Abstract
Background
Soil salinity is a major abiotic stress that limits agriculture productivity worldwide. Salicornia europaea is well adapted to extreme saline environments with more than 1,000 mM NaCl in the soil, so it could serve as an important model species for studying halophilic mechanisms in euhalophytes. To obtain insights into the molecular basis of salt tolerance, we present here the first extensive transcriptome analysis of this species using the Illumina HiSeq™ 2000.
Principal Findings
A total of 41 and 39 million clean reads from the salt-treated (Se200S) and salt-free (SeCKS) tissues of S. europaea shoots were obtained, and de novo assembly produced 97,865 and 101,751 unigenes, respectively. Upon further assembly with EST data from both Se200S and SeCKS, 109,712 high-quality non-redundant unigenes were generated with a mean unigene size of 639 bp. Additionally, a total of 3,979 differentially expressed genes (DEGs) were detected between the Se200S and SeCKS libraries, with 348 unigenes solely expressed in Se200S and 460 unigenes solely expressed in SeCKS. Furthermore, we identified a large number of genes that are involved in ion homeostasis and osmotic adjustment, including cation transporters and proteins for the synthesis of low-molecular compounds. All unigenes were functionally annotated within the COG, GO and KEGG pathways, and 10 genes were validated by qRT-PCR.
Conclusion
Our data contains the extensive sequencing and gene-annotation analysis of S. europaea. This genetic knowledge will be very useful for future studies on the molecular adaptation to abiotic stress in euhalophytes and will facilitate the genetic manipulation of other economically important crops.
Introduction
Along with environmental deterioration and anthropogenic influence, soil salinity is one of the major abiotic stresses in plant and crop systems, causing yield losses of primary crops worldwide [1], [2]. Much efforts have been made to enhance salt tolerance of economically important plants for future agricultural demand [3]. Many salt stress-induced genes are part of a complicated cascade of molecular networks and are thought to play important roles in responding to environment stress [4], [5]. These genes are involved in many cellular and physiological processes such as signal perception and transduction [6], [7], photosynthesis, energy metabolism [8], membrane trafficking and protein biosynthesis, folding and decay [9], [10]. The isolation and identification of these functional genes, using the tools of molecular biology, is necessary for understanding the mechanisms of salt tolerance in plants [11]–[13]. Several essential genes involved in different molecular processes are reportedly involved in the plant response to salt stress, including AtHKT1;1 [14], [15], PutHKT2;1 [16], AtSOS1 [17], [18], AtNHX1 [19], Ca2+/calmodulin-dependent protein phosphatase [20], flavoprotein AtHAL3 [21], plasma membrane (PM) H+-ATPase and transcription factor Alfin1 [22], [23]. However, only a few salt-tolerance genes have been previously identified, and most of the studies on plant salt tolerance have focused on model plants such as Arabidopsis, rice and tobacco. Research on salt-tolerant genes in halophytes is scarce, even though some euhalophytes have the specific ability to cope with high levels of salinity using unique mechanisms [24]–[30].
Salicornia europaea L., one of the most salt-accumulating euhalophytes, can withstand concentrations of more than 1,000 mM NaCl in the soil and is widely distributed in coastal and inland salt marshes around the world [24], [31]. It is an important species for soil desalination because it hyper-accumulates salt in weakly saline soil, with salt reaching 50% of the dry weight of shoots. In contrast to glycophytes, which are negatively affected by increasing salinity beyond a threshold of approximately 50 mM, S. europaea survives in a high saline habitat. The optimal salinity for maximum growth is in the range of 100 to 200 mM, and its growth is significantly hindered if the soil salinity is outside this range [31]. An ion transport assay performed in the presence of various salts has revealed that Na+, Cl−, K+ and Ca2+ were absorbed and distributed into the tissue of S. europaea. The root K+ content reached 90 meq l−1, while in the soil, it never exceeded 5 meq l−1. The Na+, Cl− and Ca2+ concentrations were generally higher in the shoots than in the roots [32]. Previous studies have determined that the fresh and dry weight of the plant reached the highest values at optimal salinity, and superoxide dismutase (SOD), catalase (CAT) and guaiacol peroxidase (GPX) activities also increased at higher salinities [25], [33]. Recently, SePSY (phytoene synthase gene) [34], SeNHX1 (Vacuolar Na+/H+ antiporters) [35], [36], SeLCY (beta-lycopene cyclase gene) [37] and SeCMO (choline monooxygenase) [38] were isolated from S. europaea, and it was shown that the expression of these genes was correlated to the salt concentration. However, information concerning gene expression and regulation as they relate to salt adaptation in S. europaea is scarce [39], [40]. Previously, it has been laborious and time-consuming to identify and characterise the genes for salt adaptation or tolerance in S. europaea because transcriptomic and genomic data for S. europaea were unavailable in public databases. For example, no genome and only fourteen EST sequences were available at the time of publication. In halophytes, large numbers of cDNA fragments from several typical species were isolated from cDNA and SSH libraries [41]–[43], but those sequences could not provide global transcriptome information for the species. Only limited studies of the transcriptome of salt-treated halophytes have been reported thus far. This lack of information is a significant obstacle to our understanding of the molecular mechanisms for salt adaptation in halophyte species and impedes the exploitation of halophytes for the restoration of saline soil.
To obtain novel insights into the molecular basis of salt adaptation in S. europaea, two-group de novo assembly data were generated from Illumina sequencing of shoot samples for salt-treated or untreated plants. Transcriptome changes resulting from salt treatment were also compared. Our objective was to uncover and characterise a core set of salt stress-related transcripts.
Materials and Methods
Plant material and growth conditions
S. europaea seeds were collected from saline soil in Fukang, Northwestern China. Seeds were sown in plastic pots (12×12 cm) filled with sands irrigated with tap water. After germination, seedlings were maintained in a greenhouse with a day/night thermoperiod of 25/20°C, a photoperiod of 16 h, relative humidity of 50±10% and weekly irrigation with half-strength hoagland nutrient solution. Two months later, the plants were irrigated with NaCl solutions with concentrations of 0, 10, 200, 400 or 800 mM NaCl. The shoots were harvested at 72 h after salt treatment and frozen immediately in liquid nitrogen for the extraction of total RNA.
cDNA library preparation and Solexa sequencing for transcriptome analysis
Total RNA was extracted from S. europaea shoots treated with 0 mM (SeCKS) and 200 mM (Se200S) NaCl using the QIAGEN RNeasy Plant Mini kit (Qiagen) according to the manufacturer's protocol. The RNA samples were used to construct two tissue-specific cDNA libraries for RNA sequencing and transcriptome analysis. According to the manufacturer Illumina's instructions, poly (A)+ RNA was isolated from 20 μg total RNA using oligo (dT) magnetic beads. Fragmentation buffer was added to break the mRNA into short fragments. Using these short fragments for templates, random hexamer-primers were used to synthesise first-strand cDNA. Second-strand cDNA was synthesised using buffer, dNTPs, Rnase H (Invitrogen) and DNA polymerase I (New England Biolabs). The resulting short fragments were purified using the QiaQuick PCR extraction kit and then resuspended in EB buffer for end repair and poly(A) addition. The short fragments were then joined to sequencing adapters. Following agarose gel electrophoresis, suitable fragments were selected for PCR amplification. The libraries could then be sequenced using the Illumina HiSeq™ 2000.
De novo assembly of sequencing reads and sequence clustering
Following cDNA library sequencing, high-quality clean reads were picked out from the raw reads of each library following removal of reads containing adaptor sequences, reads with an N (unknown bases in a read) percentage higher than 5% and low-quality reads (>50%of the bases with a quality score Q-value ≤5) using perl scripts. Transcriptome de novo assembly was carried out with the short-reads assembly programme Trinity [44]. We first combined reads with a certain length of overlap to form contigs with a zero N value (no unknown bases). Then, the reads were mapped back to contigs with paired-end reads. This approach detected contigs from the same transcript as well as the distances between these contigs. Next, Trinity connected the contigs using N to represent unknown sequences between each contig pair to form scaffolds. Finally, unigenes were generated with zero N values in the sequence that could not be extended on either end. In the final step, BLASTX alignment (e value <0.00001) between the recovered unigenes and protein databases such as nr, Swiss-Prot, KEGG (Kyoto Encyclopedia of Genes and Genomes) and COG (Clusters of Orthologous Groups) was performed, and the best aligning results were used to determine the sequence direction of the unigenes. If the results from different databases conflicted with one another, a priority order of nr, Swiss-Prot, KEGG and COG was followed when deciding the unigene sequence direction. When a unigene was unaligned in all of the databases, the programme ESTScan was used to decide its sequence direction [45]. For unigenes with verified sequence directions, we provided their sequences in the 5′ to 3′ orientation; for those without direction, we provided their sequences as determined by the assembly software. All the obtained data were submitted to DDBJ/EMBL/GenBank database.
Unigene functional annotation
The individual unigenes were analysed by searching the protein databases nr, Swiss-Prot, KEGG and COG (e-value<0.00001) with the BLASTX algorithm (http://www.ncbi.nlm.nih.gov/) to retrieve functionally annotated proteins with the highest sequence similarity to the unigenes on our list. The COG database was used to predict and to classify possible functions of our unigenes. The KEGG pathways database was used for inner-cell metabolic pathways annotation and to determine the potential complicated biological behaviours of the genes on our list. The Blast2GO programme was used to understand the GO functional annotations derived from the molecular function, cellular location and biological processes of all of our unigenes [46].
Differential gene expression ofS. europaea between salt-free and salt-treated shoots
To determine the DEGs (differentially expressed genes) between SeCKS and Se200S, gene expression level analysis was performed using the RPKM (Reads per kb per million reads) method and the formula RPKM = 106C/10−3NL, where C is the number of reads that uniquely align to a unigene, N is the total number of reads that uniquely align to all unigenes, and L is the base number in the Coding sequence (CDS) of a unigene [47]. The RPKM method eliminates the influence of different gene lengths and sequencing levels on the calculation of gene expression. Therefore, calculated gene expression can be directly used for comparing the differences in gene expression between samples. We compared the transcriptome profiles of SeCKS and Se200S for S. europaea to detect DEGs between both samples using the statistical method FDR (False discovery rate) and the ratio of RPKMs for the two samples (FDR ≤0.001 and |log2 ratio| ≥1). DEGs were then used to carry out GO functional and KEGG Pathway analysis.
RT-PCR and qRT-PCR used to validate and analyse the unigenes
RNA was extracted from the shoots and roots of 0, 10, 200, 400, and 800 mM NaCl-treated plants. cDNA was synthesised from 2 μg of RNase-free, DNaseI-treated total RNA with 500 ng of 18 mer oligo-dT primers and M-MLV reverse transcriptase (Promega). The primers for RT-PCR and quantitative RT-PCR were designed using the sequences determined for the differentially expressed unigenes using the programme Primer 5.0. The α-tubulin gene was used as an endogenous control. Sequences of the primers are given in Additional File S1. Quantitative RT-PCR was performed using the CFX96 real-time PCR detection system (Bio-Rad), a 25-µl reaction system and the SYBR Premix Ex Taq Kit (TaKaRa Corp. Beijing, China) according to the manufacturer's protocol. The following cycling programme was used: 95°C for 60 sec, followed by 40 cycles of 10 sec at 95°C, 30 sec at 55°C and 30 sec at 72°C. All products were subjected to melting curve analysis between 55°C and 95°C to determine the specificity of the PCR reaction. The relative quantitative method (ΔΔCt) was used to evaluate the quantitative variation.
Results and Discussion
A number of researchers have proven that transcriptome sequencing of RNA/cDNA provides a rapid and cost-effective way to generate whole-transcriptome sequences from many types of biological samples and to identify differentially expressed genes. In particular, transcriptomes from a number of plant species, including Arabidopsis thaliana, rice and wheat, were investigated for various purposes [48]–[50]. To the best of our knowledge, although S. europaea is one of most salt-tolerant halophytes, only a limited numbers of genes from this organism have been cloned and characterised [24], [51]. Transcriptome sequencing of S. europaea is an important step for gene identification and the elucidation of the molecular mechanisms underlying salt adaptation and hyper-accumulation in S. europaea.
De novo assembly and quantitative assessment of Illumina ESTs
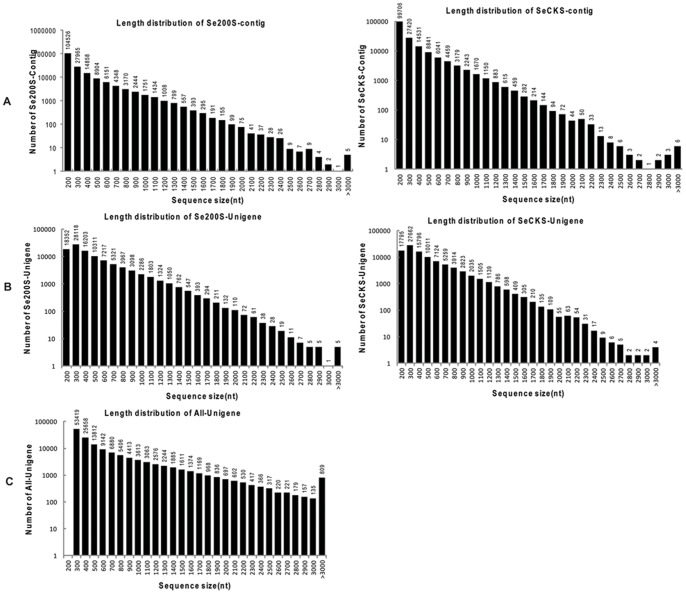
As a first step to gain insight into transcriptional regulation of S. europaea adaptation to saline habitats, we used the RNA-Seq technique to generate two tissue-specific whole-transcriptome profiles of S. europaea shoots treated with either 0 mM or 200 mM NaCl. After cDNA enrichment and Illumina sequencing, low quality and adapter sequences were eliminated. A total of 41 million and 39 million clean reads were generated from the SeCKS and Se200S shoot cDNA libraries with 97.21 and 97.24% Q20 percentages, respectively. The total length of the reads was 7.21 gigabases (GB). The reads from two libraries were clustered into 172,174 and 179,292 contigs by the programme Trinity. Contig sizes ranged from 100 bp to 3,000 bp, and the average contig sizes were 267 bp for SeCKS and 271 bp for Se200S (Figure 1A). To reduce sequence redundancy, all contigs were further assembled into 97,865 and 101,751 unigenes in the SeCKS and Se200S cDNA libraries, with 424 bp and 438 bp average lengths, respectively, and the size distribution ranged from 200 bp to 3,000 bp (Figure 1B). Unigenes from SeCKS and Se200S were deposited at DDBJ/EMBL/GenBank as the TSA accession GAHZ00000000 and GAIA00000000 respectively. To acquire the longest non-redundant sequences possible, unigenes from SeCKS and Se200S were further assembled, and 109,712 non-redundant unigenes, termed All-Unigene, were generated. The All-Unigene size distribution demonstrated that most sequences (64,411; 58.71%) were less than 500 bp in length; 23.66% (25,961) were between 500 and 1,000 bp; 16.89% (18,532) were between 1,000 and 3,000 bp; and 0.74% (808) were more than 3,000 bp (Figure 1C, table 1). The unigene size distribution revealed that shorter fragments were reduced and longer fragments were generated as a result of further assembly. All of these unigene sequences can be accessed in Additional File S1: Additional File S2.z01, Additional File S2.z02 and Additional File S2.
Figure 1. Size distribution of the contigs and unigenes generated byde novo assembly.
(A) Size distribution of contigs. The x-axis represents contig size, and the y-axis represents numbers of contigs of a certain length. (B) Size distribution of unigenes. The x-axis represents unigene size, and the y-axis represents the number of unigenes with a certain length. (C) Size distribution of All-Unigenes. The x-axis represents All-Unigenes size, and the y-axis represents the number of All-Unigenes with a certain length.
Table 1. Summary of sequencing and assembly results.
| sample | number/percent | 100–200nt | 200–300nt | 300–400nt | 400–500nt | > = 500nt | N50 | Mean | No. | Length (nt) |
| Se200S-Contig | number | 104,526 | 27,965 | 14,858 | 8,904 | 23,039 | 372 | 271 | 179,292 | 48,616,772 |
| percent | 58.30% | 15.60% | 8.29% | 4.97% | 12.85% | |||||
| SeCKS-Contig | number | 99,706 | 27,420 | 14,531 | 8,841 | 21,676 | 363 | 267 | 172,174 | 45,962,606 |
| percent | 57.91% | 15.93% | 8.44% | 5.13% | 12.59% | |||||
| Se200S-Unigene | number | 18,352 | 28,118 | 16,203 | 10,311 | 28,767 | 547 | 438 | 101,751 | 44,551,677 |
| percent | 18.04% | 27.63% | 15.92% | 10.13% | 28.27% | |||||
| SeCKS-Unigene | number | 17,795 | 27,662 | 15,796 | 10,011 | 26,601 | 523 | 424 | 97,865 | 41,490,134 |
| percent | 18.18% | 28.27% | 16.14% | 10.23% | 27.18% | |||||
| All-Unigene | number | 0 | 34,417 | 18,808 | 11,186 | 45, 301 | 780 | 636 | 109,712 | 69,795,900 |
| percent | 0 | 31.37% | 17.14% | 10.20% | 41.29% |
N50: 50% of the assembled bases were incorporated into sequence with N50 length or longer.
Mean: Mean length of assembled sequences.
To confirm the sequencing quality of our transcriptome data and sequence assembly, 18 unigenes were randomly selected for RT-PCR analysis. In this analysis, 13 out of 18 primer pairs amplified a product of the expected length (Additional File S3). The unigene sequences were then confirmed by sequencing the PCR products.
Functional annotation and analysis
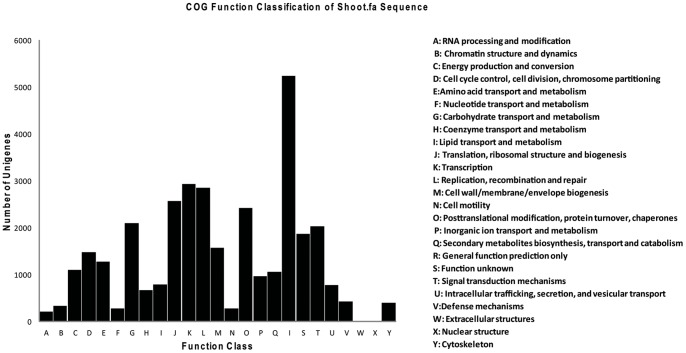
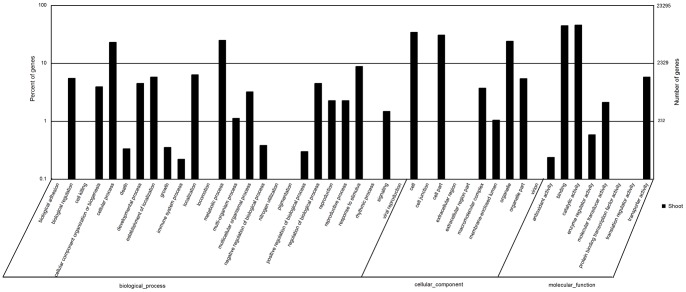
All-Unigene sequences of S. europaea were aligned on the basis of similarities to the NCBI non-redundant protein databases nr, Swiss-Prot, KEGG and COG (e-value <0.00001). These alignments retrieved proteins with the highest sequence similarity to the S. europaea unigenes along with their functional annotations. Among the 109,712 high-quality All-Unigene sequences from S. europaea, 50,767 (46.27%) had significant BLAST matches in the NCBI non-redundant protein database and 36,156 (32.96%) aligned to Swiss-prot protein sequence database. Of the group, 20,473 (18.66%) aligned with the KEGG database and were assigned to 120 KEGG pathways (Additional File S4). Most of these unigenes were sorted to metabolic pathways (6,303 unigenes), biosynthesis pathways for secondary metabolites (3,507 unigenes) and pathways involved in plant-pathogen interactions (1,665 unigenes) (Additional File S5). Of All-Unigenes, 33,636 (16.2%) unigenes aligned to the COG database and were classified into 25 functional categories (Figure 2). After Blast2GO analysis, 35,268 (32.15%), 43,713 (39.84%) and 21,334 (19.45%) unigenes were classified into 44 terms from three ontologies involved in biological processes, cellular components and molecular function, respectively (Figure 3).
Figure 2. Histogram presentation of COGs classification.
A total of 33,636 (16.2%) unigenes out of the All-Unigenes set were aligned to the COG database and classified into 25 functional-categories.
Figure 3. Histogram presentation of GO classification.
A total of 35,268 (32.15%), 43,713 (39.84%) and 21,334 (19.45%) unigenes of S. europaea were classified into 44 terms from three ontologies involved in biological processes, cellular components and molecular function, respectively. The right y axis indicates the number of genes in a category. The left y axis indicates the percentage of a specific category of genes in the main category.
Differential genes expression ofS. europaea between salt-free and salt-treated shoots
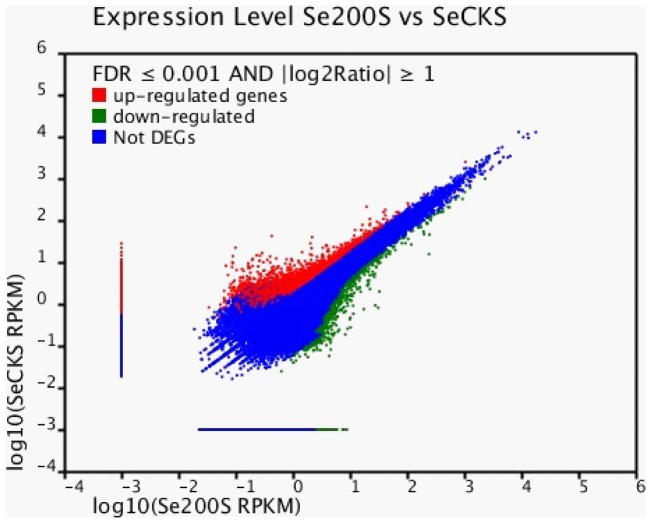
To identify genes with differential expression under different salt conditions, we compared the transcriptome profiles of SeCKS and Se200S. The transcriptome profile of SeCKS was defined as control to get the up- or down-regulated unigenes in Se200S. The results showed that 3,979 out of 109,712 total unigenes were differentially expressed in shoots of S. europaea under salt-free and salt-treated conditions, including 2,275 unigenes up-regulated genes (FDR<0.001) and 1,704 down-regulated genes (FDR<0.001). In addition, there were 348 of 2,275 unigenes detected only in Se200S, while 460 of 1,704 unigenes were only expressed in SeCKS (Additional File S6, Figure 4).
Figure 4. Se200S-vs-SeCKS differentially expressed genes.
DEGs were filtered using FDR ≤0.001 and Log2Ratio ≤1 as a threshold. Red spots represent upregulated genes, and green spots indicate downregulated genes. Blue represents those genes that did not show obvious changes between the Se200S and SeCKS samples.
Go annotation and statistical analyses demonstrated that differentially expressed genes were classified into three GO ontologies and 35 terms. Of these, 1,469 were in the category of biological processes, 1,972 were in the category of cellular components and 880 were in the category of molecular function (Additional File S7). In the molecular function category, 880 DEGs mapped to seven terms including antioxidant activity, binding, catalytic activity, enzyme regulator activity, molecular transducer activity, protein binding transcription factor activity and transporter activity. For further clarification of gene function, 797 DEGs were enriched in 199 terms, with a p-value as good as or better than 1 (Additional File S8). The overwhelming majority of the genes with differential expression between salt-free and salt-treated conditions were related to transmembrane transporter activity (49 genes, or 6.1%), iron ion binding (36 genes, or 4.5%), transition metal ion binding (106 genes, or 13.3%), ion binding (173, or 21.7%), cation binding (166 genes, or 20.8%), metal ion transmembrane transporter activity (9 genes, or 1.1%) and active transmembrane transporter activity (36 genes, or 4.5%). The differentially expressed transporter and ion binding genes are presumed to be important for maintaining and re-establishing the ion homeostasis of the cytoplasm. ATPase-related genes were also differentially expressed with a high frequency and were included in five terms as follows: ATPase activity (36 genes, or 4.5%), ATPase activity, coupled (33 genes, or 4.1%), ATPase activity, coupled to transmembrane movement of substances (17 genes, or 2.1%), ATPase activity, coupled to movement of substances (17 genes, or 2.1%), and ATPase activity, coupled to transmembrane movement of ions and phosphorylative mechanisms (3 genes, or 0.4%). One unigene (Unigene30025_All) classified into the symporter activity term was upregulated dramatically in Se200S and annotated as a Na+ symporter family member protein from Populus trichocarpa. Three differentially expressed unigenes (Unigene21877_All, Unigene22273_All and Unigene39816_All) were categorised in the antiporter activity term and were identified as a Na+/H+ antiporter, TT12-2 MATE transporter and a Na+/H+ exchanger 4, respectively. The discovery of these differentially expressed genes at the whole transcriptome level is expected to clarify the molecular mechanisms of ion absorption and transmembrane transport by S. europaea.
Living organisms are complex systems that are flexible and adaptive to their surroundings. At the molecular level, different genes generally cooperate with each other to exercise their biological functions in intricate networks of molecular reactions. In our study, KEGG pathway analysis helped us to understand the biological function of these (DEGs) under salt-free and salt-treated conditions. In this analysis, 1,094 DEGs mapped to 101 pathways (FDR ≤0.05) with most mapping to metabolic pathways (322 DEGs), biosynthesis pathways for secondary metabolites (186 DEGs), pathways involved in plant-pathogen interactions (59 DEGs), protein processing pathways in the endoplasmic reticulum (40 DEGs), starch and sucrose metabolism pathways (30 DEGs) and ABC transporter pathways (6 DEGs) (Additional File S9).
Quantitative real-time PCR validation
The salt tolerance mechanisms of S. europaea are not yet clear. However, it is possible that the survival of this plant depends on the accumulation of high levels of ions in the tissue for the maintenance of osmotic balance, with the accumulation of Na+ being higher than other ions under salt treatment. K+ and Ca2+ uptakes were also promoted by moderate concentrations of NaCl but were reduced at higher salt concentrations [52]. In this research, we isolated many transporter and ion binding genes that were presumed to be effector molecules for coping with salt stress.
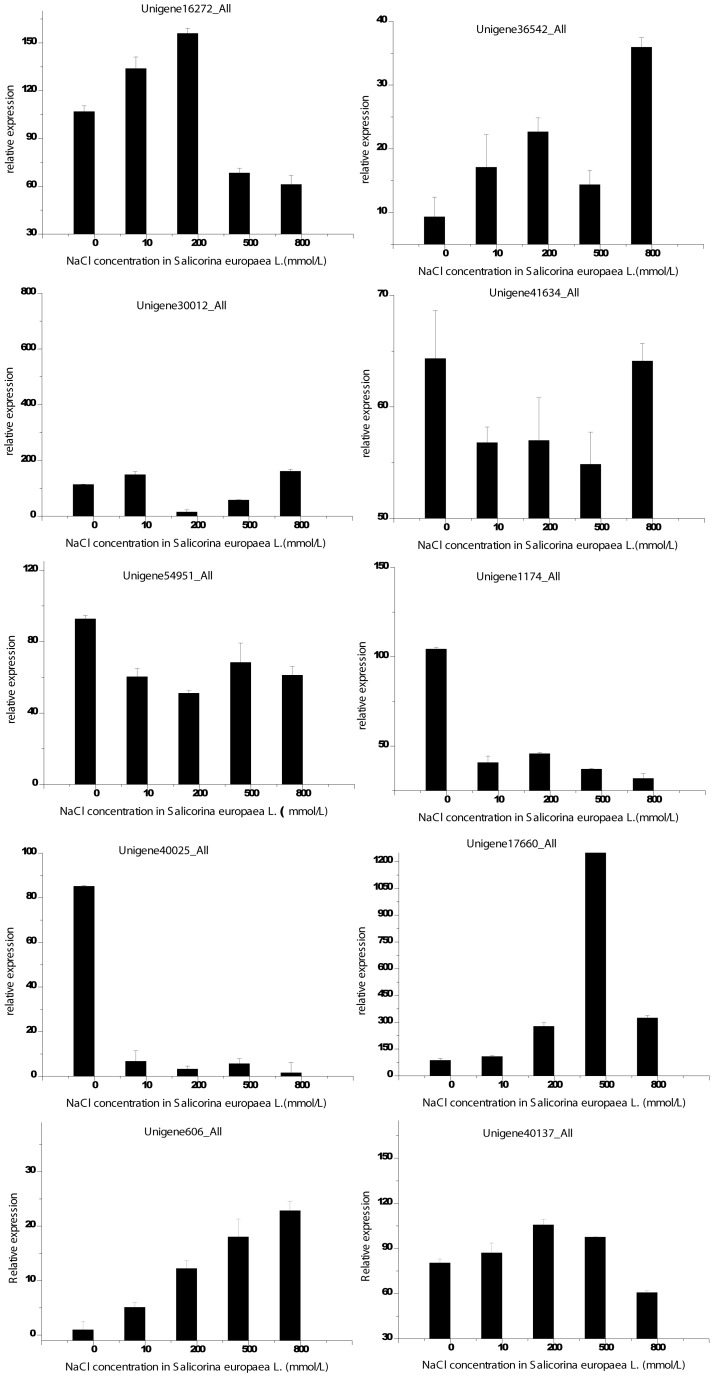
Five cDNAs were homologous to HKT genes from Populus trichocarpa and Suaeda salsa, as determined by protein database searches. HKT genes are reported to be involved in Na+ homeostasis under salt stress in several plant species [53]. The sequence analysis of five cDNAs showed reduced similarity to each other at the nucleotide level, and we have speculated that these HKT genes are new members of the HKT family and perform distinct functions for salt adaptation in S. europaea. Unigene16272_All is an HKT gene homologous to the hkt1-like gene of Populus trichocarpa, a sodium transporter. qPCR results revealed that the expression of this gene was upregulated with increasing salt concentration, whereas it decreased beyond 200 mM NaCl (Figure 5). Different members of the HKT family exhibit different ion selectivity and transport mechanisms [54], [55]. The role of the HKT gene in S. europaea is unclear, and further study is being conducted in our laboratory to clarify the function of the different HKT members of S. europaea in a saline habitat.
Figure 5. Validation of Transcriptome Profiling expression with QRT-PCR.
The transcript abundance from Transcriptome Profiling data is shown above each gene. Relative transcript levels are calculated by real-time PCR using alpha-tubulin as the standard. Three biological replicates were performed, and the data shown are typical results.
Vacuolar compartmentation of Na+ is an important mechanism for salt adaptation in plants. This process is undertaken by vacuolar membrane Na+/H+ exchangers (NHX) and is driven by the intracellular electrochemical gradient of protons [56], [57]. At the cellular level, Na+ accumulation in vacuoles reduces the toxicity of Na+ ions in the cytoplasm and in sensitive organelles and also lowers the osmotic potential of the vacuoles to preserve cellular turgor pressure and cell expansion under saline stress. Therefore, the translocation and storage of Na+ inside vacuoles of the shoot are thought to be important mechanisms for sustained growth under saline conditions by halophytes such as S. europaea [58]. Here, we identified four NHX genes that are homologous to genes in Arabidopsis thaliana, Mesembryanthemum crystallinum, Chrysanthemum morifolium and Oryza sativa. As determined by qRT-PCR analysis, Unigene36542_All (Na+/H+ exchanger 4) was upregulated gradually with increasing NaCl concentration, with the exception of 500 mM NaCl (Figure 5). In Arabidopsis, AtNHX4 localises to vacuoles, transports Na+ out of the vacuolar lumen and into the cytosol and functions in Na+ homeostasis in plant cells [59], [60]. The functions of NHX4 in euhalophytes are still unknown, however, and merit further exploration. We have suggested that Na+/H+ exchanger 4 plays a crucial role in the maintenance of the normal growth of S. europaea under saline conditions by sequestering Na+ into vacuoles.
Plants can cope with osmotic stress by synthesising low molecular weight hydrophilic proteins [58] such as osmotin. In S. europaea, Unigene30012_All is homologous to an osmotin protein belonging to a member of the PR5 group, and it was differentially expressed in salt-treated and salt-free shoots (Figure 5). Previous studies determined that osmotin up-regulation and overexpression may enhance plant tolerance to NaCl stress [61], [62]. In contrast to glycophytes, the osmotin gene in S. europaea was expressed at its lowest level at the highest Na+ concentration (200 mM) and, remarkably, increased in expression in response to both low and high Na+ treatments below 200 mM (Figure 5). This expression helped to increase cellular turgor pressure at lower Na+ levels and improved osmoregulation of the cytoplasm at higher Na+ levels. Proline and other amino acids are ubiquitous osmoregulation compounds in plants. The synthesis and transport of these amino acids promote salt tolerance in most plants [63]–[65]. The genes proline transporter (ProT, Unigene41634_All) and amino acid permease (AAP, Unigene54951_All) were identified as homologous to genes in Populus trichocarpa. Similar to osmotin, the ProT gene was upregulated in the absence of salt and at the highest salt concentration but remained low in 10–500 mM NaCl. The AAP gene was highly transcribed in salt-free conditions (Figure 5). Previous studies have reported that ProT accumulation is increased under salt stress in most plants [64], [66], [67]. This finding suggests that a salt-free environment is deleterious for halophytes and that these plants will synthesise and transport various osmoprotectants to maintain cell turgor pressure under osmotic stress [68], [69]. In addition, the genes vinorine synthase (Unigene1174_All) and salt-induced protein (Unigene40025_All) were dramatically upregulated under salt-free conditions (Figure 5). These results suggest that S. europaea is likely to synthesise, transport and accumulate low-molecular weight organic compounds such as osmotin, proline, other amino acids, simple sugars, disaccharides, alkaloid and stress protein in the cytosol and the organelles to improve cellular turgor pressure and cell expansion in the shoots under salt-free conditions.
On the other hand, previous studies showed that NaCl treatments suitable for growth, such as 200 mM NaCl, caused a significant increase in plant biomass in some halophytes, while salt-free and high-salt conditions could decrease plant height and biomass [40], [74]. Plant growth and development are regulated by Gibberellin (GAs), which is modulated by the various genes involved in GAs biosynthesis and deactivation. In particular, GAs biosynthesis is tightly controlled by two gene families that catalyse bioactive GAs formation, the GA 20-oxidases (GA20ox) and the GA 3-oxidases (GA3ox). The GA 2-oxidases (GA2ox), on the other hand, catalyse the deactivation of bioactive GAs. In Arabidopsis, the cellular concentration of bioactive GAs was reduced via an increase in GA2ox7 transcript levels [72], resulting in an accumulation of DELLA, an inhibitor of plant growth [72], [73]. Based on transcriptome sequence analysis of S. europaea, we found that several GAs genes were regulated in various ways by 200 mM NaCl. Unigene21299_All and Unigene5033_All are homologues of gibberellin 3-oxidase and gibberellin 20-oxidase from Populus trichocarpa, and GA20ox and GA3ox were detected at a higher level than GA2ox (Unigene19515_All) in plants treated with 200 mM salt. In addition, there are two DELLA domain GRAS family transcription factors and DELLA protein GAI that were down-regulated in plants treated with 200 mM salt, albeit at a higher level than in salt-free plants. We propose a model whereby GAs are upregulated in halophytes under appropriate salt concentrations (200 mM) for normal growth of S. europaea, accompanied by negative regulation of the growth-repressing DELLA proteins.
Plants protect cells and subcellular systems from salt damage caused by ROS using antioxidant enzymes such as superoxide dismutase (SOD), catalase (CAT), peroxidase (POX) and glutathione reductase [70]. In S. europaea, Unigene17660_All is a homologue of a peroxidase from Spinacia oleracea that was gradually upregulated after NaCl treatment but decreased in concentration with 800 mM NaCl treatment (Figure 5). According to the results of a previous study, peroxidase activity increases to protect plants against stress [71]. In this study, we identified many differentially expressed genes. However, many have no homology to database sequences and no known function. These proteins may play a critical role in S. europaea adaption to saline conditions. To examine the validity of these genes, two genes (Unigene606_All and Unigene40137_All) were selected and qRT-PCR was performed. The analysis showed that the expression of each gene was influenced significantly by different NaCl solutions (Figure 5).
Conclusions
Totals of 41 million and 39 million clean reads from the tissues Se200S and SeCKS from S. europaea shoots were obtained using the Illumina HiSeq™ 2000. The assembled sequences represent a substantial part of the transcriptome of this plant. Transcriptome comparisons identified the DEGs that play significant roles in response to salt-free and salt-treated conditions. These results will facilitate the discovery of specific stress-resistant genes in S. europaea, aid the analysis of expression profiles of salt-tolerance-related genes, prompt studies on the molecular mechanisms of salt resistance, and lead to the development of novel plant cultivars through genetic engineering. More importantly, this study creates a new method for large-scale identification of resistance genes from native wild plants and for the conservation of germplasm resources from rare and endangered plants.
Supporting Information
Primers used for experimental validation and gene expression profile analysis.
(DOCX)
Contains: Additional File S2.z01– All-Unigene sequences of S. europaea shoots. Sequences with no gap and with a length longer than 200 bp were selected from the assembly results. Additional File S2.z02– All-Unigene sequences of S. europaea shoots. Sequences with no gap and with a length longer than 200 bp were selected from the assembly results. Additional File S2– All-Unigene sequences of S. europaea shoots. Sequences with no gap and with a length longer than 200 bp were selected from the assembly results.
(ZIP)
Primers and amplification results of 18 unigenes used to confirm the sequencing quality. The table shows sequences and annealing temperatures (Tm) and production of primers.
(XLS)
Functional annotation of All-Unigenes, including GO, COG, and KEGG analysis. All-Unigene sequences were searched against protein databases (Nr, SwissProt, KEGG, COG, and GO) using BLASTX (E-value ≤10–5).
(ZIP)
Summary of All-Unigenes enriched in KEGG pathways. Pathway ID and gene number are given in the table. The q-value for all of these pathways was less than or equal to 0.05.
(XLSX)
Summary and functional annotation of identified DEGs. Unigenes with an absolute value of |log2Ratio| ≥1 and FDR ≤0.001 were identified as DEGs. GO and KEGG analyses of DEGs were based on a cutoff E-value of less than or equal to 10–5.
(XLS)
GO categories of DEGs between Salt-Free and Salt-Treated Shoots of S. europaea. DEGs were divided into three major categories: molecular function, cellular component and biological process. The gene numbers and gene ID are listed in this file.
(XLS)
Summary of DEGs GO Molecular Function. GO functional classification annotation provides a gene list and gene number for every specific GO term. Additional File S9 Summary of DEGs enriched in KEGG pathways. Pathways and backbone gene numbers are given in the table. The q-values for all pathways are less than or equal to 0.05.
(XLSX)
Summary of DEGs enriched in KEGG pathways. Pathways and backbone gene numbers are given in the table. The q-values for all pathways are less than or equal to 0.05.
(XLS)
Funding Statement
This work was supported by the programme of 100 Distinguished Young Scientists in the Chinese Academy of Sciences, Natural Science Foundation in China (Grant No. 31270660), the Youth Science Foundation of Xinjiang Uygur Autonomous Region of China (Grant No. 2011211B47), the Open Fund of the State Key Laboratory of Crop Stress Biology for Arid Areas (CSBAA2011-04), and the Doctor Western-funded Projects of Chinese Academy of Sciences (XBBS 201201). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhu JK (2001) Plant salt tolerance. Trends in plant science 6: 66–71. [DOI] [PubMed] [Google Scholar]
- 2. Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Current Opinion in Biotechnology 16: 123–132. [DOI] [PubMed] [Google Scholar]
- 3. Flowers T (2004) Improving crop salt tolerance. Journal of Experimental botany 55: 307–319. [DOI] [PubMed] [Google Scholar]
- 4.Roy SJ, Tucker EJ, Tester M (2011) Genetic analysis of abiotic stress tolerance in crops. Current opinion in plant biology. [DOI] [PubMed] [Google Scholar]
- 5.Chinnusamy V, Zhu J, Zhu JK (2006) Salt stress signaling and mechanisms of plant salt tolerance. Genetic engineering: 141–177. [DOI] [PubMed] [Google Scholar]
- 6. Zhu JK (2002) Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53: 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cockburn W, Whitelam GC, Broad A, Smith J (1996) The participation of phytochrome in the signal transduction pathway of salt stress responses in Mesembryanthemum crystallinum L. Journal of Experimental botany. 47: 647–653. [Google Scholar]
- 8. Liska AJ, Shevchenko A, Pick U, Katz A (2004) Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella as revealed by homology-based proteomics. Plant physiology 136: 2806–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant physiology 134: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barkla B, Pantoja O (2011) Plasma membrane and abiotic stress. The Plant Plasma Membrane: 457–470. [Google Scholar]
- 11. Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal 31: 279–292. [DOI] [PubMed] [Google Scholar]
- 12. Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, et al. (2001) Gene expression profiles during the initial phase of salt stress in rice. The Plant Cell Online 13: 889–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, et al. (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant physiology 135: 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, et al. (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proceedings of the National Academy of Sciences of the United States of America 98: 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berthomieu P, Conejero G, Nublat A, Brackenbury WJ, Lambert C, et al. (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. Embo Journal 22: 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ardie SW, Xie L, Takahashi R, Liu S, Takano T (2009) Cloning of a high-affinity K+ transporter gene PutHKT2;1 from Puccinellia tenuiflora and its functional comparison with OsHKT2;1 from rice in yeast and Arabidopsis. Journal of Experimental botany 60: 3491–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Halfter U, Liu J, Ishitani M, Xiong L, Stevenson B, et al. (1999) The role of Arabidopsis SOS genes in plant salt tolerance. Plant Biology (Rockville) 1999: 123–123. [Google Scholar]
- 18. Shi H, Ishitani M, Kim C, Zhu J-K (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Plant Biology (Rockville) 2000: 91–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258. [DOI] [PubMed] [Google Scholar]
- 20. Pardo JM, Reddy MP, Yang SL, Maggio A, Huh GH, et al. (1998) Stress signaling through Ca2+/calmodulin-dependent protein phosphatase calcineurin mediates salt adaptation in plants. Proceedings of the National Academy of Sciences of the United States of America 95: 9681–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Espinosa-Ruiz A, Belles JM, Serrano R, Culianez-Macia FA (1999) Arabidopsis thaliana AtHAL3: a flavoprotein related to salt and osmotic tolerance and plant growth. Plant Journal 20: 529–539. [DOI] [PubMed] [Google Scholar]
- 22. Zhang JS, Xie C, Li ZY, Chen SY (1999) Expression of the plasma membrane H+-ATPase gene in response to salt stress in a rice salt-tolerant mutant and its original variety. Theoretical and Applied Genetics 99: 1006–1011. [Google Scholar]
- 23. Winicov I, Bastola DR (1999) Transgenic overexpression of the transcription factor Alfin1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants. Plant physiology 120: 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytologist 179: 945–963. [DOI] [PubMed] [Google Scholar]
- 25. Ungar IA (1996) Effect of salinity on seed germination, growth, and ion accumulation of Atriplex patula (Chenopodiaceae). American Journal of Botany 83: 604–607. [Google Scholar]
- 26. Guo Y, Jia W, Song J, Wang D, Chen M, et al. (2012) Thellungilla halophila is more adaptive to salinity than Arabidopsis thaliana at stages of seed germination and seedling establishment. Acta Physiologiae Plantarum 34: 1287–1294. [Google Scholar]
- 27. Lv S, Nie L, Fan P, Wang X, Jiang D, et al. (2012) Sodium plays a more important role than potassium and chloride in growth of Salicornia europaea. Acta Physiologiae Plantarum 34: 503–513. [Google Scholar]
- 28. Lv S, Jiang P, Chen X, Fan P, Wang X, et al. (2012) Multiple compartmentalization of sodium conferred salt tolerance in Salicornia europaea. Plant Physiology and Biochemistry 51: 47–52. [DOI] [PubMed] [Google Scholar]
- 29. Wang D, Wang H, Han B, Wang B, Guo A, et al. (2012) Sodium instead of potassium and chloride is an important macronutrient to improve leaf succulence and shoot development for halophyte Sesuvium portulacastrum. Plant Physiology and Biochemistry 51: 53–62. [DOI] [PubMed] [Google Scholar]
- 30. Kachout SS, Ben Mansoura A, Mechergui R, Leclerc JC, Rejeb MN, et al. (2012) Accumulation of Cu, Pb, Ni and Zn in the halophyte plant Atriplex grown on polluted soil. Journal of the Science of Food and Agriculture 92: 336–342. [DOI] [PubMed] [Google Scholar]
- 31. Ozawa T, Wu J, Fujii S (2007) Effect of inoculation with a strain of Pseudomonas pseudoalcaligenes isolated from the endorhizosphere of Salicornia europea on salt tolerance of the glasswort. Soil science and plant nutrition 53: 12–16. [Google Scholar]
- 32. Riehl TE, Ungar IA (1982) Growth and ion accumulation in Salicornia europaea under saline field conditions. Oecologia 54: 193–199. [DOI] [PubMed] [Google Scholar]
- 33. Aghaleh M, Niknam V, Ebrahimzadeh H, Razavi K (2011) Effect of salt stress on physiological and antioxidative responses in two species of Salicornia (S. persica and S. europaea). Acta Physiologiae Plantarum 33: 1261–1270. [Google Scholar]
- 34. Han H, Li Y, Zhou S (2008) Overexpression of phytoene synthase gene from Salicornia europaea alters response to reactive oxygen species under salt stress in transgenic Arabidopsis. Biotechnology letters 30: 1501–1507. [DOI] [PubMed] [Google Scholar]
- 35. Yang X, Ji J, Wang G, Yang S, Zhao Q, et al. (2011) Over-expressing Salicornia europaea (SeNHX1) gene in tobacco improves tolerance to salt. African Journal of Biotechnology 10: 16452–16460. [Google Scholar]
- 36. Zhou S, Chen X, Zhang X, Li Y (2008) Improved salt tolerance in tobacco plants by co-transformation of a betaine synthesis gene BADH and a vacuolar Na+/H+ antiporter gene SeNHX1. Biotechnology letters 30: 369–376. [DOI] [PubMed] [Google Scholar]
- 37. Chen X, Han H, Jiang P, Nie L, Bao H, et al. (2011) Transformation of β-lycopene cyclase genes from Salicornia europaea and Arabidopsis conferred salt tolerance in Arabidopsis and tobacco. Plant and Cell Physiology 52: 909–921. [DOI] [PubMed] [Google Scholar]
- 38.Wu S, Su Q, An L (2010) Isolation of choline monooxygenase (CMO) gene from Salicornia europaea and enhanced salt tolerance of transgenic tobacco with CMO genes. [PubMed] [Google Scholar]
- 39. Aghaleh M, Niknam V, Ebrahimzadeh H, Razavi K (2009) Salt stress effects on growth, pigments, proteins and lipid peroxidation in Salicornia persica and S. europaea. Biologia Plantarum 53: 243–248. [Google Scholar]
- 40. Wang X, Fan P, Song H, Chen X, Lil X, et al. (2009) Comparative Proteomic Analysis of Differentially Expressed Proteins in Shoots of Salicornia europaea under Different Salinity. Journal of Proteome Research 8: 3331–3345. [DOI] [PubMed] [Google Scholar]
- 41. Taji T, Sakurai T, Mochida K, Ishiwata A, Kurotani A, et al. (2008) Large-scale collection and annotation of full-length enriched cDNAs from a model halophyte, Thellungiella halophila. BMC plant biology 8: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen SH, Guo SL, Wang ZL, Zhao JQ, Zhao YX, et al. (2007) Expressed sequence tags from the halophyte Limonium sinense. Mitochondrial DNA 18: 61–67. [DOI] [PubMed] [Google Scholar]
- 43. Kore-eda S, Cushman MA, Akselrod I, Bufford D, Fredrickson M, et al. (2004) Transcript profiling of salinity stress responses by large-scale expressed sequence tag analysis in Mesembryanthemum crystallinum . Gene 341: 83–92. [DOI] [PubMed] [Google Scholar]
- 44. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644–U130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. 1999: 138–148. [PubMed] [Google Scholar]
- 46. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 47. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 48. Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, et al. (2009) Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. The Plant Journal 58: 1068–1082. [DOI] [PubMed] [Google Scholar]
- 49. Kyndt T, Denil S, Haegeman A, Trooskens G, De Meyer T, et al. (2012) Transcriptome analysis of rice mature root tissue and root tips in early development by massive parallel sequencing. Journal of Experimental botany 63: 2141–2157. [DOI] [PubMed] [Google Scholar]
- 50.Xu H, Gao Y, Wang J (2012) Transcriptomic Analysis of Rice (Oryza sativa) Developing Embryos Using the RNA-Seq Technique. Plos One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ushakova S, Kovaleva N, Gribovskaya I, Dolgushev V, Tikhomirova N (2005) Effect of NaCl concentration on productivity and mineral composition of Salicornia europaea as a potential crop for utilization NaCl in LSS. Advances in Space Research 36: 1349–1353. [Google Scholar]
- 52. Moghaieb REA, Saneoka H, Fujita K (2004) Effect of salinity on osmotic adjustment, glycinebetaine accumulation and the betaine aldehyde dehydrogenase gene expression in two halophytic plants, Salicornia europaea and Suaeda maritima. Plant science 166: 1345–1349. [Google Scholar]
- 53. Schachtman DP, Schroeder JI (1994) Structure and Transport Mechanism of a High-Affinity Potassium Uptake Transporter From Higher-Plants. Nature 370: 655–658. [DOI] [PubMed] [Google Scholar]
- 54. Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, et al. (2001) Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant Journal 27: 129–138. [DOI] [PubMed] [Google Scholar]
- 55. Su H, Balderas E, Vera-Estrella R, Golldack D, Quigley F, et al. (2003) Expression of the cation transporter McHKT1 in a halophyte. Plant Molecular Biology 52: 967–980. [DOI] [PubMed] [Google Scholar]
- 56. Blumwald E, Aharon GS, Apse MP (2000) Sodium transport in plant cells. Biochimica Et Biophysica Acta-Biomembranes 1465: 140–151. [DOI] [PubMed] [Google Scholar]
- 57. Fukuda A, Yazaki Y, Ishikawa T, Koike S, Tanaka Y (1998) Na+/H+ antiporter in tonoplast vesicles from rice roots. Plant and Cell Physiology 39: 196–201. [Google Scholar]
- 58. Rodríguez-Rosales MP, Gálvez FJ, Huertas R, Aranda MN, Baghour M, et al. (2009) Plant NHX cation/proton antiporters. Plant signaling & behavior 4: 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bassil E, Ohto MA, Esumi T, Tajima H, Zhu Z, et al. (2011) The Arabidopsis Intracellular Na+/H+ Antiporters NHX5 and NHX6 Are Endosome Associated and Necessary for Plant Growth and Development. Plant Cell 23: 224–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li HT, Liu H, Gao XS, Zhang HX (2009) Knock-out of Arabidopsis AtNHX4 gene enhances tolerance to salt stress. Biochemical and Biophysical Research Communications 382: 637–641. [DOI] [PubMed] [Google Scholar]
- 61. Subramanyam K, Sailaja KV, Rao DM, Lakshmidevi K (2011) Ectopic expression of an osmotin gene leads to enhanced salt tolerance in transgenic chilli pepper (Capsicum annum L.). Plant Cell Tissue and Organ Culture 105: 181–192. [Google Scholar]
- 62.Subramanyam K, Arun M, Mariashibu TS, Theboral J, Rajesh M, et al.. (2012) Overexpression of tobacco osmotin (Tbosm) in soybean conferred resistance to salinity stress and fungal infections. Planta: 1–17. [DOI] [PubMed] [Google Scholar]
- 63. Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN (1982) Living With Water-Stress – Evolution of Osmolyte Systems. Science 217: 1214–1222. [DOI] [PubMed] [Google Scholar]
- 64. Rentsch D, Hirner B, Schmelzer E, Frommer WB (1996) Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell 8: 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ashraf M, Foolad MR (2007) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environmental and Experimental Botany 59: 206–216. [Google Scholar]
- 66. Ueda A, Shi WM, Sanmiya K, Shono M, Takabe T (2001) Functional analysis of salt-inducible proline transporter of barley roots. Plant and Cell Physiology 42: 1282–1289. [DOI] [PubMed] [Google Scholar]
- 67. Waditee R, Hibino T, Tanaka Y, Nakamura T, Incharoensakdi A, et al. (2002) Functional characterization of betaine/proline transporters in betaine-accumulating mangrove. Journal of Biological Chemistry 277: 18373–18382. [DOI] [PubMed] [Google Scholar]
- 68. Sokhansanj A, Noori SAS, Niknam V (2006) Comparison of bacterial and plant genes participating in proline biosynthesis with osmotin gene, with respect to enhancing salinity tolerance of transgenic tobacco plants. Russian Journal of Plant Physiology 53: 110–115. [Google Scholar]
- 69. Ueda A, Kathiresan A, Inada M, Narita Y, Nakamura T, et al. (2004) Osmotic stress in barley regulates expression of a different set of genes than salt stress does. Journal of Experimental botany 55: 2213–2218. [DOI] [PubMed] [Google Scholar]
- 70. Foyer C, Lelandais M, Galap C, Kunert KJ (1991) Effects of Elevated Cytosolic Glutathione-Reductase Activity on The Cellular Glutathione Pool and Photosynthesis in Leaves Under Normal and Stress Conditions. Plant physiology 97: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Torabi S, Niknam V (2011) Effects of Iso-osmotic Concentrations of NaCl and Mannitol on some Metabolic Activity in Calluses of Two Salicornia species. In Vitro Cellular & Developmental Biology-Plant 47: 734–742. [Google Scholar]
- 72. Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2008) The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant Journal 56: 613–626. [DOI] [PubMed] [Google Scholar]
- 73. Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2004) dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant Journal 37: 720–729. [DOI] [PubMed] [Google Scholar]
- 74. Riehl TE, Ungar IA (1982) Growth and ion Accumulation in Salicornia-Europaea Under Saline field Conditions. Oecologia 54: 193–199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for experimental validation and gene expression profile analysis.
(DOCX)
Contains: Additional File S2.z01– All-Unigene sequences of S. europaea shoots. Sequences with no gap and with a length longer than 200 bp were selected from the assembly results. Additional File S2.z02– All-Unigene sequences of S. europaea shoots. Sequences with no gap and with a length longer than 200 bp were selected from the assembly results. Additional File S2– All-Unigene sequences of S. europaea shoots. Sequences with no gap and with a length longer than 200 bp were selected from the assembly results.
(ZIP)
Primers and amplification results of 18 unigenes used to confirm the sequencing quality. The table shows sequences and annealing temperatures (Tm) and production of primers.
(XLS)
Functional annotation of All-Unigenes, including GO, COG, and KEGG analysis. All-Unigene sequences were searched against protein databases (Nr, SwissProt, KEGG, COG, and GO) using BLASTX (E-value ≤10–5).
(ZIP)
Summary of All-Unigenes enriched in KEGG pathways. Pathway ID and gene number are given in the table. The q-value for all of these pathways was less than or equal to 0.05.
(XLSX)
Summary and functional annotation of identified DEGs. Unigenes with an absolute value of |log2Ratio| ≥1 and FDR ≤0.001 were identified as DEGs. GO and KEGG analyses of DEGs were based on a cutoff E-value of less than or equal to 10–5.
(XLS)
GO categories of DEGs between Salt-Free and Salt-Treated Shoots of S. europaea. DEGs were divided into three major categories: molecular function, cellular component and biological process. The gene numbers and gene ID are listed in this file.
(XLS)
Summary of DEGs GO Molecular Function. GO functional classification annotation provides a gene list and gene number for every specific GO term. Additional File S9 Summary of DEGs enriched in KEGG pathways. Pathways and backbone gene numbers are given in the table. The q-values for all pathways are less than or equal to 0.05.
(XLSX)
Summary of DEGs enriched in KEGG pathways. Pathways and backbone gene numbers are given in the table. The q-values for all pathways are less than or equal to 0.05.
(XLS)