Abstract
Five strains (1126-1H-08T, 51B-09, 986-08, 1084B-08 and 424-08) were isolated from diseased rainbow trout. Cells were Gram-negative rods, 0.7 µm wide and 3 µm long, non-endospore-forming, catalase and oxidase positive. Colonies were circular, yellow-pigmented, smooth and entire on TGE agar after 72 hours incubation at 25°C. They grew in a temperature range between 15°C to 30°C, but they did not grow at 37°Cor 42°C. Based on 16S rRNA gene sequence analysis, the isolates belonged to the genus Flavobacterium. Strain 1126-1H-08T exhibited the highest levels of similarity with Flavobacterium oncorhynchi CECT 7678T and Flavobacterium pectinovorum DSM 6368T (98.5% and 97.9% sequence similarity, respectively). DNA–DNA hybridization values were 87 to 99% among the five isolates and ranged from 21 to 48% between strain 1126-1H-08T, selected as a representative isolate, and the type strains of Flavobacterium oncorhynchi CECT 7678T and other phylogenetic related Flavobacterium species. The DNA G+C content of strain 1126-1H-08T was 33.2 mol%. The predominant respiratory quinone was MK-6 and the major fatty acids were iso-C15∶0 and C15∶0. These data were similar to those reported for Flavobacterium species. Several physiological and biochemical tests differentiated the novel bacterial strains from related Flavobacterium species. Phylogenetic, genetic and phenotypic data indicate that these strains represent a new species of the genus Flavobacterium, for which the name Flavobacterium plurextorum sp. nov. was proposed. The type strain is 1126-1H-08T ( = CECT 7844T = CCUG 60112T).
Introduction
The genus Flavobacterium is the type genus of the family Flavobacteriaceae accommodating Gram-negative, non-endospore-forming, aerobic, oxidase-positive, non-fermenting, predominantly gliding, yellow-pigmented bacteria [1], [2]. The genus, initially described to accommodate seven species, has considerably expanded with the description of many new species. Currently it includes 99 species, many of them described during the last five years. [3]. Members of the genus Flavobacterium can been isolated from a number of diverse habitats such as soil, water, sludge, plants, food products such as fish, meat, poultry, milk or lactic acid beverages [2], [4]. Most species are non-pathogenic, although a number of species have been associated with different clinical infections, being freshwater fish the animals most prone to flavobacterial infections [5]. Some Flavobacterium species, mainly Flavobacterium columnare, Flavobacterium branchiophilum and Flavobacterium psychrophilum, are well-recognized fish pathogens responsible for important economic losses in the fish farming industry [6], [7]. However, several other species such as Flavobacterium hydatis, Flavobacterium jhonsoniae, Flavobacterium succinicans, Flavobacterium chilense, Flavobacterium araucananum or Flavobacterium oncorhynchi have been also associated with infections in fish [1], [4], [5], [8]–[10]. Additionally, a number of new Flavobacterium species also have been described from the water of aquaculture facilities [11]–[13]. This plethora of Flavobacterium species could reproduce the diversity of flavobacteria associated with fish or fish surrounding environments. Some of these species could be considered commensal and opportunistic pathogenic bacteria [4], which point out the necessity for an accurate identifications of those strains of Flavobacterium spp. isolated from fish or fish farm environments. However, such identifications are extremely difficult based exclusively on biochemical criteria [4], [8], [14] and must be complemented with chemotaxomic and genetic methods [4], [5].
In this article, we report the phenotypic, genotypic and phylogenetic characterization of five novel Flavobacterium-like strains isolated from diseased trout. Based on the presented findings, a new species of the genus Flavobacterium, Flavobacterium plurextorum sp. nov., is proposed.
Materials and Methods
The present work does not include any experimental infections trial with farmed trout, just trout exclusively were used to identify microbiologically the etiological agent of the bacterial septicemia. Therefore, we did not consult with the IACUC and no specific national regulations for these procedures are available. Nevertheless, in order to ensure the welfare and ameliorate suffering of trout during transportation to the laboratory and euthanasia, trout were handled according to guidelines of relevant international organisms such as OIE (http://www.oie.int/doc/ged/D7821.PDF) and AVMA (https://www.avma.org/KB/Policies/Documents/euthanasia.pdf) and they were further necropsied under aseptic conditions. In addition, these procedures were approved by the responsible of animal welfare of the UCM Animal Health Department. The trout were sacrificed for the purpose of the study and the sacrifice was approved by the Technical Manager (Mari Angel Palacios, DVM, PhD) of the fish farm located in the west of Spain.
Trout and Strain Isolation
A clinical episode of septicemia occurred in a rainbow trout (Oncorhynchus mykiss) farm located in the central region of Spain. Affected trout were submitted by the Technical Manager of the fish farm to the Animal Health Surveillance Centre (VISAVET) of the Universidad Complutense (Madrid, Spain) for a confirmatory microbiological diagnosis.
Five Gram-negative, rod-shaped bacteria were isolated from liver (strains 986-08 and 424-08), gills (strains 1084B-08 and 51B-09) and eggs (1126-1H-08T) of five different trout. The strains were recovered in two different years (2008 and 2009) and they were isolated on tryptone glucose extract agar (TGE; Difco) after incubation at 25°C for 72 hours under aerobic conditions.
Phylogenetic Analysis
A large continuous sequence (approximately 1,400 bases) of the 16S rRNA gene of five strains was determined bidirectionally using universal primers pA (5′- AGAGTTTGATCCTGGCTCAG, positions 8–27, Escherichia coli numbering) and pH* (5′-AAGGAGGTGATCCAGCCGCA, positions 1541–1522, E. coli numbering) as described previously [10], and subjected to a comparative analysis. The identification of the phylogenetic relatives and calculations of pair-wise 16S rRNA gene sequence similarities were achieved, using the EzTaxon-e server [15]. The 16S rRNA gene sequences of the type strains of all validly published species of the genus Flavobacterium were retrieved from GenBank and aligned with the newly determined sequences using the program SeqTools [16]. Phylogenetic trees were constructed according to three different algorithms: neighbour-joining [17], using the programs SeqTools and TREEVIEW [18]; maximum-likelihood, using the PHYML software [19]; and maximum-parsimony, using the software package MEGA (Molecular Evolutionary Genetics Analysis) version 5.0 [20]. Genetic distances for the neighbour-joining and the maximum-likelihood algorithms were calculated by the Kimura two-parameter [21] and close-neighbour-interchange (search level = 2, random additions = 100) was applied in the maximum-parsimony analysis. The stability of the groupings was estimated by bootstrap analysis (1000 replications).
Genomic DNA G+C Content Determination and DNA-DNA Hybridizations
The G+C content of the genomic DNA of a representative strain (1126-1H-08T) was determined from the mid-point value (Tm) of the thermal denaturation profile [22], obtained with a Perkin-Elmer UV-Vis Lambda 20 spectrophotometer at 260 nm.
Genomic DNA-DNA hybridizations were carried out between strains 1126-1H-08T, 986-08, 424-08, 1084B-08 and 51B-09, and between strain 1126-1H-08T and the type strains of the closest phylogenetically related species. DNA was extracted and purified by the method of Marmur [22]. Hybridization studies were carried out, using the membrane method of Johnson [23], described in detail by Arahal et al. [24]. The hybridization experiments were carried out under optimal conditions, at a temperature of 44°C, which is within the limits of validity for the membrane method [25]. The percentages of hybridization were calculated as described by Johnson [26]. Three independent determinations were carried out for each experiment and the results reported as mean values. The type strains of species F. aquidurense CCUG 59847T, F. araucananum CCUG 61031T, F. hydatis DSM 2063T, F. pectinovorum CCUG 58916T, F. frigidimaris CCUG 59364T, F. chungangense CCUG 58910T and F. oncorhynchi CECT 7678T were included in this study.
Chemotaxonomic Characteristics
Respiratory quinones of strain 1126-1H-08T were extracted from 100 mg of freeze-dried cell material, using the two stage method described by Tindall [27], [28], and further separated by thin layer chromatography on silica gel and analyzed, using HPLC, by the identification service of the DSMZ (Braunschweig, Germany).
For cell fatty acid-fatty acid methyl ester (CFA-FAME) analyses, strain 1126-1H-08T was grown on Columbia II agar base (BBL 4397596) with 5% horse blood, at 30°C for 30–48 h, under aerobic conditions. The CFA-FAME profile was determined using gas chromatography (Hewlett Packard HP 5890) and a standardized protocol similar to that of the MIDI Sherlock MIS system [29], described previously [10]. CFAs were identified and the relative amounts were expressed as percentages of the total fatty acids of the respective strains.
Morphological, Physiological and Biochemical Characteristics
The minimal standards for the description of new taxa in the family Flavobacteriaceae [30] were followed for the phenotypic characterization of the strains. Gram-staining was performed as described by Smibert & Krieg [31]. Oxidase activity was determined by monitoring the oxidation of tetramethyl-p-phenylenediamine on filter paper and catalase activity was determined, using 3% H2O2 solution [31]. Hydrolysis of L-tyrosine (0.5%, w/v), lecithin (5%, w/v) [31], esculin (0.01% esculin and 0.05% ferric citrate, w/v), gelatin (4%; w/v), starch (0.2%, w/v), and casein [50% skimmed milk (Difco), v/v] were tested using nutrient agar as basal medium [30]. DNase test agar (Difco) was used for the DNase assay. Hydrolysis of urea (1%, w/v) was tested as described by Bowman et al. [32]. Growth in brain heart infusion broth was assessed at 15, 25, 30, 37 and 42°C, with 3.0, 4.5 and 6.5% added NaCl, and under anaerobic (with 4–10% CO2) and micro-aerobic (with 5–15% O2 and 5–12% CO2) conditions, using GasPak Plus and CampyPak Plus systems (BBL), respectively. Growth was tested on MacConkey (bioMérieux), nutrient (Difco) and trypticase-soy (bioMérieux) agar plates. The presence of gliding motility, using the hanging drop technique, and the production of flexirubin-type pigments and extracellular glycans were assessed, using the KOH and Congo red tests, respectively [1]. The strains were further biochemically characterized using the API 20NE and API ZYM systems (bioMérieux) according to the manufacturer’s instructions, except that incubation temperature was 25°C. The type strains of species F. aquidurense CCUG 59847T, F. araucananum CCUG 61031T, F. hydatis DSM 2063T, F. pectinovorum CCUG 58916T, F. frigidimaris CCUG 59364T, F. chungangense CCUG 58910T and F. oncorhynchi CECT 7678T were included in this study as references for the investigation of the phenotypic properties of the trout strains, using the same laboratory conditions.
PGFE Typing
The five strains were characterized by pulsed-field gel electrophoresis (PFGE), after digestion of their genomic DNAs with the restriction enzymes Bsp120I and XhoI, according to the specifications of Chen et al. [33]. DNA fragments were resolved in a 1% agarose gel with a pulse-field gel electrophoresis apparatus, CHEF-DR III (Bio-Rad), at 6V/cm for 40 hours, with switching times ramped from 0.1 to 12 s at 14°C, with an angle of 120°. The gels were stained for 30 min with Syber-Safe and photographed under UV light (Gel-Doc, Bio-Rad). Strains differing in at least one band were considered different.
Results and Discussion
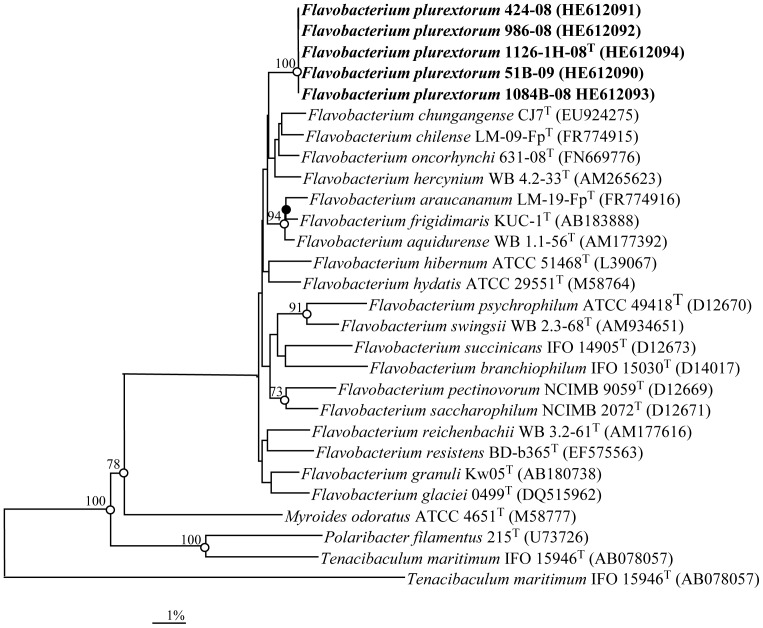
16S rRNA gene sequences were determined for the five trout strains, displaying 100% 16S rRNA sequence similarity among them. Sequence searches showed that the 16S rRNA gene sequence of the strains were most similar to those of species of the genus Flavobacterium, exhibiting the highest levels of similarity with the sequence of the type strains of Flavobacterium oncorhynchi CECT 7678T and Flavobacterium pectinovorum DSM 6368T (98.5% and 97.9% sequence similarity, respectively). In addition, strains exhibited 16S rRNA gene sequence similarities greater than 97.0% with other seventeen other Flavobacterium species. It is clear from the phylogenetic analysis (Fig. 1) that the trout strains held a clear affiliation to the genus Flavobacterium and represented a distinct sub-lineage clustering with a cluster of four species that included F. pectinovorum, F. chilense, F. oncorhynchi and F. hercynium. However, their position within this sub-group was not supported by significant bootstrap values. The GenBank accession numbers for the 16S rRNA gene sequences of five strains sequenced in this study are shown in Fig. 1.
Figure 1. Phylogenetic tree based on 16S rRNA gene sequence comparisons, obtained with the neighbour-joining algorithm, showing the relationships of Flavobacterium plurextorum sp. nov. with related species.
Flexibacter flexilis ATCC 23079T was used as an outgroup. Bootstrap values (expressed as a percentage of 1,000 replications) greater than 70% are given at the nodes. Solid circles indicate that the corresponding nodes (groupings) are also obtained on the maximum-likelihood tree. Open circles indicate that the corresponding nodes (groupings) are also obtained on the maximum-likelihood and parsimony trees. Sequence accession numbers are indicated in brackets. Bar, 1% sequence divergence.
Genomic DNA–DNA hybridizations between the trout strains yielded binding values of 87 to 100%. Flavobacterium species with 16S rRNA gene sequence similarities to the sequences of the trout strains lower than 98.0% correlated with levels of genomic DNA-DNA relatedness always lower than 70% [9]–[11], [34]–[36]. For that reason, DNA-DNA hybridizations were carried out only between strain 1126-1H-08T and the type strains of the phylogenetically closest related species; i.e., those species with 16S rRNA gene sequence similarities greater than 97.5%. The levels of DNA-DNA relatedness for strain 1126-1H-08T with respect to F. aquidurense CCUG 59847T, F. araucananum CCUG 61031T, F. hydatis DSM 2063T, F. pectinovorum CCUG 58916T, F. frigidimaris CCUG 59364T, F. chungangense CCUG 58910T and F. oncorhynchi CECT 7678T ranged between 21 and 48%. These values were below the 70% cut-off point for species delineation [37], [38] and clearly confirmed that the trout strains belong to a distinct genomic species of the genus Flavobacterium. The DNA G+C content of strain 1126-1H-08T was 33.2 mol%, a value consistent with those of the genus Flavobacterium [1], [30].
Chemotaxonomic characteristics of strain 1126-1H-08T were in accordance with those of members of the genus Flavobacterium [5], [6]: the major quinone was MK-6 (95%) with minor amounts of MK-5 (5%). The predominant cell fatty acids of strain 1126-1H-08T were iso-C15∶0 (19%) and C15∶0 (15%). Strain 1126-1H-08T also contained moderate or small amounts of C16∶1 ω7c (10%), C15∶1 ω6c (9%), iso-C15∶0 3-OH, C17∶1 ω6c, isoG-C15∶1 (6%/each), iso-C17∶0 3-OH (5%), iso-C17∶1 ω9c, C15∶0 3-OH, C16∶0 3-OH (3%/each), isoaldehyde-C15∶0, C16∶0, iso-C16∶0 3-OH, unknown fatty acids with an equivalent chain length of 11.5 (2%/each) and C17∶1 ω8c, iso-C16∶0, C12∶1, aldehyde-C14∶0, anteiso-C15∶0 and unknown fatty acids with an equivalent chain lengths of 14.8 and 12.5 (1%/each) (Table 1).
Table 1. Cellular fatty acid compositions of Flavobacterium plurextorum 1126-1H-08T and its closest phylogenetic neighbours.
| Fatty acid | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Saturated | ||||||||
| C12∶1 | 1 | tr | – | – | – | – | tr | – |
| C14∶0 | tr | – | tr | 1.1 | tr | tr | – | tr |
| C15∶0 | 15 | 13.5 | 11.9 | 5.5 | 5.6 | 6.9 | 20.6 | 15.7 |
| C16∶0 | 2 | 1.6 | 1.1 | 2.3 | 2.8 | 2.2 | tr | 2.9 |
| Hydroxy | ||||||||
| C15∶0 2OH | – | – | 1.1 | – | tr | – | – | – |
| C15∶0 3OH | 3 | 3.3 | 1.9 | – | – | – | 1.8 | – |
| iso-C15∶0 3OH | 6 | 7.8 | 6.9 | 7.7 | 8.6 | 5.8 | 7.1 | 5.8 |
| C16∶0 3OH | 3 | – | 1.1 | 3.5 | 4.5 | 1.4 | – | 2.5 |
| iso-C16∶0 3OH | 2 | 1.0 | 2.1 | 1.6 | 1.9 | tr | 2.1 | 1.5 |
| iso-C17∶0 3OH | 5 | 8.2 | 7.3 | 5.9 | 10.3 | 5.1 | 7.0 | |
| Branched | ||||||||
| C14∶0 aldehyde | 1.0 | – | – | – | – | – | – | tr |
| iso-C15∶0 | 19 | 26.1 | 14.6 | 28.2 | 23.5 | 28.0 | 24.8 | 25.5 |
| anteiso-C15∶0 | 1.0 | 1.3 | 3.0 | 3.2 | tr | 4.3 | 2.5 | 1.9 |
| iso-C15∶0 aldehyde | 2.0 | 3.2 | 1.2 | 1.3 | tr | 1.3 | 2.3 | 2.0 |
| iso-C15∶1 G | 6.0 | 2.9 | 7.4 | 3.7 | 5.8 | 7.2 | 5.0 | 5.0 |
| iso-C16∶0 | 1 | – | 1.1 | 1.0 | tr | 1.0 | – | 1.1 |
| iso-C16∶1 H | tr | – | 1.0 | 1.0 | tr | – | – | tr |
| iso-C17∶1 ω9c | 3 | 6.0 | 5.2 | 4.3 | 4.1 | 6.0 | 1.1 | 2.9 |
| Unsaturated | ||||||||
| C15∶1 ω6c | 9 | 12.3 | 10.1 | 4.1 | 2.9 | 5.5 | 12.2 | 7.6 |
| C16∶1 ω7c | 10 | 3.7 | 11.2 | 19.2 | 15.7 | 18.1 | 2.2 | 9.8 |
| C17∶1 ω6c | 6 | 5.9 | 6.4 | 3.5 | 2.5 | 3.2 | 6.2 | 2.4 |
| C17∶1 ω8c | 1 | 1.0 | 1.5 | – | tr | tr | 1.3 | tr |
| Summed feature 1 a | – | – | – | 2.0 | 1.7 | 1.4 | – | 1.7 |
| Unidentified fatty acid b | ||||||||
| ECL 11.541 | 2 | 1.4 | tr | tr | tr | tr | 1.2 | 1.1 |
| ECL 12.555 | 1 | – | tr | – | – | – | 1.1 | tr |
| ECL 14.809 | 1 | – | – | – | – | – | – | – |
| ECL 16.580 | – | – | tr | – | 1.1 | – | – | tr |
Taxa: 1, F. plurextorum 1126-1H-08T; 2, F. pectinovorum CCUG 58916T; 3, F. aquidurense CCUG 59847T; 4, F. frigidimaris CCUG 59364T; 5, F. hydatis DSM 2063T; 6, F. araucananum CCUG 61031T; 7, F. chungangense CCUG 58910T; 8, F. oncorhynchi CECT 7678T.
Values are percentages of total fatty acids; fatty acids representing less than 1% in all strains were omitted. tr = trace amount, i.e., <1%. - = not detected.
CFA values for type strains other than F. plurextorum 1126-1H-08T were taken from the CCUG culture collection (http://www.ccug.se/). Strains were cultivated on the same medium and growth conditions.
Summed features represent groups of two or three fatty acids that cannot be separated by GLC with the MIDI system. Summed feature 1 comprised iso-C17∶1 I/C16∶0 DMA.
ECL, equivalent chain length.
The trout strains exhibited identical physiological and biochemical characteristics. Cells were Gram-negative rods, 0.7 µm wide and 3 µm long, non-endospore-forming, and non-gliding. Strains grew well under aerobic conditions and grew weakly under micro-aerobic conditions. Strains grew at 15–30°C with optimal growth at approximately 25°C, while no growth was observed at 37°C or 42°C. Growth occurred on trypticase-soy and nutrient agars but not on Marine agar after incubation at 25°C for 72 hours. Colonies were circular, yellow-pigmented, smooth and entire on TGE agar after 72 hours incubation at 25°C. Colonies are non-hemolytic on Columbia agar after 72 hours incubation at 25°C. Diffusible flexirubin-type pigments were produced and congo red was not absorbed by colonies. Growth did not occur in brain heart infusion broth containing 3, 4.5 and 6.5% NaCl. Catalase and oxidase were produced and nitrate and nitrite were reduced. Starch and tyrosine were degraded but DNA, gelatin, casein or agarose were not. A brown pigment was not produced on tyrosine agar. Aesculin was hydrolyzed but not urea, lecithin and arginine. Indole and H2S were not produced. Acid was not produced from D-glucose. Arabinose, D-glucose, mannose, N-acetyl-glucosamine, and maltose were used as sole carbon and energy sources but not citrate, mannitol, gluconate, caprate, adipate, and malate. Activities for alkaline phosphatase, leucine arylamidase, N-acetyl-β-glucosaminidase, α-glucosidase, acid phosphatase, and naphthol-AS-BI-phophohydrolase were detected. Esterase C4, valine arylamidase, β-galactosidase, ester lipase C8, lipase C14, cystine arylamidase, α-chymotrypsin, trypsin, α-galactosidase, β-glucuronidase, β-glucosidase, α-mannosidase and α-fucosidase were not detected.
The phenotypic characteristics that differentiated the trout strains from phylogenetically related species are shown in Table 2. The new species also can be also differentiated from the clinically relevant fish pathogens F. columnare, F. psycrophilum and F. branchiophilum, by the inability of these three species to grow in trypticase-soy agar and to hydrolyze aesculin [4]. Other species isolated from diseased fish such as F. hydatis, F. jonshoniae and F. succinicans are motile (gliding), degrade DNA and produce acid from carbohydrates [4], while the new species exhibited opposite results for those tests. Moreover, the new species can be readily differentiated from F. chilense and F. araucananum because the latter species are motile (gliding), grow in 3% NaCl and assimilate mannitol [9] and from F. oncorhynchi which produces β-galactosidase while the new species give opposite results for this test [10].
Table 2. Characteristics that differentiate Flavobacterium plurextorum sp. nov. from closely related Flavobacterium species based in the 16S rRNA tree topology.
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Growth on Marine agar | − | − | − | + | − | − | − | − |
| Growth at 30°C | + | + | + | − | + | + | + | + |
| Hydrolysis of: | ||||||||
| L- tyrosine | + | − | + | − | + | − | − | + |
| DNA | − | − | − | − | + | − | + | − |
| Urea | − | − | + | − | − | − | − | − |
| Nitrate reduction | + | + | − | − | + | + | + | + |
| Assimilation of: | ||||||||
| Arabinose | + | + | + | + | − | + | + | + |
| Mannitol | − | − | − | + | − | − | − | − |
| N-acetyl-glucosamine | + | + | + | + | + | + | - | + |
| Production of: | ||||||||
| Valine arylamidase | − | + | + | + | + | + | + | − |
| α-Glucosidase | + | − | − | + | + | + | − | + |
| β-Glucosidase | − | + | − | + | − | + | + | − |
| N-Acetyl-β-glucosaminidase | + | − | − | + | + | − | − | + |
Taxa: 1, F. plurextorum 1126-1H-08T; 2, F. pectinovorum CCUG 58916T; 3, F. aquidurense CCUG 59847T; 4, F. frigidimaris CCUG 59364T; 5, F. hydatis DSM 2063T; 6, F. araucananum CCUG 61031T; 7, F. chungangense CCUG 58910T; 8, F. oncorhynchi CECT 7678T.
Data are from this study.
+, positive reaction; −, negative reaction.
After PFGE typing, the trout strains were characterized by 3 different restriction profiles with the enzymes Bsp120I (Fig. 2) and XhoI (not shown). Strains 986-08 and 1084B-08 exhibited indistinguishable restriction profiles with both enzymes and strain 51B-09 could not be characterized because its DNA systematically was autodegraded.
Figure 2. PFGE patterns generated after Bsp120I macrorestriction of Flavobacterium plurextorum sp. nov.
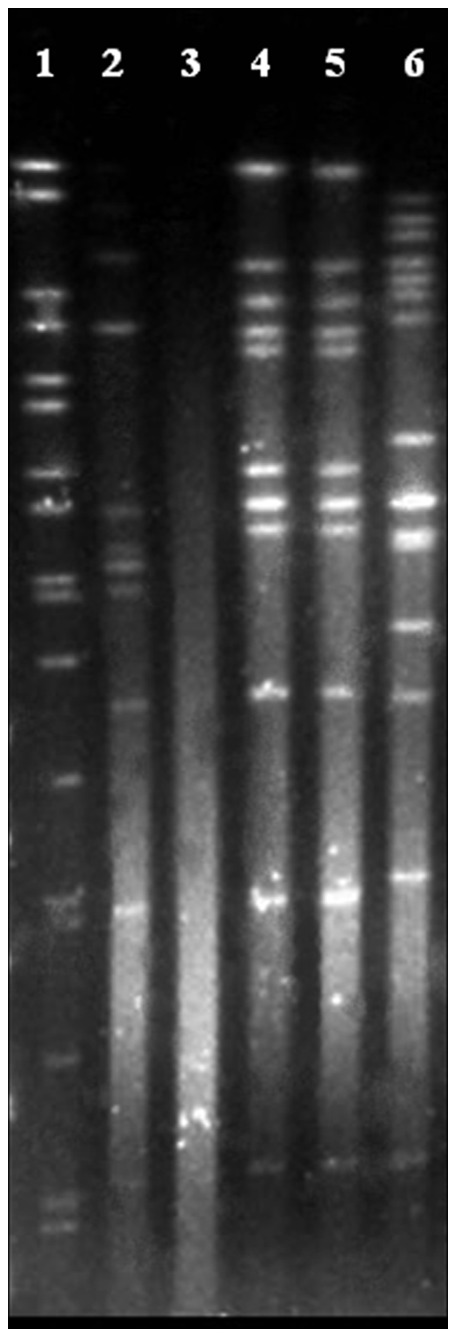
Lane 1, DNA molecular size marker; Lanes 2 to 6, strains 1126-1H-08T, 51B-09, 986-08, 1084B-08 and 424-08, respectively.
Flavobacteria are known to belong to the microbiota of fish and fish eggs [4], [5]. Therefore, although two strains were isolated from internal organs, the other three were recovered from gills and eggs which suggest that the new species could be saprophytic or commensal and able to colonize fish, and produce disease under stressful conditions or other predisposing circumstances such as coinfections with other bacteria or viruses, poor farming conditions or environmental disorders [4], [39]. This assumption should be confirmed by experimental infection trials. Nevertheless, the formal description of Flavobacterium plurextorum and the availability of tests to facilitate its identification from other Flavobacterium species associated with fish disease or isolated from diseased fish will aid laboratories in its recognition and identification in the future, and to improve the knowledge of its distribution and possible association with disease.
Conclusion
The phylogenetic, genotypic and phenotypic results of the present polyphasic study demonstrated that the new strains isolated from rainbow trout represented a novel species of the genus Flavobacterium, for which the name Flavobacterium plurextorum sp. nov. is proposed (plu.rex.to’rum. L. comp. pl. plures, more, several, many; L. pl. n. exta -orum, entrails; N.L. gen. pl. n. plurextorum, of several internal organs). Detailed description of the morphological, physiological and biochemical characteristics of this species were indicated above. The type strain is 1126-1H-08T ( = CECT 7844T = CCUG 60112T).
Acknowledgments
The authors thank Professor J. P. Euzéby of the Ecole Nationale Vétérinaire in Toulouse for advice concerning the Latin species name and A. Casamayor (VISAVET) for technical assistance in PFGE analysis and Kent Molin (CCUG) for the analyses of CFAs.
Funding Statement
This work was funded by projects CENIT 2007-2010 (ACUISOST) of the Spanish Office for Science and Technology (CDETI), CGL2010-19303 of the Spanish Ministry of Science and Innovation and P10-CVI-6226 from the Junta de Andalucía. ERBM was supported by funding of Västra Götaland Region projects VGFOUREG-30781, 83080 and 157801. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bernardet JF, Segers P, Vancanneyt M, Berthe F, Kersters K, et al. (1996) Cutting a Gordian knot: emended classification and description of the genus Flavobacterium emended description of the family Flavobacteriaceae and proposal of Flavobacterium hydatis nom nov (basonym Cytophaga aquatilis Strohl and Tait 1978). Int J Syst Bacteriol 46: 128–148. [Google Scholar]
- 2.Bernardet JF, Nakagawa Y (2006) An introduction to the family Flavobacteriaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes: a Handbook on the Biology of Bacteria, 3rd ed, vol, 7, New York: Springer. 455–480.
- 3.Euzéby JP (1997) List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet. Int J Syst Bacteriol 47 590–592. Available: http://wwwbacterionet Accessed (April 13, 2013). [DOI] [PubMed]
- 4.Bernardet JF, Bowman JP (2006) The genus Flavobacterium. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes: a Handbook on the Biology of Bacteria, 3rd ed. Vol, 7. New York: Springer. 481–531.
- 5.Bernardet JF, Bowman JP (2011) Genus I. Flavobacterium Bergey, Harrison, Breed, Hammer and Huntoon 1923, 97AL emend. Bernardet, Segers, Vancanneyt, Berthe, Kersters and Vandamme 1996, 139. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, et al. editors. Bergey’s Manual of Systematic Bacteriology, 2nd ed. Vol, 4. New York; Springer. 112–155.
- 6.Roberts RJ (2012) The Bacteriology of Teleosts. In Fish Pathology, 4 ed. UK: Wiley-Blackell. 339–382.
- 7.Starliper CE, Schill WB (2012) Flavobacterial diseases: columniaris disease, coldwater disease and bacterila gill disease. In: Woo TTK, Bruno DW, editors. Fish Diseases and Disorders, 2nd ed, Vol. 3. UK: CAB International. 606–631.
- 8. Flemming L, Rawlings D, Chenia H (2007) Phenotypic and molecular characterization of fish-borne Flavobacterium johnsoniae-like isolates from aquaculture systems in South Africa. Res Microbiol 158: 18–30. [DOI] [PubMed] [Google Scholar]
- 9. Kämpfer P, Lodders N, Martin K, Avendaño-Herrera R (2012) Flavobacterium chilense sp nov and Flavobacterium araucananum sp. nov. two novel species isolated from farmed salmonid in Chile. Int J Syst Evol Microbiol 62: 1402–1408. [DOI] [PubMed] [Google Scholar]
- 10. Zamora L, Fernández-Garayzábal JF, Svensson-Stadler LA, Palacios MA, Domínguez L, et al. (2012) Flavobacterium oncorhynchi sp. nov. a new species isolated from rainbow trout (Oncorhynchus mykiss). Syst Applied Microbiol 35: 86–91. [DOI] [PubMed] [Google Scholar]
- 11. Chen WM, Huang WC, Young CC, Sheu SY (2013) Flavobacterium tilapiae sp. nov. isolated from a freshwater pond and emended descriptions of Flavobacterium defluvii and Flavobacterium johnsoniae. . Int J Syst Evol Microbiol 63: 827–834. [DOI] [PubMed] [Google Scholar]
- 12.Sheu SY, Lin YS, Chen WM. (2012) Flavobacterium squillarum sp. nov., isolated from a freshwater shrimp culture pond and emended descriptions of Flavobacterium haoranii, F. cauense, F. terrae and F. aquatile. Int J Syst Evol Microbiol (in press) doi: 10.1099/ijs.0.046425-0. [DOI] [PubMed]
- 13. Sheu SY, Chiu TF, Young CC, Arun AB, Chen WM (2011) Flavbacterium macrobrachii sp. nov. isolated from a freshwater shrimp culture pond. Int J Syst Evol Microbiol. 61: 1402–1407. [DOI] [PubMed] [Google Scholar]
- 14. Ilardi P, Avendaño-Herrera R (2008) Isolation of Flavobacterium-like bacteria from diseased salmonids cultured in Chile. Bull Eur Assoc Fish Pathol 28: 176–185. [Google Scholar]
- 15.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, et al. (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62: 716–721. Available: http://eztaxon-e.ezbiocloud.net Accessed May 29, 2013. [DOI] [PubMed]
- 16.Rasmussen SW (2002) SEQtools a software package for analysis of nucleotide and protein sequences. Available: http://wwwseqtoolsdk Accessed May 27, 2013.
- 17. Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 18. Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers Comput Appl Biosci. 12: 357–358. [DOI] [PubMed] [Google Scholar]
- 19. Guindon S, Gascuel O (2003) A simple fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 20. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 22. Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microoganisms. J Mol Biol 3: 208–219. [Google Scholar]
- 23.Johnson JL (1994) Similarity analysis of DNAs. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR, editors. Methods for General and Molecular Bacteriology. Washington DC: American Society for Microbiology. 655–681.
- 24. Arahal DR, García MT, Vargas C, Canovas D, Nieto JJ, et al. (2001) Chromohalobacter salexigens sp. nov. a moderately halophilic species that includes Halomonas elongata DSM 3043 and ATCC 33174. Int J Syst Evol Microbiol 51: 1457–1462. [DOI] [PubMed] [Google Scholar]
- 25. De Ley J, Tijtgat R (1970) Evaluation of membrane filter methods for DNA-DNA hybridization. A Van Leeuw J Microb 36: 461–474. [DOI] [PubMed] [Google Scholar]
- 26.Johnson JL (1994) Similarity analysis of DNAs. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR, editors. Methods for General and Molecular Bacteriology, Washington DC: American Society for Microbiology. 655–681.
- 27. Tindall BJ (1990) A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol 13: 128–130. [Google Scholar]
- 28. Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. . FEMS Microbiol Letts 66: 199–202. [Google Scholar]
- 29.Sasser M (2001) Identification of bacteria by gas chromatography of cellular fatty acids MIDI. Available: http://wwwmicrobialidcom/PDF/TechNote_101pdf Accessed May 27, 2013.
- 30. Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52: 1049–1070. [DOI] [PubMed] [Google Scholar]
- 31.Smibert RM, Krieg NR (1994) Phenotypic Characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR, editors. Methods for General and Molecular Bacteriology, Washington DC: American Society for Microbiology. 607–653.
- 32. Bowman JP, Cavanagh J, Austin JJ, Sanderson K (1996) Novel Psychrobacter species from Antarctic ornithogenic soils. Int J Syst Bacteriol 46: 841–848. [DOI] [PubMed] [Google Scholar]
- 33. Chen YC, Davis MA, Lapatra SE, Cain KD, Snekvik KR, et al. (2008) Genetic diversity of Flavobacterium psychrophilum recovered from commercially raised rainbow trout Oncorhynchus mykiss (Walbaum) and spawning coho salmon O. kisutch (Walbaum). J Fish Dis 31: 765–73. [DOI] [PubMed] [Google Scholar]
- 34. Lim CS, Oh YS, Lee JK, Park AR, Yoo JS, et al. (2011) Flavobacterium chungbukense sp. nov. isolated from soil. Int J Syst Evol Microbiol 61: 2734–2739. [DOI] [PubMed] [Google Scholar]
- 35. Xu M, Xin Y, Tian J, Dong K, Yu Y, et al. (2011) Flavobacterium sinopsychrotolerans sp. nov. isolated from a glacier. Int J Syst Evol Microbiol 61: 20–24. [DOI] [PubMed] [Google Scholar]
- 36. Yoon JH, Park S, Kang SJ, Oh SJ, Myung SC, et al. (2011) Flavobacterium ponti sp. nov.isolated from seawater. Int J Syst Evol Microbiol 61: 81–85. [DOI] [PubMed] [Google Scholar]
- 37. Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, et al. (1987) International Committee on Systematic Bacteriology Report of the ad hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int J Syst Bacteriol 37: 463–464. [Google Scholar]
- 38. Stackkebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44: 846–849. [Google Scholar]
- 39. Georgiadis MP, Gardner IA, Hedrick RP (2001) The role of epidemiology in the prevention, diagnosis, and control of infectious diseases of fish. Prev Vet Med 48: 287–302. [DOI] [PubMed] [Google Scholar]