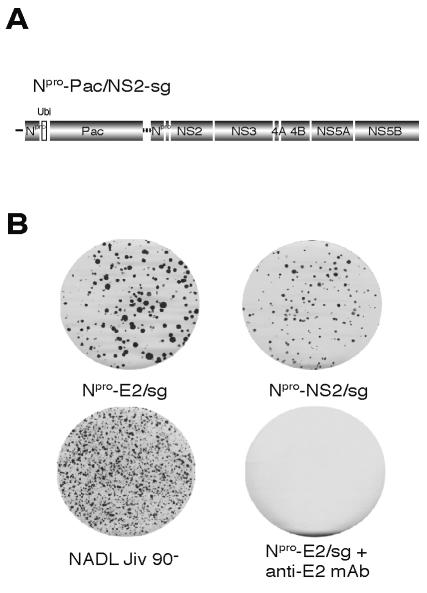
It was possible that the bicistronic genomes were unable to produce virus because of a functional defect in their structural proteins. To test the ability of the capsid and envelope proteins to package BVDV RNAs, their ability to encapsidate RNA in trans was investigated. It is known that defective interfering genomes can be incorporated into virions in the presence of a helper virus expressing the BVDV structural proteins (39). We used a modified subgenome, Npro-PAC/NS2-sg, for a trans packaging assay (Fig. 4A). Npro-PAC/NS2-sg is a bicistronic replicon containing the puromycin N-acetyltransferase (PAC) gene in the first cistron and the NADL Jiv90− NS genes (p7 to NS5B) in the second cistron. In the context of this noncytopathic BVDV replicon, puromycin N-acetyltransferase confers resistance to puromycin and provides a dominant selectable marker to detect the presence of the modified subgenomic RNA in virions. Replication of Npro-PAC/NS2-sg was confirmed by immunoprecipitation with anti-NS3 antibodies (data not shown).
FIG. 4.
Bicistronic genomes can package a modified defective interfering genome. (A) Schematic representation of Npro-PAC/NS2-sg, a bicistronic genome that expresses ubiquitin (Ubi, open box) and the dominant selectable marker puromycin N-acetyltransferase (PAC) from the first cistron and the NADL Jiv 90− NS proteins from the second cistron (p7-NS5B). Puromycin selection was used to establish a population of Npro-PAC/NS2-sg-expressing MDBK cells. (B) These cells were then electroporated with the indicated bicistronic constructs and the media were harvested 48 h postelectroporation. The supernatants were assayed for infectivity by determining their ability to transfer puromycin resistance to naïve cells. Drug-resistant foci in the newly infected cells were visualized by staining with crystal violet after 10 days of puromycin selection. Incubation of supernatants with anti-E2 antibodies abolished infectivity.
An Npro-PAC/NS2-sg-expressing MDBK cell population was created by electroporation of the RNA and selection for puromycin-resistant cells. Puromycin-resistant cells were then transfected with Npro-E2/sg, Npro-NS2/sg, or NADL Jiv90−. Supernatants were harvested 48 h postelectroporation and used to infect naïve cells, which were then grown in the presence of puromycin (Fig. 4B). Both NADL Jiv90− and the bicistronic constructs were able to encapsidate the Npro-PAC/NS2-sg RNA, resulting in supernatants that transferred puromycin resistance to naive cells. Differences in colony morphology and number were noticed, likely resulting from a slight decrease in trans-packaging efficiency by the bicistronic RNAs compared to wild-type virus. Incubation of the culture supernatants with an anti-E2 monoclonal antibody prevented the appearance of puromycin-resistant foci (Fig. 4B). These results illustrated that the structural proteins encoded by the bicistronic genomes were functional and able to package viral RNA into particles whose infectivity could be neutralized by an envelope-specific antibody. These results also suggested that the NS proteins encoded by Npro-PAC/NS2-sg could rescue the bicistronic genomes, at least allowing them to produce viral particles that contained the Npro-PAC/NS2-sg genome. In the case of Npro-NS2/sg, which differs from wild-type virus only in the separation of NS2 and NS3 by the EMCV IRES, this result suggested that the uncleaved NS2-3 provided by Npro-PAC/NS2-sg was integral in restoring infectious virus production.
Expression of uncleaved NS2-3 by a bicistronic genome rescues infectious particle production.
We investigated whether the introduction of uncleaved NS2-3 into a bicistronic genome would restore particle production. To this end, the bicistronic construct Npro-E2/NS2-sg was created. Npro-E2/NS2-sg contains the NADL structural genes up to E2 in the first cistron and the NS genes (p7 to NS5B) in the second cistron (Fig. 5A). This construct differs from Npro-NS2/sg in the movement of p7 and NS2 from the first cistron to the second cistron and the inclusion of Jiv 90 in NS2. Electroporation of MDBK cells with Npro-E2/NS2-sg RNA resulted in a viral titer of 1.7 × 107 infectious units (IU)/ml in the supernatant (Fig. 5A). The infectivity of the supernatant was confirmed by immunofluorescence for NS3 in the inoculated cells (Fig. 5C). These data suggested that expression of uncleaved NS2-3 in the second cistron of a bicistronic RNA restored efficient virion production.
FIG.5.
Expression of NS2-3 restores virion production. (A) Schematic representation of Npro-E2/sg, Npro-E2/NS2-sg, Npro-E2/NS2-Ubi-sg, and Npro-NS2/NS2-Ubi-sg. Jiv90 (black bar) in Npro-E2/NS2-sg and ubiquitin (open box) in Npro-E2/NS2-Ubi-sg and Npro-NS2/NS2-Ubi-sg are shown. Titers of virus were determined by plaque assay or focus-forming assay. (B) Protein expression. MDBK cells electroporated with the indicated RNA were labeled with [35S]methionine for 6 h beginning at 12 h postelectroporation. Lysates were immunoprecipitated with NS3-specific antiserum G40 and separated by SDS-10% PAGE. (C) Determination of infectious virus production. Media from cells transfected with the indicated genome were harvested after 48 h and used to infect naïve cells. Infected cells were fixed after 30 h and stained with an NS3-specific antibody (monoclonal antibody 184). Fields are magnified 10 times.
It appeared that the expression of uncleaved NS2-3 was responsible for the ability of Npro-E2/NS2-sg to produce infectious virus. We next investigated whether promoting complete cleavage of NS2-3 in Npro-E2/NS2-sg would again abolish virus production. Npro-E2/NS2-Ubi-sg was created to test this hypothesis. In this genome, a ubiquitin monomer was placed between NS2 and NS3 in the Npro-E2/NS2-sg background, resulting in complete cleavage at this site. To exclude any negative effects of ubiquitin on the function of NS2, a second construct, Npro-NS2/NS2-Ubi-sg, was made. Here, p7 and NS2 were duplicated in the first cistron while the second cistron remained identical to that of Npro-E2/NS2-Ubi-sg. In these constructs, the Npro-E2/NS2-sg background was modified to contain Jiv90− NS2 because cleavage of ubiquitin by the cellular enzyme ubiquitin carboxy terminal hydrolase produces discrete NS3 in these constructs.
Immunoprecipitation with NS3-specific antiserum showed no production of NS2-3 in Npro-E2/NS2-Ubi-sg or Npro-NS2/NS2-Ubi-sg-transfected cells (Fig. 5B). Culture supernatants from cells electroporated with the ubiquitin constructs did not contain infectious virus, as shown by immunofluorescence of inoculated cells (Fig. 5C) or by plaque assay (data not shown). These results showed that a bicistronic construct that expressed NS2-3 could make infectious particles (Npro-E2/NS2-sg), while those that lacked the uncleaved protein could not (Npro-E2/NS2-Ubi-sg and Npro-NS2/NS2-Ubi-sg).
Determination of the minimal polyprotein regions and functions sufficient for trans rescue of an uncleaved NS2-3-deficient genome.
We next investigated which regions of the genome were involved in uncleaved NS2-3 activity. In order to facilitate genetic manipulation, a trans-complementation system was established. Constructs were made to express NS2-3 in a variety of contexts, allowing assessment of the contributions of p7, NS4A, and the NS3 protease activity to NS2-3 function (Fig. 6A). In these genomes, the first cistron of Npro-E2/sg was replaced with a cassette encoding Npro-p7-NS2-3-4A (p7-NS2-3-4A/sg), Npro-Ubi-NS2-3-4A (Ubi-NS2-3-4A/sg), Npro-p7-NS2-3 (p7-NS2-3/sg), or Npro-p7-NS2-3(S1842A) [p7-NS2-3(S1842A)/sg].
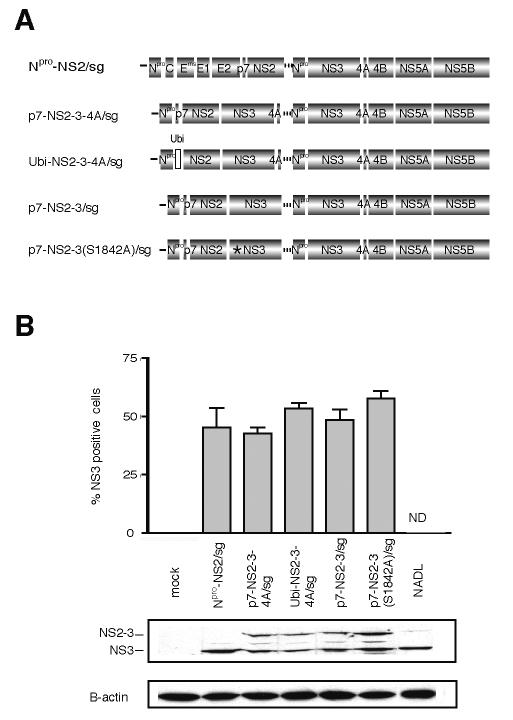
FIG. 6.
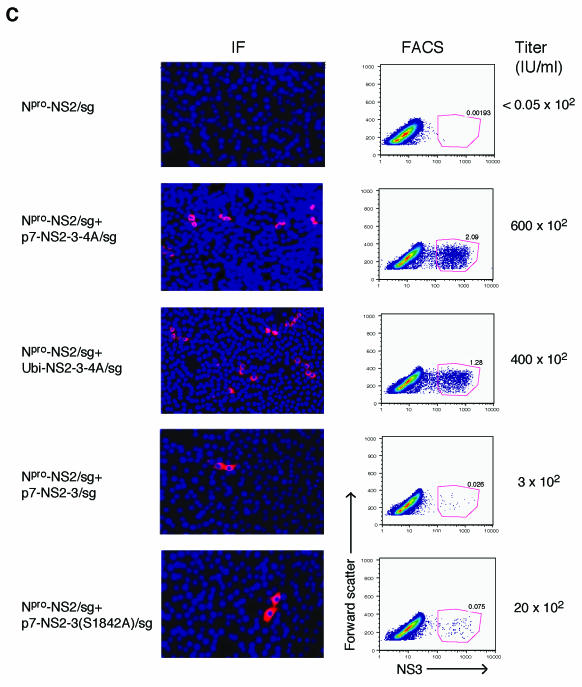
Minimal genomic regions that can rescue an uncleaved NS2-3 defect in trans. (A) Schematic representation of the genomes of Npro-NS2/sg, p7-NS2-3-4A/sg, Ubi-NS2-3-4A/sg, p7-NS2-3/sg, and p7-NS2-3(S1842A)/sg. The separate cistrons are driven by the BVDV IRES and the EMCV IRES as described for Fig. 2. The first cistron of each genome expresses the proteins shown as well as portions of the upstream and downstream open reading frames to allow correct processing (see Materials and Methods for details). Ubiquitin (open box) in Ubi-NS2-3-4A/sg and the serine-to-alanine mutation in a catalytic residue of the NS3 protease (S1842, star) in p7-NS2-3(S1842A)/sg are shown. (B) Replication of bicistronic genomes. Replication efficiencies were compared by determining NS3 levels in electroporated cells by flow cytometry (upper panel) and Western blot (lower panel). For flow cytometry, cells were fixed 30 h postelectroporation and stained with an NS3-specific antibody (monoclonal antibody 184). The percentage of electroporated cells expressing NS3 as determined by flow cytometry in two separate experiments is plotted. ND, not done. For Western blotting, cells were lysed 24 h postelectroporation and separated by SDS-10% PAGE, and the blot was probed with NS3-specific antiserum G40. The positions of NS2-3 and NS3 are indicated. β-Actin was used as a loading control. (C) Titers of virus produced by trans-complementation. MDBK cells were electroporated with Npro-NS2/sg alone or in combination with the indicated RNAs. Supernatants were harvested at 48 h postelectroporation and used to infect naïve cells. The appearance of NS3 in the newly infected cells was assayed at 30 h postinfection by immunofluorescence (left panel) and flow cytometry (FACS, right panel). The trans-complementation assay was repeated several times, and the titers were calculated by flow cytometry from two separate experiments. Flow cytometry titers were similar to those estimated by counting immunofluorescence-positive cells. Representative immunofluorescence and flow cytometry data are shown. For immunofluorescence, NS3-positive cells are red and nuclei are stained blue with Hoechst 33342. Fields are magnified 10 times. For flow cytometry, 105 cells were counted and forward scatter versus NS3 expression of the live cell subset was plotted. The NS3-positive cells are enclosed in the red gate, and the percentage of positive cells is indicated. The top row shows the supernatant assay of a representative single electroporation, Npro-NS2/sg.
The replication efficiencies of the NS2-3-expressing bicistronic genomes were compared by flow cytometry to count NS3-positive cells 30 h after electroporation (Fig. 6B, upper panel). Levels of NS2-3 expression in transfected cells were compared by Western blotting of lysates collected 24 h postelectroporation (Fig. 6B, lower panel). None of the NS2-3 expression constructs showed replication or protein levels significantly lower than that of the parental construct, p7-NS2-3-4A/sg. Electroporation of the NS2-3-expressing genomes alone was not expected to produce infectious virus, as these genomes lack envelope proteins. Accordingly, infection of naïve cells with supernatants collected 48 h after electroporation, and subsequent staining for NS3, revealed no detectable virus (Fig. 6C, data not shown).
The NS2-3 expression constructs were tested for their ability to rescue particle production by Npro-NS2/sg; 5 μg of each RNA was electroporated into MDBK cells alone or in combination with Npro-NS2/sg and the media were harvested after 48 h. Supernatants were used to infect naïve cells and the presence of virus was determined by immunostaining for NS3 at 30 h postinfection. The visualization of infected cells by immunofluorescence and flow cytometry is shown; viral titers were determined from the percentage of infected cells by flow cytometry as described in Materials and Methods (Fig. 6C).
The initial construct, p7-NS2-3-4A/sg, was able to rescue particle production from Npro-NS2/sg, resulting in a titer of 6 × 104 IU/ml (Fig. 6C). We next determined if p7 and NS4A were required in cis with NS2-3 for its function. To investigate the importance of p7 in cis, Ubi-NS2-3-4A/sg was created. This construct contains no p7 but has a ubiquitin monomer upstream of NS2 in the first cistron to produce its amino terminus. Coelectroporation of Ubi-NS2-3-4A/sg with Npro-NS2/sg resulted in the appearance of infectious virus in the supernatant at a titer of 4 × 104 IU/ml (Fig. 6C). To determine whether the presence of NS4A in cis affected the ability of NS2-3 to function, p7-NS2-3/sg was created. When p7-NS2-3/sg was coelectroporated with Npro-NS2/sg, a titer of 3 × 102 IU/ml resulted (Fig. 6C). These results showed that NS2-3 could be complemented in trans and that the presence of NS4A but not p7 in cis was required for optimal function of the protein. To rule out the possibility that recombination between the coelectroporated genomes and not trans-complementation was responsible for the recovered virus, the supernatants were passaged on MDBK cells. Recombinant genomes would be expected to increase in titer during passaging. After the first passage, however, virus was undetectable, suggesting that recombinants were not present.
We next investigated whether the protease activity of NS2-3 was required for its activity in infectious virus production. The NS3 protease involvement was investigated by mutating the active site serine (residue 1842) to alanine in the context of p7-NS2-3-4A [p7-NS2-3(S1842A)-NS4A/sg]. When this RNA was coelectroporated with Npro-NS2/sg, no infectious virus was detected (data not shown). Interpretation of the protease mutation in this context was difficult, however, as the NS3 protease activity is required to generate its own carboxy terminus. We speculated that the retention of NS4A on the carboxy terminus of NS2-3 might interfere with its function. A second construct was therefore generated in which NS4A was deleted [p7-NS2-3(S1842A)/sg]. When p7-NS2-3(S1842A)/sg was coelectroporated with Npro-NS2/sg, an intermediate level of virus was seen in the supernatant at a titer of 2 × 103 IU/ml (Fig. 6C). These results suggested that, apart from its role in cleaving NS2-3 from NS4A, the protease activity of NS3 was not absolutely required for NS2-3 function in morphogenesis.
DISCUSSION
Although the functions of many of the pestivirus gene products have been elucidated, NS2, NS2-3, NS4B, and NS5A are still largely uncharacterized. In this study we intended to investigate the functions of uncleaved NS2-3. We first investigated whether uncleaved NS2-3 had a distinct function that could not be fulfilled by the individual proteins. Uncleaved NS2-3 was specifically abolished in the NADL Jiv 90−/Ubi genome and the bicistronic construct Npro-NS2/sg. Surprisingly, when these constructs were transfected into cells, infectious virus production was abolished. Virus production could be rescued by restoring NS2-3 expression either in the context of the bicistronic genome (Npro-E2/NS2-sg) or in trans (p7-NS2-3-4A/sg and Ubi-NS2-3-4A/sg). These results showed that uncleaved NS2-3 plays a critical role in virion production for which NS2 and NS3 alone are not sufficient.
This essential function for NS2-3 is supported by the observation that the uncleaved protein is maintained in all autonomous BVDV genomes despite the increased processing of NS2-3 in many cytopathic BVDV strains. Preservation of the protein is achieved either by incomplete processing at the NS2-3 junction or by duplication of the uncleaved protein elsewhere in the genome. The importance of NS2-3 encoded by genomic duplications is seen in the JaCp strain. This cytopathic BVDV contains an insertion of a portion of light chain 3 of microtubule-associated proteins 1A and 1B (LC3*) upstream of NS3, causing the complete release of NS3 from the polyprotein (28). The genome also encodes a duplication of NS2 up to the amino-terminal part of NS4B. Meyers et al. investigated the function of this duplication by engineering a JaCp-like genome that contained the LC3* insertion between NS2 and NS3 but did not express the duplicated region. Like our NS2-3-deficient genomes, this construct did not produce infectious virus after transfection into cells (28).
The Osloss strain of BVDV is likely an example of the selective pressure towards incomplete NS2-3 processing. Like NADL Jiv 90−/Ubi, Osloss expresses a ubiquitin monomer between NS2 and NS3. Unlike NADL Jiv 90−/Ubi, however, Osloss NS2-3 is not completely cleaved by ubiquitin carboxy-terminal hydrolase (27, 38). Complete processing may be prevented by two or more mutations in ubiquitin or by the Osloss polyprotein background (9, 27). A second BVDV strain, CP14, contains unmodified ubiquitin sequences between NS2 and NS3 (38). Although processing of NS2 from NS3 is complete when the proteins are expressed in vitro, the extent of processing in the full-length viral polyprotein is not known. We speculate that the CP14 polyprotein sequence may influence NS2-3 processing in this virus to prevent complete cleavage, but this remains to be tested.
After defining a role for NS2-3 in the viral life cycle, we determined the minimal regions of the genome that would rescue an NS2-3 defect in trans. A similar experiment is performed in nature when portions of the BVDV genome are duplicated to compensate for complete processing of NS2-3 induced by a cellular insertion. The duplications in these cytopathic genomes encompass all of the proteins NS2, NS3, and NS4A (2-4, 28, 31, 32). The importance of NS4A was also revealed in our trans-complementation experiments. Whereas deletion of p7 from the p7-NS2-3-4A expression cassette had little effect on its ability to rescue Npro-NS2/sg particle production, constructs that lacked NS4A in cis with NS2-3 were dramatically decreased in efficiency [p7-NS2-3/sg and p7-NS2-3(S1842A)/sg]. This may indicate a requirement for the protease domain of NS3 to interact with its cofactor, NS4A, in order for NS2-3 to function optimally. Although NS4A is expressed from the second cistron in all the bicistronic constructs, this protein may be sequestered by NS3 encoded immediately upstream, and therefore be unavailable for interaction with NS2-3. Our attempts to overexpress NS4A in trans and overcome this deficiency were unsuccessful (data not shown).
Nonstructural protein involvement in assembly is not unprecedented in the family Flaviviridae. Studies of yellow fever virus, a flavivirus, have shown that a basic motif in NS2A is important for the release of infectious virus (20). Remarkably, second site mutations in the helicase domain of NS3 were found to suppress mutations in this motif and restore virus production, suggesting a required interaction between NS2A and NS3 in flavivirus morphogenesis (20). In the pestiviruses, p7 has been shown to be essential for virion formation. This small hydrophobic protein immediately precedes NS2 in the genome; its mechanism of action in morphogenesis is not known (18).
The viral protease is also known to be essential for assembly of the flaviviruses; the NS2B-3 protease cleaves the capsid protein from its membrane anchor, allowing it to assemble into virions. The processing of capsid is also a prerequisite for signal peptidase cleavage of the downstream envelope protein, prM, from the polyprotein (1, 24, 35, 36). Here we report that, although the pestivirus NS3 protease cofactor, NS4A, seems to play a role in NS2-3 action, inhibition of the catalytic activity of the protease did not completely abolish virion production. Although the ability of the protease mutant to trans-complement Npro-NS2/sg was reduced, the deletion of NS4A may have contributed to this phenotype.
The precise role of uncleaved NS2-3 in infectious pestivirus production is unknown. The possibilities include critical interactions with other viral or cellular components required for assembly events. Unfortunately, little is known about these processes for the pestiviruses, particularly the early events in nucleocapsid assembly and budding. NS2-3 could participate in any of these processes, but one possible model, based on an interaction between NS2-3 and the capsid protein, is shown in Fig. 7 and outlined below.
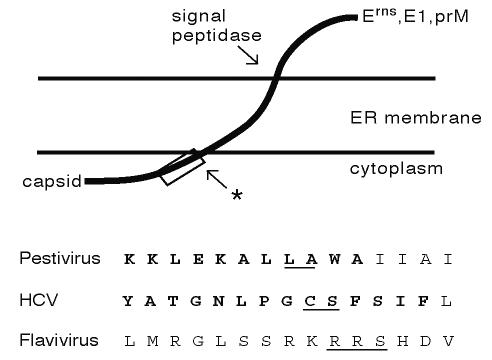
FIG. 7.
Flaviviridae capsid protein carboxy termini contain viral protease cleavage sites. The membrane topology of the capsid-envelope protein junction of the pestiviruses (Erns), hepatitis C virus (E1), and flaviviruses (prM) is diagrammed (top). The site of signal peptidase cleavage of the envelope proteins from capsid is shown. A second cleavage takes place in the region of the open box (arrow with star). This cleavage is mediated by the NS2B-3 proteases (flaviviruses), signal peptide peptidase (hepatitis C virus), and an unknown enzyme (pestiviruses). The exact site of cleavage in the pestiviruses and hepatitis C virus is not known. Sequences of the region of the capsid protein carboxy termini outlined by the open box above are shown in detail (bottom). Representative sequences from the three genera are shown (pestivirus, NADL; flavivirus, yellow fever virus; hepatitis C virus, genotype 1a [19]). Conserved amino acids are shown in bold. The recognition sites of the appropriate viral proteases are underlined. While none of the flavivirus protease recognition sites are absolutely conserved, they generally consist of two basic amino acids before the cleavage site and a residue with a short side chain immediately following it.
In the flaviviruses, the NS2B-3 protease cleaves at a consensus site that separates the carboxy terminus of the mature capsid protein from the signal sequence of the downstream prM protein (Fig. 7). In the pestiviruses, although the exact carboxy terminus of the capsid protein is unknown, residues identical to the NS3 protease cleavage site (Leu-Ala) are found preceding a hydrophobic region that likely serves as a signal sequence for Erns (Fig. 7). In hepatitis C virus, a potential NS3-4A protease cleavage site (Cys-Ser) is also found near the capsid protein carboxy terminus. Although there is no evidence that the pestiviral or hepaciviral (14) proteases cleave at these sites (for hepatitis C virus, signal peptide peptidase is believed to be responsible [25]), it is striking that residues involved in protease substrate recognition are present in the same location for all three genera. Studies of hepatitis C virus have shown that the NS3-4A protease is potently inhibited by peptides mimicking its amino-terminal cleavage products (34). Although it is not known if this is the case for the flavivirus and pestivirus proteases, they presumably also have affinity for these cleavage site residues irrespective of cleavage events.
Interaction of the BVDV NS2-3 protease with the capsid protein carboxy terminus may allow it to function in capsid protein folding, multimerization, or some other step in nucleocapsid assembly. Such an interaction would not necessarily require the serine nucleophile of the protease active site. Since NS3 is rather hydrophilic and NS2 is hydrophobic, it has been postulated that the intracellular locations of NS2-3 and its cleavage products are different (42). NS2-3 may therefore act as a scaffold to recruit the virion components to the site of nucleocapsid assembly. The ability of NS3 to bind RNA, its putative ability to interact with the capsid protein, and the observation that NS2 binds cellular chaperones (33) are in keeping with this hypothesis.
The evidence presented here indicates that uncleaved NS2-3 is indispensable for the assembly or egress of BVDV particles. The optimal activity of NS2-3 in this role requires a close genetic association with the protease cofactor NS4A but does not appear to absolutely require the catalytic activity of the protease. Investigations into the mechanisms of action of NS2-3 in pestivirus assembly are ongoing.
Acknowledgments
We thank James Fan, Jin-Hwan Han, Jack Hietpas, Pat Holst, Hernan Jaramillo, Merna Torres, and Anesta Webson for lab support and technical assistance. We are grateful to many colleagues for helpful discussions during the course of this work and to Arash Grakoui, Brett Lindenbach, Margaret MacDonald, Matthew Paulson, Glenn Randall, Donna Tscherne, Tim Tellinghuisen, and Shihyun You for critical reading of the manuscript.
This work was supported by grants from the Public Health Service (CA57973). C.L.M. was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (PGSA-232691).
REFERENCES
- 1.Amberg, S. M., and C. M. Rice. 1999. Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J. Virol. 73:8083-8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baroth, M., M. Orlich, H. J. Thiel, and P. Becher. 2000. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology 278:456-466. [DOI] [PubMed] [Google Scholar]
- 3.Becher, P., M. Orlich, and H. J. Thiel. 2001. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J. Virol. 75:6256-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher, P., H. J. Thiel, M. Collins, J. Brownlie, and M. Orlich. 2002. Cellular sequences in pestivirus genomes encoding gamma-aminobutyric acid (A) receptor-associated protein and Golgi-associated ATPase enhancer of 16 kilodaltons. J. Virol. 76:13069-13076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens, S. E., C. W. Grassmann, H. J. Thiel, G. Meyers, and N. Tautz. 1998. Characterization of an autonomous subgenomic pestivirus RNA replicon. J. Virol. 72:2364-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branza-Nichita, N., D. Durantel, S. Carrouée-Durantel, R. A. Dwek, and N. Zitzmann. 2001. Antiviral effect of N-butyldeoxynojirimycin against bovine viral diarrhea virus correlates with misfolding of E2 envelope proteins and impairment of their association into E1-E2 heterodimers. J. Virol. 75:3527-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collett, M. S., R. Larson, C. Gold, D. Strick, D. K. Anderson, and A. F. Purchio. 1988. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology 165:191-199. [DOI] [PubMed]
- 8.Collett, M. S., R. Larson, S. K. Belzer, and E. Retzel. 1988. Proteins encoded by bovine viral diarrhea virus: the genomic organization of a pestivirus. Virology 165:200-208. [DOI] [PubMed] [Google Scholar]
- 9.De Moerlooze, L., C. Lecomte, S. Brown-Shimmer, D. Schmetz, C. Guiot, D. Vandenbergh, D. Allaer, M. Rossius, G. Chappuis, D. Dina, et al. 1993. Nucleotide sequence of the bovine viral diarrhoea virus Osloss strain: comparison with related viruses and identification of specific DNA probes in the 5′ untranslated region. J. Gen. Virol. 74:1433-1438. [DOI] [PubMed] [Google Scholar]
- 10.Deregt, D., S. A. Masri, H. J. Cho, and H. B. Ohmann. 1990. Monoclonal antibodies to the p80/125 gp53 proteins of bovine viral diarrhea virus: their potential use as diagnostic reagents. Can. J. Vet. Res. 54:343-348. [PMC free article] [PubMed] [Google Scholar]
- 11.Elbers, K., N. Tautz, P. Becher, D. Stoll, T. Rumenapf, and H. J. Thiel. 1996. Processing in the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J. Virol. 70:4131-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Pragai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frolov, I., M. S. McBride, and C. M. Rice. 1998. cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA 4:1418-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray, E. W., and P. F. Nettleton. 1987. The ultrastructure of cell cultures infected with border disease and bovine virus diarrhoea viruses. J. Gen. Virol. 68:2339-2346. [DOI] [PubMed] [Google Scholar]
- 16.Greiser-Wilke, I., K. E. Dittmar, B. Liess, and V. Moennig. 1991. Immunofluorescence studies of biotype-specific expression of bovine viral diarrhoea virus epitopes in infected cells. J. Gen. Virol. 72:2015-2019. [DOI] [PubMed] [Google Scholar]
- 17.Grummer, B., M. Beer, E. Liebler-Tenorio, and I. Greiser-Wilke. 2001. Localization of viral proteins in cells infected with bovine viral diarrhoea virus. J. Gen. Virol. 82:2597-2605. [DOI] [PubMed] [Google Scholar]
- 18.Harada, T., N. Tautz, and H. J. Thiel. 2000. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 74:9498-9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 20.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kummerer, B. M., D. Stoll, and G. Meyers. 1998. Bovine viral diarrhea virus strain Oregon: a novel mechanism for processing of NS2-3 based on point mutations. J. Virol. 72:4127-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa.
- 24.Lobigs, M. 1993. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc. Natl. Acad. Sci. USA 90:6218-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendez, E., N. Ruggli, M. S. Collett, and C. M. Rice. 1998. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J. Virol. 72:4737-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyers, G., T. Rumenapf, and H.-J. Thiel. 1989. Ubiquitin in a togavirus. Nature 341:491. [DOI] [PubMed] [Google Scholar]
- 28.Meyers, G., D. Stoll, and M. Gunn. 1998. Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle. J. Virol. 72:4139-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moennig, V., and P. G. Plagemann. 1992. The pestiviruses. Adv. Virus Res. 41:53-98. [DOI] [PubMed] [Google Scholar]
- 30.Purchio, A. F., R. Larson, and M. S. Collett. 1984. Characterization of bovine viral diarrhea virus proteins. J. Virol. 50:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi, F., J. F. Ridpath, and E. S. Berry. 1998. Insertion of a bovine SMT3B gene in NS4B and duplication of NS3 in a bovine viral diarrhea virus genome correlate with the cytopathogenicity of the virus. Virus Res. 57:1-9. [DOI] [PubMed] [Google Scholar]
- 32.Qi, F., J. F. Ridpath, T. Lewis, S. R. Bolin, and E. S. Berry. 1992. Analysis of the bovine viral diarrhea virus genome for possible cellular insertions. Virology 189:285-292. [DOI] [PubMed] [Google Scholar]
- 33.Rinck, G., C. Birghan, T. Harada, G. Meyers, H. J. Thiel, and N. Tautz. 2001. A cellular J-domain protein modulates polyprotein processing and cytopathogenicity of a pestivirus. J. Virol. 75:9470-9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinkuhler, C., G. Biasiol, M. Brunetti, A. Urbani, U. Koch, R. Cortese, A. Pessi, and R. De Francesco. 1998. Product inhibition of the hepatitis C virus NS3 protease. Biochemistry 37:8899-8905. [DOI] [PubMed] [Google Scholar]
- 35.Stocks, C. E., and M. Lobigs. 1995. Posttranslational signal peptidase cleavage at the flavivirus C-prM junction in vitro. J. Virol. 69:8123-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stocks, C. E., and M. Lobigs. 1998. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J. Virol. 72:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tautz, N., T. Harada, A. Kaiser, G. Rinck, S. Behrens, and H. J. Thiel. 1999. Establishment and characterization of cytopathogenic and noncytopathogenic pestivirus replicons. J. Virol. 73:9422-9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tautz, N., G. Meyers, and H. J. Thiel. 1993. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology 197:74-85. [DOI] [PubMed] [Google Scholar]
- 39.Tautz, N., H. J. Thiel, E. J. Dubovi, and G. Meyers. 1994. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by an internal deletion. J. Virol. 68:3289-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiel, H. J., R. Stark, E. Weiland, T. Rumenapf, and G. Meyers. 1991. Hog cholera virus: molecular composition of virions from a pestivirus. J. Virol. 65:4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiland, F., E. Weiland, G. Unger, A. Saalmuller, and H. J. Thiel. 1999. Localization of pestiviral envelope proteins E(rns) and E2 at the cell surface and on isolated particles. J. Gen. Virol. 80:1157-1165. [DOI] [PubMed] [Google Scholar]
- 42.Wiskerchen, M., and M. S. Collett. 1991. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology 184:341-350. [DOI] [PubMed] [Google Scholar]
- 43.Xu, J., E. Mendez, P. R. Caron, C. Lin, M. A. Murcko, M. S. Collett, and C. M. Rice. 1997. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J. Virol. 71:5312-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]