Abstract
Short-term assays have suggested that RNA interference (RNAi) may be a powerful new method for intracellular immunization against human immunodeficiency virus type 1 (HIV-1) infection. However, RNAi has not yet been shown to protect cells against HIV-1 in long-term virus replication assays. We stably introduced vectors expressing small interfering RNAs (siRNAs) directed against the HIV-1 genome into human T cells by retroviral transduction. We report here that an siRNA directed against the viral Nef gene (siRNA-Nef) confers resistance to HIV-1 replication. This block in replication is not absolute, and HIV-1 escape variants that were no longer inhibited by siRNA-Nef appeared after several weeks of culture. These RNAi-resistant viruses contained nucleotide substitutions or deletions in the Nef gene that modified or deleted the siRNA-Nef target sequence. These results demonstrate that efficient inhibition of HIV-1 replication through RNAi is possible in stably transduced cells. Therefore, RNAi could become a realistic gene therapy approach with which to overcome the devastating effect of HIV-1 on the immune system. However, as is known for antiviral drug therapy against HIV-1, antiviral approaches involving RNAi should be used in a combined fashion to prevent the emergence of resistant viruses.
The introduction of small interfering RNA (siRNA) into mammalian cells can activate RNA interference (RNAi), resulting in sequence-specific degradation of the targeted RNA. RNAi may be a powerful new method for intracellular immunization against human immunodeficiency virus type 1 (HIV-1) infection. It has been demonstrated in short-term assays that HIV-1 replication can be inhibited by siRNAs directed against viral or cellular targets (1, 6, 10, 11, 14, 15, 17, 19, 20). To demonstrate that RNAi can protect cells against HIV-1 in long-term virus replication assays and to study the evolution of the inhibited virus, we stably introduced vectors expressing siRNAs directed against eight HIV-1 targets into human T cells (Fig. 1A). We selected target sequences in the infectious HIVLAI molecular clone that are highly conserved among virus isolates (13), which warrants a widespread application of the potential RNAi therapy. Furthermore, we selected targets within the structured HIV-1 RNA genome that are highly accessible as determined with antisense DNA oligonucleotide arrays (2, 19a). There is recent evidence that the efficacy of siRNAs is similarly influenced by secondary structure in the target transcript (3, 12). These two selection criteria forced us in some cases to accept a suboptimal design of the siRNA molecule. The selected targets included essential replication signals such as the primer-binding site (Fig. 1A, PBS-A and PBS-B) and the polypurine tract (PPT), a segment in the overlap between the Tat and the Rev genes (TatRevA and TatRevB), and part of the 5′ untranslated leader region (UTR-A and UTR-B). Furthermore, we selected an siRNA targeting the viral Nef gene (siRNA-Nef) that was previously demonstrated to provide efficient silencing in a transient-transfection system (10).
FIG. 1.
siRNA targeting of HIV-1. (A) SupT1 cells were stably transduced with the empty pRETRO-SUPER retroviral vector (4, 5) (control) or with vectors expressing siRNAs against eight target sequences in the HIV genome: PBS-A, GTGGCGCCCGAACAGGGACTT; PBS-B, TGGCGCCCGAACAGGGACTT; PPT, GGGGGGACTGGAAGGGCTA; TatRev-A, GCCTTAGGCATCTCCTATG;TatRev-B, CCTATGGCAGGAAGAAGCG; UTR-A, GCGGAGGCTAGAAGGAGAG; UTR-B, GGCTAGAAGGAGAGAGATG; and Nef, GTGCCTGGCTAGAAGCACA. (B) Cells were infected with HIVLAI (800 pg of CA-p24 in a 5-ml culture), and virus replication was monitored by determining the CA-p24 level in the culture supernatant. (C) Predicted structure of the siRNA-Nef transcript. The sequence targeting the Nef gene is shown in bold.
The pRETRO-SUPER vector (5) expresses the siRNAs from the human H1 polymerase III promoter as a small hairpin. These vectors were stably transduced into the human T-cell line SupT1. This cell line allows the rapid and massive spread of HIVLAI, as is apparent for the control SupT1 cells transduced with the empty pRETRO-SUPER vector (Fig. 1B). SupT1 T cells expressing a specific siRNA were infected with equal amounts of HIVLAI. Fast virus replication was observed in all SupT1 cultures except those with cells expressing the siRNA against Nef (Fig. 1C). The replication of HIVLAI was profoundly reduced in cells expressing siRNA-Nef compared with that in the control cells transduced with the empty vector. We did not observe any growth retardation of the siRNA-Nef cells, indicating that the decrease in HIV-1 replication in these cells is not due to a nonspecific cell toxicity problem.
A second independently prepared batch of siRNA-Nef-transduced cells showed the same resistance phenotype against both low and intermediate doses of HIVLAI (Fig. 2A, top and middle panels, respectively). However, the block in viral replication was not absolute, and a high dose of input virus allowed a low level of replication (Fig. 2A, bottom panels). We demonstrated efficient replication of HIVLAI on a new batch of control cells transduced with the empty vector (Fig. 2A, right panels). As a further control for the specificity of the observed inhibition, we used an HIV-1 variant in which the Nef gene is replaced by the rtTA gene (HIVrtTA) (18, 21). HIVrtTA lacks the Nef target sequence and replicated efficiently in both the control cells and the cells that express the siRNA against Nef (Fig. 2A). These results demonstrate the sequence-specific inhibition of HIV-1 replication by the RNAi targeting of Nef gene sequences that are present in the unspliced viral RNA genome and all spliced subgenomic HIV-1 transcripts.
FIG. 2.
Sequence-specific inhibition of HIV-1 replication. (A) SupT1 cells stably transduced with the siRNA-Nef vector (left panels) or the empty vector (right panels) were infected with wild-type HIVLAI (closed circles) or HIVrtTA with the Nef gene deleted (open circles). The virus input levels were 800 pg of CA-p24 (top panels), 4,000 pg of CA-p24 (middle panels), and 8,000 pg of CA-p24 (bottom panels) in 5-ml cultures. Virus spread was monitored by determining the CA-p24 level in the culture supernatant. (B) siRNA-Nef and control cells were transfected with 10 μg of DNA encoding wild-type HIVLAI or HIVrtTA with the Nef gene deleted as described previously (7). Virus production in the culture supernatant at 2 and 3 days after transfection was measured by a CA-p24 enzyme-linked immunosorbent assay.
siRNA-mediated gene silencing may inhibit virus replication at a number of stages of the HIV-1 replication cycle. It is most likely that siRNA-Nef is able to target the newly synthesized viral transcripts in HIV-infected cells. To directly test this possibility, we transfected the set of SupT1 cells with the proviral HIVLAI DNA and measured virus production after 2 and 3 days (Fig. 2B). The cells that actively produce siRNA-Nef showed a >10-fold reduction in virus production compared with that in the control cells. Furthermore, this inhibition of gene expression was specific for wild-type HIV-1, because no such effect was observed for the HIVrtTA variant with the Nef gene deleted.
Only one out of eight siRNA constructs tested can effectively control a spreading HIV-1 infection. This siRNA against Nef was also effective when transfected as an oligonucleotide in a transient-transfection assay (10). The other siRNAs tested did not inhibit HIVLAI, even though the sequences perfectly matched the viral transcript. Presently, we do not know why siRNA-Nef is such an effective inhibitor of HIV-1 replication. One reason may be that this siRNA targets both the unspliced and all spliced forms of HIV-1 RNA. However, the inactive siRNA-PBS and siRNA-PPT constructs also have this property. Thus, there may be additional features of the siRNA (e.g., expression level and intracellular location) or the target RNA (e.g., masking by viral and/or cellular proteins within the cell or virion particle) that determine the overall efficiency of inhibition. We have already mentioned that the selection of highly conserved and highly accessible target sites within the HIV-1 RNA genome had a negative impact on the design of some siRNAs. More-detailed studies are needed to resolve some of these issues.
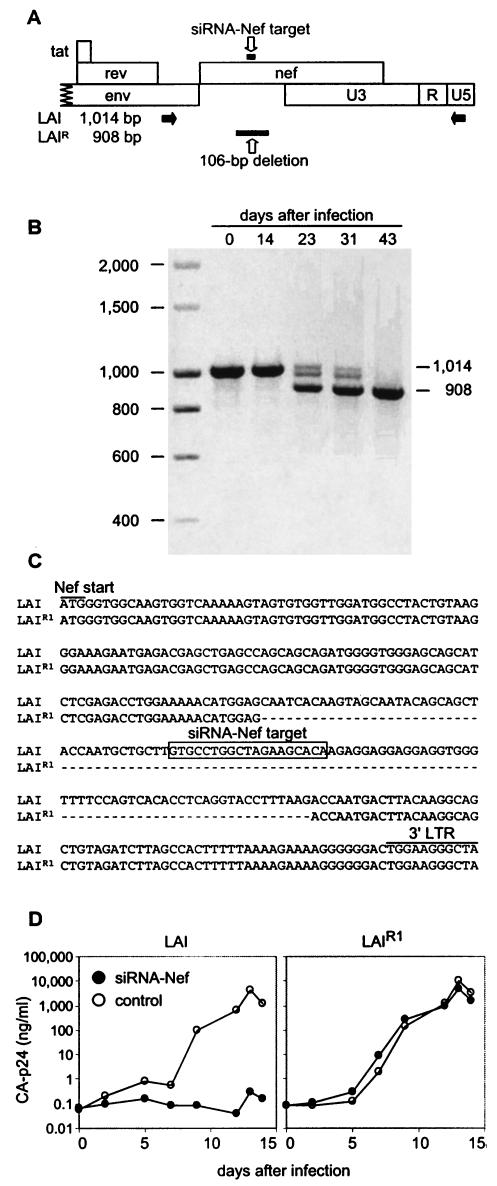
We cultured the siRNA-Nef-transduced cells for more than 8 months and frequently assayed HIVLAI and HIVrtTA replication. The cells maintained a constant level of resistance to HIVLAI replication, demonstrating the stable expression of the siRNA-Nef gene. The nearly complete resistance of these T cells to HIV-1 replication allowed the selection of HIV-1 escape mutants that are no longer sensitive to RNAi against the Nef target sequence. To select an escape virus, we massively infected the nonpermissive cells that express siRNA-Nef, and a relatively fast replicating variant was obtained after 23 days. The targeted Nef gene was PCR amplified (Fig. 3A) at each passage, and DNA gel analysis of the PCR product indicated the loss of viral sequences around day 23 (Fig. 3B). Sequence analysis revealed a deletion of 106 bp within the Nef gene that removes the siRNA-Nef target sequence (Fig. 3C). The deletion of the target sequence provides a simple explanation for the observed resistance phenotype. To verify this explanation, we used equal amounts of the evolved HIVLAI (LAIR1) and the wild-type virus (LAI) to infect control SupT1 cells and cells that express siRNA-Nef. Whereas LAI is selectively inhibited in the siRNA-Nef-expressing cells (Fig. 3D, left panel), no such effect is apparent for the resistant LAIR1 variant (Fig. 3D, right panel).
FIG. 3.
HIVLAI develops resistance against siRNA-Nef by deleting the Nef target sequence. SupT1 cells expressing siRNA-Nef were massively infected with HIVLAI to overcome the siRNA-mediated inhibition of replication. The virus was passaged repeatedly onto fresh SupT1-siRNA-Nef cells. Cells were initially infected with a high virus dose that could be reduced gradually. (A) Schematic of the 3′ end of the HIVLAI proviral genome. Indicated are the positions of the siRNA-Nef-targeted sequence, the PCR primers (black arrows), and the 106-bp deletion observed after prolonged virus culture. (B) At each passage, the proviral DNA present in infected cells was PCR amplified with primers that amplify the complete Nef gene as a 1,014-bp fragment. At 23 days after infection, a fast-replicating variant containing a 106-bp deletion in the Nef gene was observed. At 43 days after infection, this siRNA-Nef-resistant virus dominated the virus population. (C) Sequences of the Nef gene in the HIVLAI virus and in the evolved siRNA-Nef-resistant virus (LAIR1) with the 106-bp deletion encompassing the siRNA-Nef target sequence. (D) Infection of SupT1 control cells and siRNA-Nef-expressing cells with wild-type LAI (left panel) or the evolved LAIR1 variant harvested at day 43 (right panel). We used equal virus input levels (400 pg of CA-p24 in a 5-ml culture).
We selected six additional resistant HIV-1 variants in independent cultures of HIVLAI on siRNA-Nef-expressing cells (Fig. 4). In four of these escape variants, the siRNA-Nef target sequence was completely or partially deleted (Fig. 4, variants LAIR2, LAIR4, LAIR5, and LAIR7). These deletions did not affect genes other than the Nef gene and did not affect important regulatory elements such as the 3′ polypurine tract or long terminal repeat sequences. For two variants (LAIR3 and LAIR6), we observed nucleotide substitutions within the siRNA-Nef target sequence. These results show that resistance to RNAi can result from both the deletion and the mutation of the siRNA-targeted sequence. In one of the cultures (LAIR3), the RNAi-resistant virus contained one nucleotide substitution after 27 days of culture, and a second mutation was acquired after 62 days. These results suggest that one nucleotide mismatch between the siRNA and its target sequence provides only partial RNAi resistance.
FIG. 4.
RNAi-resistant HIV-1 variants. HIVLAI variants resistant to siRNA-Nef were selected in independent cultures and analyzed as described in the legend to Fig. 3. (A) PCR amplification of the Nef gene on the indicated day (LAI, input HIVLAI virus; R1, LAIR1 escape virus shown in Fig. 3 at day 43; R2 to R7, independent HIVLAI escape variants); (B) Nef target sequence in the evolved RNAi-resistant viruses. In LAIR5, nucleotides 179 to 241 of the Nef gene are deleted. In LAIR7, we observed the deletion of nucleotides 44 to 268 and a T269A substitution.
Antiviral therapy based on siRNA has been proposed previously (9) for the rapidly replicating and highly cytolytic poliovirus, and this study also reported an escape virus with a point mutation in the middle of the targeted sequence. This RNAi-resistant poliovirus variant was likely present in the initial virus population (9). In contrast, the HIV-1 experiments in the present study were initiated with a molecularly cloned viral genome. Furthermore, we observed diverse deletions and substitutions in the resistant viruses. Therefore, we can conclude that the selected RNAi-resistant HIV-1 variants emerged de novo. The combined analysis of siRNA escape viruses in both studies confirms that the antiviral effect is potent and sequence specific.
LAIR6 is the only RNAi-resistant HIV-1 variant with a silent codon change and thus produces the wild-type Nef protein. Nonsilent nucleotide substitutions in LAIR3 result in two amino acid changes (A56T and L58R). The LAIR1 and LAIR2 variants have a deletion that results in a frameshift and a large C-terminal truncation of the Nef protein. The in-frame deletions in LAIR4, LAIR5, and LAIR7 result in the absence of 2, 31, and 75 amino acids in the Nef protein, respectively. The complete inactivation of the accessory Nef gene has a relatively minor impact on the replication fitness of HIV-1 in vitro but significantly attenuates replication in vivo (8). It therefore seems less likely that HIV-1 will acquire resistance in vivo by the deletion of the Nef sequence. The virus may instead evolve resistance by accumulating silent point mutations in the Nef target sequence. Also, when targeting essential viral genes, the virus may become resistant as a result of silent mutations. The targeting of cellular genes could therefore be advantageous. Obvious cellular targets that have an immediate impact on HIV-1 replication are the CD4 receptor and the CCR5/CXCR4 coreceptors. As was recently demonstrated, siRNAs specific for these receptors do indeed inhibit HIV-1 replication (17, 19, 20). Although the suppression of CD4 and CXCR4 may be restricted by their normal role in the immune system and cell migration, respectively, CCR5 seems dispensable for normal life (16). Unfortunately, not all HIV strains require CCR5, and the inhibition of CCR5 may result in the selection of HIV-1 variants that use CXCR4 as a coreceptor. We therefore propose that HIV-1 should be targeted by multiple siRNAs against essential HIV-1 elements, either genes or regulatory sequences. Although the targeting of a single HIV-1 sequence can result in the strong inhibition of viral replication, it is likely followed by viral escape. Analogous to the current clinical use of combinations of antiviral drugs that target the reverse transcriptase and protease enzymes, antiviral approaches using RNAi should also be administered in a combined fashion to prevent viral escape.
Acknowledgments
We thank S. P. Heynen for the CA-p24 enzyme-linked immunosorbent assay.
This work was supported by grants from the Technology Foundation STW, the NWO-VICI program, and the Center for Biomedical Genetics.
REFERENCES
- 1.Arteaga, H. J., J. Hinkula, I. Dijk-Hard, M. S. Dilber, B. Wahren, B. Christensson, A. J. Mohamed, and C. I. Edvard Smith. 2003. Choosing CCR5 or Rev siRNA in HIV-1. Nat. Biotechnol. 21:230-231. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout, B., M. Ooms, N. Beerens, H. Huthoff, E. Southern, and K. Verhoef. 2002. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem. 277:19967-19975. [DOI] [PubMed] [Google Scholar]
- 3.Bohula, E. A., A. J. Salisbury, M. Sohail, M. P. Playford, J. Riedemann, E. M. Southern, and V. M. Macaulay. 2003. The efficacy of small interfering RNAs targeted to the type 1 insulin-like growth factor receptor (IGF1R) is influenced by secondary structure in the IGF1R transcript. J. Biol. Chem. 278:15991-15997. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cells 2:243-247. [DOI] [PubMed] [Google Scholar]
- 6.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das, A. T., B. Klaver, B. I. F. Klasens, J. L. B. van Wamel, and B. Berkhout. 1997. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 71:2346-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, V. A. Lawson, S. Crowe, A. Maerz, S. Sonza, J. Learmont, J. S. Sullivan, A. Cunningham, D. Dwyer, D. Dowton, and J. Mills. 1995. Genomic structure of an attenuated quasi species of HIV-1 from blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 9.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 10.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joost Haasnoot, P. C., D. Cupac, and B. Berkhout. 2003. Inhibition of virus replication by RNA interference. J. Biomed. Sci. 607-616. 10: [DOI] [PubMed]
- 12.Kretschmer-Kazemi, F. R., and G. Sczakiel. 2003. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 31:4417-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuiken, C., B. Foley, E. Freed, B. Hahn, P. A. Marx, F. McCutchan, J. W. Mellors, S. Wolinsky, and B. Korber. 2002. HIV sequence compendium. LA-UR no. 03-3564. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 14.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 15.Li, M. J., G. Bauer, A. Michienzi, J. K. Yee, N. S. Lee, J. Kim, S. Li, D. Castanotto, J. Zaia, and J. J. Rossi. 2003. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol. Ther. 8:196-206. [DOI] [PubMed] [Google Scholar]
- 16.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 17.Martinez, M. A., B. Clotet, and J. A. Este. 2002. RNA interference of HIV replication. Trends Immunol. 23:559-561. [DOI] [PubMed] [Google Scholar]
- 18.Marzio, G., K. Verhoef, M. Vink, and B. Berkhout. 2001. In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc. Natl. Acad. Sci. USA 98:6342-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 19a.Ooms, M., K. Verhoef, E. Southern, H. Huthoff, and B. Berkhout. Probing alternative foldings of the HIV-1 leader RNA by antisense oligonucleotide scanning arrays. Nucleic Acids Res.in press. [DOI] [PMC free article] [PubMed]
- 20.Qin, X. F., D. S. An, I. S. Y. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhoef, K., G. Marzio, W. Hillen, H. Bujard, and B. Berkhout. 2001. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J. Virol. 75:979-987. [DOI] [PMC free article] [PubMed] [Google Scholar]