Abstract
DNA repair is a key determinant in the cellular response to therapy and tumor repair status could play an important role in tailoring patient therapy. Our goal was to evaluate the mRNA of 13 genes involved in different DNA repair pathways (base excision, nucleotide excision, homologous recombination, and Fanconi anemia) in paraffin embedded samples of triple negative breast cancer (TNBC) compared to luminal A breast cancer (LABC). Most of the genes involved in nucleotide excision repair and Fanconi Anemia pathways, and CHK1 gene were significantly less expressed in TNBC than in LABC. PARP1 levels were higher in TNBC than in LABC. In univariate analysis high level of FANCA correlated with an increased overall survival and event free survival in TNBC; however multivariate analyses using Cox regression did not confirm FANCA as independent prognostic factor. These data support the evidence that TNBCs compared to LABCs harbour DNA repair defects.
Introduction
Triple negative breast cancer (TNBC) defines a clinical subset of breast cancer negative for estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor type 2 (HER2). It accounts for about 15% of breast tumors and is characterized by an aggressive clinical course with therapeutic resistance, high rates of local and systemic relapse, and poor survival. This is probably due to the intrinsic biology of this type of tumor as well as to the absence of specific targeted treatments such as hormonal therapy used for ER positive tumors and trastuzumab/lapatinib for HER2 over-expressing tumors [1], [2].
TNBC encompasses more than one subtype [3], [4]. Morphologically they include metaplastic, adenoid cystic and medullary like and at a molecular level they display different mRNA profiles such as basal and claudin type [5], [6]. Cumulating evidence suggest that the triple negative phenotype on clinical assays enriches for basal-like cancer, but no complete overlap exists between the two groups [7], [8]. Indeed, 25% of the TNBCs are not basal-like on gene expression profile and similarly there are basal-like tumors that are not triple negative by immunohistochemistry.
Most of the tumors that develop in women with germline mutations in the BRCA1 breast cancer susceptibility gene are TNBC [9], [10]. BRCA1 has a crucial role in the repair of double-strand breaks and its mutation leads to cancer predisposition and genomic instability [11], [12]. BRCA1 related TNBCs share several pathological features with sporadic TNBCs and cluster within the basal subtype by gene expression profile [13]. Thus it is not surprising that there are data suggesting that a proportion of sporadic basal-like breast cancers may have a dysfunctional BRCA1 pathway, due to gene promoter methylation or transcriptional inactivation [13], [14]. Deficits in other DNA-repair pathways, such as base excision repair (BER) inactivation [15], MTMG promoter methylation [16] and lack of hOGG1 [17] have been reported in TNBCs. These tumors also exhibit higher DNA copy alterations and loss of heterozygosity than other breast cancer types, suggesting higher genomic instability [18]–[20]. These latter data, together with the reported association between BRCA1 deficiency and TNBC, suggest that DNA repair alterations may be important for the development of this tumor type. If this is the case, there could have important prognostic and predictive implications, such as the response to the currently used anticancer agents and to novel targeted agents, as the poly-ADP-ribose polymerase (PARP) inhibitors [21].
While tests for the evaluation of DNA repair defects in primary cells or in clinical samples have been described [22], [23], to our knowledge validated functional assays for the quantification of the tumor DNA repair capabilities to be used in clinical setting do not exist. There are however some biomarkers that, even if not completely validated [24]–[27], have been used as surrogate of DNA repair functionality, i.e. the mRNA and/or expression levels of key proteins involved in DNA repair pathways. Aim of our study was to evaluate in a multiparametric manner the mRNA expression level of different key player of DNA repair pathways (i.e. base excision repair-BER-, nucleotide excision repair-NER- and homologous recombination- HR- and Fanconi anemia- FA) in TNBCs compared with LABCs and to correlate with clinico-pathological patient characteristics.
Materials and Methods
Patients and samples
150 formalin-fixed paraffin embedded core breast cancer samples, diagnosed as triple negative (80 cases) and luminal A (70 cases), were retrospectively collected from patients who came to the medical observation at the Breast Care Unit, A.O. Istituti Ospitalieri di Cremona, Italy. Ethical permission for the study was obtained from the A.O. Istituti Ospitalieri di Cremona (Italy) ethical committee. Written informed consent was obtained from patients, if applicable. All the samples were anonymized by a pathologist staff member and none of the researcher conducting the gene expression analysis had access to disclosed clinico-pathological data. The patients characteristics are summarized in Table 1.
Table 1. Baseline patients' demographic.
| CLINICAL PARAMETERS | TNBC (N = 80) | LABC (N = 70) |
| Follow-up time – years | ||
| Median (range) | 5.5 (0.1–16.0) | 3.16 (0.6–17.3) |
| Mean ± SD | 6.7±4.7 | 5.9±5.5 |
| Age – years | ||
| Median (range) | 59.5 (33.1–91.3) | 66.6 (36.5–84.5) |
| Mean ± SD | 60.2±14.4 | 63.1±11.5 |
| Age – no. (%) | ||
| <65 | 46 (57.5) | 31 (44.3) |
| ≥65 | 34 (42.5) | 39 (55.7) |
| Menopausal status – no. (%) | ||
| Pre- | 19 (23.7) | 12 (17.1) |
| Post- | 59 (73.8) | 58 (82.9) |
| not known | 2 (2.5) | 0 (0.0) |
| Clinical tumor size – no. (%) | ||
| ≤20 mm | 16 (20.0) | 21 (30.0) |
| >20 mm | 36 (45.0) | 12 (17.1) |
| not known | 28 (35.0) | 37 (52.9) |
| Tumor size at surgery – no. (%) | ||
| ≤20 mm | 36 (45.0) | 42 (60.0) |
| >20 mm | 22 (27.5) | 11 (15.7) |
| not known | 22 (27.5) | 17 (24.3) |
| Pathological T staging – no. (%) | ||
| T1 | 41 (51.3) | 48 (68.6) |
| T2 | 20 (25.0) | 11 (15.7) |
| T3 | 2 (2.5) | 1 (1.4) |
| T4 | 4 (5.0) | 3 (4.3) |
| not known | 13 (16.2) | 7 (10.0) |
| Pathological N staging – no. (%) | ||
| N0 | 37 (46.3) | 66 (94.3) |
| N+ | 18 (22.5) | 0 (0.0) |
| not known | 25 (31.2) | 4 (5.7) |
| Metastatic status – no. (%) | ||
| M0 | 60 (75.0) | 68 (97.1) |
| M1 | 4 (5.0) | 0 (0.0) |
| not known | 16 (20.0) | 2 (2.9) |
| Pathological staging – no. (%) | ||
| Stage I | 28 (35.0) | 48 (68.6) |
| Stage IIA | 13 (16.3) | 11 (15.7) |
| Stage IIB | 5 (6.2) | 1 (1.4) |
| Stage IIIA | 4 (5.0) | 0 (0.0) |
| Stage IIIB | 4 (5.0) | 3 (4.3) |
| Stage IV | 4 (5.0) | 0 (0.0) |
| not known | 22 (27.5) | 7 (10.0) |
| Basal histotype – no. (%) | ||
| IDC | 66 (82.5) | 23 (32.9) |
| ILC | 5 (6.2) | 16 (22.9) |
| Other | 7 (8.8) | 30 (42.8) |
| not known | 2 (2.5) | 1 (1.4) |
| Basal tumor grade – no. (%) | ||
| 1 | 1 (1.2) | 6 (8.6) |
| 2 | 14 (17.5) | 47 (67.1) |
| 3 | 64 (80.1) | 15 (21.4) |
| not known | 1 (1.2) | 2 (2.9) |
| Basal Ki67 – no. (%) | ||
| Median (range) | 37 (3–90) | 7.5 (1–16) |
| Mean ± SD | 44.6±26.4 | 7.3±3.3 |
| <10 | 3 (3.7) | 50 (71.4) |
| ≥10 | 75 (93.8) | 20 (28.6) |
| <20 | 15 (18.7) | 70 (100.0) |
| ≥20 | 63 (78.8) | 0 (0.0) |
| not known | 2 (2.5) | 0 (0.0) |
| Neoadjuvant treatment – no. (%) | ||
| Done | 35 (43.8) | 19 (27.2) |
| not done | 38 (47.5) | 50 (71.4) |
| not known | 7 (8.7) | 1 (1.4) |
| Surgical treatment – no. (%) | ||
| Mastectomy | 21 (26.3) | 14 (20.0) |
| Breast conservation | 48 (60.0) | 54 (77.1) |
| not known | 11 (13.7) | 2 (2.9) |
| Vital status – no. (%) | ||
| Alive | 53 (66.2) | 69 (98.6) |
| Dead | 27 (33.8) | 1 (1.4) |
SD: standard deviation; IDC: invasive ductal carcinoma; ILC: invasive lobular carcinoma.
Definition of molecular classes
Tumour grade was evaluated using the modified Bloom and Richardson method. Immunohistochemical evaluation was performed on paraffin-embedded tumour samples obtained at diagnosis. Estrogen receptor, progesterone receptor, HER2 staining were carried out at the Pathology Unit of the A.O. Istituti Ospitalieri di Cremona, Italy. The immunohistochemical method used for routine markers is fully described elsewhere [28]. Tumors were classified as: luminal A (hormone receptor positive: estrogen receptor + and/or progesterone receptor + and HER2–), and triple-negative (ER-ve, PR-ve, HER2-ve) [29].
RNA and real time
RNA was extracted from core biopses with the High Pure FFPE RNA Micro Kit (Roche). RNA concentrations were then measured by NanoDrop spectrophotometer and retro-transcription to cDNA was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystem) in a 96well plate, one for each hystological subgroup. 15ug are preamplified with TaqMan Preamp Master mix (Applied Biosystem) and with the pool of primers used later for real time PCR reactions. Genes selected have a key role in the BER (PARP1); in the NER pathway (ERCC1, XPA, XPD, XPF and XPG); in the FA pathway (BRCA1, FANCA, FANCC, FANCD2, FANCF and PALB2); in DNA damage checkpoint pathway (Chk1). Optimal primer pairs (Table 2) were chosen, spanning splice junctions, using PRIMER-3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and the specificity was verified by detecting single-band amplicons of the PCR products. The EP Motion robot (Eppendorf) dispensed 10 ng of cDNA in triplicate on a 384 wells plate, one for each gene and for each subgroup. The reaction was performed with Sybr Green PCR master mix (Applied Biosystem) and the curve of dissociation was evaluated. A standard curve for each specific gene was included in each plate for an absolute quantification of the copy of RNA. Samples were then normalized using the absolute copy number of the housekeeping gene (ciclophilin A). The two different sample plates (TNBC and LABC) were calibrated using inter-run calibrators samples (four different breast cancer cell lines) to correct for run to run technical variation. The calibration procedure was performed on a gene per gene basis.
Table 2. Real Time PCR primers.
| GENE | PRIMER F | PRIMER R |
| CICLOPHILIN A | GACCCAACACAAATGGTTCC | TTTCACTTTGCCAAACACCA |
| PARP1 | AAGAAATGCAGCGAGAGCAT | CCAGTGTGGGACTTTTCCAT |
| ERCC1 | GGCCTATGAGCAGAAACCAG | TTCACGGTGGTCAGACATTC |
| XPA | GAACCACTTTGATTTGCCAAC | TTGTTTTGCCTCTGTTTTGG |
| XPF | GAGAAATCTTTTTGTGAGGAAACTG | CAACTTCAGGTTTGTGCTGTTC |
| XPG | AAGCCATCAAAACTGCCTTC | TCGTTTTCTCTTCGAACTTGG |
| XPD | GTGGCCATCAGCTCCAAAT | CAGCAGGAGGTTCCCATAGT |
| BRCA1 | GCCAGAAAACACCACATCAC | CAGTGTCCGTTCACACACAA |
| FANCA | GAGGTTCTTCAGTCATACCCTGA | TCTCTCTGCATCTGAACAGCA |
| FANCC | CCAGCCAGAGTTCTTTGAGG | CGAAGCCAGAGGCAGACTAC |
| FANCD2 | CCCATCTGCTATGATGATGAA | CGTATTTGCTGAGGGGATATG |
| FANCF | GCTAGTCCACTGGCTTCTGG | GGTGGCGGCTAGTCACTAAA |
| PALB2 | TGGGACCCTTTCTGATCAAC | GGGGCATCAAAAATTGGTTT |
| CHK1 | GGTCACAGGAGAGAAGGAAT | TCTCTGACCATCTGGTTCAGG |
Data and statistical analysis
The Mann-Whitney U test was chosen to compare clinicopathological and gene expression data between the two groups of TNBCs and LABCs. The median is taken as valid proxy for expression level of all genes. Relationships between family gene expression levels were examined using the Spearman's rank correlation. Each gene distribution was split into three expression levels (low, intermediate and high level) defined by the two tertiles calculated on patients who did not die at the end of the study. For further testing, each gene distribution was dichotomized (low and high level) using the median gene expression value as the cut-off point. Consistently, patients were divided into different subgroups based on different gene expression level. Univariate comparisons for all categorical variables were performed by Pearson's chi-squared test with Yates's correction for continuity or Fisher's exact test, when appropriate. Overall survival (OS) and event-free survival (EFS) were classified as outcome measures and defined as the length of time from the start of neoadjuvant chemotherapy or the date of curative surgery (in patients treated with only adjuvant therapy) or the date of diagnosis (in patients diagnosed with metastases) to the last follow-up date or death (irrespective of the cause) or to the first relapse or progression event. OS and EFS curves were plotted with the Kaplan-Meier method. The log-rank test and Cox proportional hazard models were used to compare time-to-event distributions between subgroups. All analyses were performed using SPSS (version 10.0– Chicago, IL) and STATISTICA software system (version 6– Tulsa, OK). All tests were performed two-sided and p values <0.05 were considered statistically significant.
Results
Patients' characteristics
We considered a cohort of 150 paraffin embedded core biopsies, 80 with a pathological diagnosis of TNBC and 70 with a diagnosis of LABC that came to the medical attention of a single medical center, the Breast Care Unit of A.O. Istituti Ospitalieri di Cremona from January 5th, 1995 to August 19th, 2008. The two groups (TNBC vs LABC), whose characteristics are depicted in Table 1, were comparable in terms of age (p = 0.2056) and menopausal status at diagnosis (p = 0.4493), surgical treatment (p = 0.3200), pathological T staging at surgery (p = 0.1404), and follow-up time (p = 0.0649).
mRNA expression of the 13 genes involved in DNA repair pathways
Genes involved in different DNA repair pathways were studied by RT-PCR and Table 3 summarizes the data for both TNBC and LABC. There was considerable variability in expression levels in the individual genes. The levels of the NER genes, ERCC1, XPA, XPF and XPD genes were significantly lower in TNBCs than in LABCs, while there was no difference for XPG. FA genes were less expressed than NER genes in TNBC. BRCA1, FANCD2, FANCF and PALB2 genes were significantly less expressed in TNBCs than in LABCs. CHK1 gene was significantly less expressed in TNBCs than in LABCs. On the contrary, PARP1 levels were higher in TNBCs than in LABCs.
Table 3. Normalized and calibrated values (mean± SD and median) of the different DNA repair genes in tumor samples.
| PATH WAY | GENE | TNBC (N = 80) | LABC (N = 70) | p value* | ||
| Mean ± SD | Median | Mean ± SD | Median | |||
| BER | PARP1 | 10.87±10.92 | 8.250 | 7.05±5.46 | 5.221 | 0.0002 |
| NER | ERCC1 | 0.71±0.66 | 0.633 | 6.87±20.77 | 1.040 | <0.0001 |
| XPA | 0.10±0.08 | 0.080 | 0.16±0.21 | 0.104 | 0.0309 | |
| XPF | 23.35±55.61 | 5.108 | 68.93±107.05 | 21.996 | <0.0001 | |
| XPG | 0.66±0.71 | 0.468 | 1.51±3.52 | 0.287 | 0.1534 | |
| XPD | 0.276±0.360 | 0.186 | 6.68±10.26 | 3.662 | <0.0001 | |
| FA | BRCA1 | 0.028±0.025 | 0.020 | 0.83±3.36 | 0.062 | <0.0001 |
| FANCA | 0.138±0.279 | 0.057 | 0.27±0.71 | 0.094 | 0.8293 | |
| FANCC | 0.005±0.007 | 0.003 | 0.002±0.050 | 0.003 | 0.3627 | |
| FANCD2 | 0.08±0.124 | 0.054 | 4.45±23.32 | 0.150 | <0.0001 | |
| FANCF | 0.026±0.027 | 0.018 | 0.451±1.170 | 0.031 | <0.0001 | |
| PALB2 | 0.453±0.563 | 0.306 | 5.91±23.03 | 2.010 | 0.0006 | |
| SEN SOR | CHK1 | 0.179±0.300 | 0.108 | 11.51±46.44 | 1.779 | <0.0001 |
p value indicate the difference of the gene expression levels between TNBC anl LABC; p value in bold are significant.
Association between gene expression and clinic-pathological characteristics
Table 4 reports the p values found between the gene expression level based on tertiles (low, intermediate and high level as specified in Materials and Methods) and some clinic-pathological characteristics in TNBC. Lower levels of XPG and FANCA were associated with higher pathological T classification at surgery (pT2-4 vs pT1); higher level of XPF was associated with no lymph node involvement (pN0 vs pN1-3), and higher levels of FANCA correlated with vital status. Using the median gene expression value as a cut-off point of categorization (low and high level), only the association of XPG and FANCA expression levels with pT classification remained statistically significant. No other associations were found among the other clinic-pathological evaluated parameters (age at diagnosis-65 years vs ≥65 years-; menopausal status-pre-menopausal vs post-menopausal-; clinical dimension at diagnosis-≤20 mm vs >20 mm-; basal histotype-ductal infiltrating carcinoma vs others-; basal grading-G1-2 vs G3-; basal Ki67-<14% vs ≥14% and <20% vs ≥20%-; clinical complete response-cCR vs not cCR-; pathological complete response-pCR vs not pCR-). When the same analysis was performed in the LABC cohort of patients, none association could be found.
Table 4. Significantp values for the association between gene expression and clinic-pathological characteristics of triple negative breast cancer patients (univariate analysis, categorization based on tertiles).
| PATHWAY | GENE | pT classification | pN classification | Vital status |
| at surgery | at surgery | at last follow up | ||
| BER | PARP1 | |||
| NER | ERCC1 | |||
| XPA | ||||
| XPF | 0.0162 | |||
| XPG | 0.0437 | |||
| XPD | ||||
| FA | BRCA1 | |||
| FANCA | 0.0366 | 0.0144 | ||
| FANCC | ||||
| FANCD2 | ||||
| FANCF | ||||
| PALB2 | ||||
| SENSOR | CHK1 |
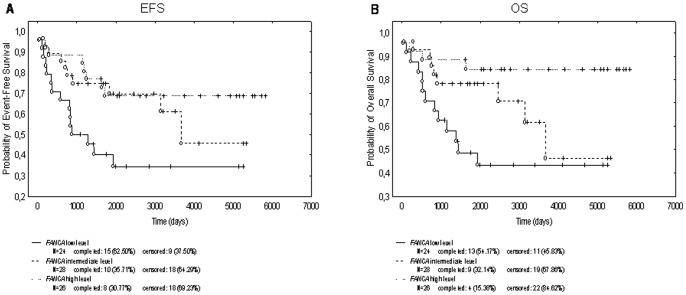
We correlated the different gene expression levels based on tertiles with OS and EFS. As depicted in Table 5, the intermediate level of XPG correlated on univariate analysis with increased OS compared to the low level. Interesting enough, the high level of FANCA correlated with an increased OS and EFS compared to the low level. Again, the intermediate level of FANCA correlated with an increase in EFS compared to the low level. The Kaplan-Meier curves (Figure 1) shows that higher level of FANCA correlated with an increased OS (p = 0.0153) and an increased EFS (p = 0.0207). Furthermore, using the median gene expression value as a cut-off point of categorization, we again found a trend (not significant) toward better OS (p = 0.0906) and EFS (p = 0.0999) in patients with high level of FANCA. Multivariate analyses using Cox regression, however, showed that the FANCA expression level was not an independent prognostic factor in TNBC patients adjusted for the potential confounding factors: age at diagnosis, surgical treatment, pathological T and N staging at surgery. As regards LABCs, OS and EFS data were not yet mature.
Table 5. Overall survival and event-free survival in triple negative breast cancer patients by expression of the different genes (univariate analysis, categorization based on tertiles).
| TNBC (N = 80) | ||||
| PATHWAY | GENE | EXPRESSION | OS | EFS |
| p value | p value | |||
| BER | PARP1 | II-I terziles | 0.3056 | 0.2334 |
| III-I terziles | 0.6140 | 0.4632 | ||
| NER | ERCC1 | II-I terziles | 0.9993 | 0.9450 |
| III-I terziles | 0.1868 | 0.3955 | ||
| XPA | II-I terziles | 0.2035 | 0.0957 | |
| III-I terziles | 0.7071 | 0.9908 | ||
| XPF | II-I terziles | 0.7813 | 0.9566 | |
| III-I terziles | 0.1088 | 0.4704 | ||
| XPG | II-I terziles | 0.0344 | 0.0674 | |
| III-I terziles | 0.2870 | 0.2902 | ||
| XPD | II-I terziles | 0.1010 | 0.0764 | |
| III-I terziles | 0.1403 | 0.2480 | ||
| FA | BRCA1 | II-I terziles | 0.4812 | 0.6162 |
| III-I terziles | 0.4196 | 0.5115 | ||
| FANCA | II-I terziles | 0.1088 | 0.0408 | |
| III-I terziles | 0.0045 | 0.0141 | ||
| FANCC | II-I terziles | 0.3579 | 0.5383 | |
| III-I terziles | 0.0850 | 0.0736 | ||
| FANCD2 | II-I terziles | 0.0845 | 0.1352 | |
| III-I terziles | 0.1554 | 0.2216 | ||
| FANCF | II-I terziles | 0.7556 | 0.9254 | |
| III-I terziles | 0.6192 | 0.7587 | ||
| PALB2 | II-I terziles | 0.5170 | 0.5381 | |
| III-I terziles | 0.2010 | 0.1617 | ||
| PARP1 | II-I terziles | 0.3056 | 0.2334 | |
| III-I terziles | 0.6140 | 0.4632 | ||
| SENSOR | CHK1 | II-I terziles | 0.7740 | 0.4396 |
| III-I terziles | 0.2325 | 0.2601 | ||
The mRNA expression distribution of all the genes was split into three groups, as described in Materials and Methods. p value in bold are significant.
Figure 1. Kaplan-Meier estimates for event-free survival (panel A) and overall survival (panel B) in TNBC patients stratifies by the different level ofFANCA expression (univariate analysis, categorization based on tertiles).
Discussion
TNBC represents a subset of breast cancer with well documented poor prognosis, whose real causes are still to be defined even if a number of adverse factors have been advocated, such as the fact that these tumors are at the diagnosis commonly of high nuclear mitotic grade, of larger tumor size and with a more aggressive expression profile (low BCL2 and high p53 and Ki67 expression) [5], [30], [31]. Several studies have demonstrated significant lower EFS in patients with TNBC compared to patients with non-TNBC; in addition, shorter OS in TNBC patients is widely reported [3], [32]–[34]. Despite the poor prognosis, these tumors are particularly chemosensitive at least in a short-term time frame [35], [36]. In fact, patients with TNBC have increased pathologic complete response (pCR) rates compared to non-TNBC patients after treatment with taxanes and antracycline agents. Wang at al reported a 38% of pCR in TNBC and 14% in non-TNBC patients treated with taxane in combination with antracycline [37]. Better response rates were also reported after treatment with alkylating agents, including cyclophosphamide, cisplatin, and carboplatin [38].
TNBC have been shown to harbour DNA repair deficiencies, including BRCA1 dysfunction, due to promoter methylation or deregulation of other genes involved in their transcriptional regulation [9], BER inactivation [15], MTMG promoter hypermethylation [16] and lack of hOGG1 [17]. Given the importance of the cellular DNA repair in determining the cellular response to different anticancer agents [39]–[43], a priori knowledge of the repair status of a given tumor could play an important role in selection of the most appropriate therapy. Attempts to investigate the functionality of a given DNA repair pathway have been undertaken and shown to be feasible. Willers et al reported the successful detection of BRCA1, FANCD2 and RAD51 foci in seven breast cancer biopsies irradiated ex vivo, suggesting the possibility to detect defects within the complex FA/BRCA DNA damage response pathway [44]. Isolation of epithelial cells from breast tumor specimens and application of specific functional DNA repair assay systems led to the detection of specific defects in double strand breaks repair [22], while homologous recombination status could be determined in ovarian cancer samples by RAD51 foci formation after in vitro treatment with a PARP inhibitor [23]. However, all these assays still need to be technically improved (ie. requirement for an automated foci scoring and setting up of tumor specific primary cultures) rendering difficult their wide spread clinical use. An alternative approach is the study of biomarkers that, even if not completely validated [24]–[27], can be considered surrogate of DNA repair functionality, i.e. the mRNA and/or expression levels of key proteins involved in DNA repair pathways.
The expression profile of a number of genes involved in the different DNA repair pathways was studied in a cohort of TNBC samples as a surrogate markers of DNA repair status and correlated with different clinic-pathological characteristics, including survival. These data were compared to the ones obtained in a group of patients with LABC, that represent the breast cancer subtype with the best prognosis.
The levels of expression of all the genes involved in the NER and FA pathways were significantly lower in TNBC as compared to LABC samples, with the exception of XPG, FANCA and FANCC whose difference did not reach a statistically significant value.
These expression data might in part explain the extremely high sensitivity of TNBC patients to chemotherapy. It has in fact been demonstrated that not only the deficiency, but also low levels of mRNA/protein involved in NER, i.e ERCC1 and XPF or FA genes are associated with increased sensitivity to alkylating agents in different cellular systems [45]. At the same time, the low expression level of these genes might explain the bad prognosis of the tumors, even if they do respond well to chemotherapy. The low DNA repair capacity would in fact increase the genomic instability of the tumor cells enabling them to accumulate much more mutations through which cells would became more aggressive. Sporadic stage I breast cancer have recently been reported to exhibite a significant deficiency of NER capacity relative to epithelial control tissue [46] by both functional unscheduled DNA synthesis and mRNA expression of NER genes. Low expression of 4 proteins – XPF, FANCD2, MLH1 and pMK2– assessed in FFPE tumor specimens by semiquantitative immunohistochemistry, was associated with shorter recurrence free survival in multivariate analysis [47], corroborating the idea that tumors with reduced DNA repair capacity will have a higher degree of genomic instability and therefore behave more aggressively.
The data on FANCA are, on the contrary, counter-intuitive and intriguing. FANCA was not differentially expressed between TNBC and LABC, but it was found to positively correlate in univariate analysis with both EFS and OS (higher level correlated with longer EFS and OS). FANCA is one of the 13 proteins cooperating in a common DNA repair pathway (for rev see [48], [49]). FANCA, higher expression correlated with an improved outcome that could be a consequence of a less malignant phenotype, due to a more stable tumor genome. This resembles what observed for the expression of ERCC1 in non small cell lung cancer (NSCLC), where it has been shown that higher ERCC1 levels were correlated with a better prognosis [50], [51], but was also higher levels of ERCC1 were a marker of resistance to a platinum containing regimens [50]. Recently, the experimental finding that ERCC1 negative NSLC samples display a higher genomic instability corroborate the more unstable and then the worst clinical history of ERCC1 low/negative tumors [52].
There are few, but contrasting data, on CHK1 expression in different breast cancer subtypes. CHK1 is a kinase that has an important role in both the maintenance of genomic instability and in the transduction pathways after DNA damage [53]. Verlinden et al. [54] reported a twofold higher expression (as assessed by RT-PCR) of CHK1 in grade 3 TNBC than in other grade 3 tumor breast subtype; while no difference CHK1 protein expression was found in different breast cancer cell lines. These discrepancies can be related to the different methodologies used (RT-PCR vs western blotting; relative vs absolute RT-PCR values), samples size and the pathological classification of the different tumor analyzed. All tumor samples analyzed expressed PARP1 mRNA, whose levels were higher in TNBC compared to LABC, reinforcing the data that neoplastic tumors express high level of PARP [55], [56].
There are a number of issues that limits the present study: i.e. its retrospective nature, the relative small sample size and the fact it analysed the mRNA expression levels of DNA repair protein rather than the quantification of a functional DNA repair status. Nevertheless, our data are the first to show the mRNA expression of multiple DNA repair genes involved in three different DNA repair pathways (BER, NER and FA) in a cohort of TNBC and LABC.
These data support the experimental evidence that TNBC harbour DNA repair defects and this could explain both the extremely chemo-sensitivity of this tumor type to chemotherapy in a short term and the worse outcome probably correlated with the inability to counteract the increased genomic instability for the lack of an efficient DNA damage status.
Funding Statement
The work was supported by the Italian Association for Cancer Research (AIRC), Associazione Ricerca in Campo Oncologico (ARCO) and the Victorian Breast Cancer Research Consortium of Australia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Carey LA (2011) Directed therapy of subtypes of triple-negative breast cancer. Oncologist 16 Suppl 171–78. [DOI] [PubMed] [Google Scholar]
- 2. Hudis CA, Gianni L (2011) Triple-negative breast cancer: an unmet medical need. Oncologist 16 Suppl 11–11. [DOI] [PubMed] [Google Scholar]
- 3. de Ruijter TC, Veeck J, de Hoon JP, van Engeland M, Tjan-Heijnen VC (2011) Characteristics of triple-negative breast cancer. J Cancer Res Clin Oncol 137: 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chacon RD, Costanzo MV (2010) Triple-negative breast cancer. Breast Cancer Res 12 Suppl 2S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perou CM (2011) Molecular stratification of triple-negative breast cancers. Oncologist 16 Suppl 161–70. [DOI] [PubMed] [Google Scholar]
- 6. Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. (2000) Molecular portraits of human breast tumours. Nature 406: 747–752. [DOI] [PubMed] [Google Scholar]
- 7. Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, et al. (2008) How basal are triple-negative breast cancers? Int J Cancer 123: 236–240. [DOI] [PubMed] [Google Scholar]
- 8. Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, et al. (2009) Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 15: 2302–2310. [DOI] [PubMed] [Google Scholar]
- 9. Turner NC, Reis-Filho JS (2006) Basal-like breast cancer and the BRCA1 phenotype. Oncogene 25: 5846–5853. [DOI] [PubMed] [Google Scholar]
- 10. Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, et al. (2008) Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol 26: 4282–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J, Powell SN (2005) The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res 3: 531–539. [DOI] [PubMed] [Google Scholar]
- 12. Hartman AR, Ford JM (2002) BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat Genet 32: 180–184. [DOI] [PubMed] [Google Scholar]
- 13. Turner N, Tutt A, Ashworth A (2004) Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 4: 814–819. [DOI] [PubMed] [Google Scholar]
- 14. Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, et al. (2007) BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 26: 2126–2132. [DOI] [PubMed] [Google Scholar]
- 15. Alli E, Sharma VB, Sunderesakumar P, Ford JM (2009) Defective repair of oxidative dna damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res 69: 3589–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fumagalli C, Pruneri G, Possanzini P, Manzotti M, Barile M, et al.. (2012) Methylation of O (6)-methylguanine-DNA methyltransferase (MGMT) promoter gene in triple-negative breast cancer patients. Breast Cancer Res Treat. [DOI] [PubMed] [Google Scholar]
- 17. Karihtala P, Kauppila S, Puistola U, Jukkola-Vuorinen A (2012) Absence of the DNA repair enzyme human 8-oxoguanine glycosylase is associated with an aggressive breast cancer phenotype. Br J Cancer 106: 344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigman VJ, Chao HH, Shabalin AA, He X, Parker JS, et al.. (2011) Basal-like Breast cancer DNA copy number losses identify genes involved in genomic instability, response to therapy, and patient survival. Breast Cancer Res Treat [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, et al. (2006) Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer 45: 1033–1040. [DOI] [PubMed] [Google Scholar]
- 20. Andre F, Job B, Dessen P, Tordai A, Michiels S, et al. (2009) Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 15: 441–451. [DOI] [PubMed] [Google Scholar]
- 21. Rowe BP, Glazer PM (2010) Emergence of rationally designed therapeutic strategies for breast cancer targeting DNA repair mechanisms. Breast Cancer Res 12: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keimling M, Kaur J, Bagadi SA, Kreienberg R, Wiesmuller L, et al. (2008) A sensitive test for the detection of specific DSB repair defects in primary cells from breast cancer specimens. Int J Cancer 123: 730–736. [DOI] [PubMed] [Google Scholar]
- 23. Mukhopadhyay A, Elattar A, Cerbinskaite A, Wilkinson SJ, Drew Y, et al. (2010) Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin Cancer Res 16: 2344–2351. [DOI] [PubMed] [Google Scholar]
- 24. Lips EH, Mulder L, Hannemann J, Laddach N, Vrancken Peeters MT, et al. (2011) Indicators of homologous recombination deficiency in breast cancer and association with response to neoadjuvant chemotherapy. Ann Oncol 22: 870–876. [DOI] [PubMed] [Google Scholar]
- 25. Rothschild SI, Gautschi O, Lara PN Jr, Mack PC, Gandara DR (2011) Biomarkers of DNA repair and related pathways: significance in non-small cell lung cancer. Curr Opin Oncol 23: 150–157. [DOI] [PubMed] [Google Scholar]
- 26. Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, et al. (2010) Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol 28: 3555–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dienstmann R, Vilar E, Tabernero J (2011) Molecular predictors of response to chemotherapy in colorectal cancer. Cancer J 17: 114–126. [DOI] [PubMed] [Google Scholar]
- 28. Generali D, Buffa FM, Berruti A, Brizzi MP, Campo L, et al. (2009) Phosphorylated ERalpha, HIF-1alpha, and MAPK signaling as predictors of primary endocrine treatment response and resistance in patients with breast cancer. J Clin Oncol 27: 227–234. [DOI] [PubMed] [Google Scholar]
- 29.Kornegoor R, Verschuur-Maes AH, Buerger H, Hogenes MC, de Bruin PC, et al.. (2011) Molecular subtyping of male breast cancer by immunohistochemistry. Mod Pathol. [DOI] [PubMed] [Google Scholar]
- 30. Irvin WJ Jr, Carey LA (2008) What is triple-negative breast cancer? Eur J Cancer 44: 2799–2805. [DOI] [PubMed] [Google Scholar]
- 31. Nishimura R, Arima N (2008) Is triple negative a prognostic factor in breast cancer? Breast Cancer 15: 303–308. [DOI] [PubMed] [Google Scholar]
- 32. Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, et al. (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13: 4429–4434. [DOI] [PubMed] [Google Scholar]
- 33. Kaplan HG, Malmgren JA (2008) Impact of triple negative phenotype on breast cancer prognosis. Breast J 14: 456–463. [DOI] [PubMed] [Google Scholar]
- 34. Rhee J, Han SW, Oh DY, Kim JH, Im SA, et al. (2008) The clinicopathologic characteristics and prognostic significance of triple-negativity in node-negative breast cancer. BMC Cancer 8: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, et al. (2007) The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 13: 2329–2334. [DOI] [PubMed] [Google Scholar]
- 36. Hugh J, Hanson J, Cheang MC, Nielsen TO, Perou CM, et al. (2009) Breast cancer subtypes and response to docetaxel in node-positive breast cancer: use of an immunohistochemical definition in the BCIRG 001 trial. J Clin Oncol 27: 1168–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang S, Yang H, Tong F, Zhang J, Yang D, et al. (2009) Response to neoadjuvant therapy and disease free survival in patients with triple-negative breast cancer. Gan To Kagaku Ryoho 36: 255–258. [PubMed] [Google Scholar]
- 38. Tan AR, Swain SM (2008) Therapeutic strategies for triple-negative breast cancer. Cancer J 14: 343–351. [DOI] [PubMed] [Google Scholar]
- 39. Damia G, D'Incalci M (2010) Genetic instability influences drug response in cancer cells. Curr Drug Targets 11: 1317–1324. [DOI] [PubMed] [Google Scholar]
- 40. Damia G, D'Incalci M (2007) Targeting DNA repair as a promising approach in cancer therapy. Eur J Cancer 43: 1791–1801. [DOI] [PubMed] [Google Scholar]
- 41. Damia G, Imperatori L, Stefanini M, D'Incalci M (1996) Sensitivity of CHO mutant cell lines with specific defects in nucleotide excision repair to different anti-cancer agents. Int J Cancer 66: 779–783. [DOI] [PubMed] [Google Scholar]
- 42. Tutt AN, Lord CJ, McCabe N, Farmer H, Turner N, et al. (2005) Exploiting the DNA repair defect in BRCA mutant cells in the design of new therapeutic strategies for cancer. Cold Spring Harb Symp Quant Biol 70: 139–148. [DOI] [PubMed] [Google Scholar]
- 43. Kennedy RD, D'Andrea AD (2006) DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol 24: 3799–3808. [DOI] [PubMed] [Google Scholar]
- 44. Willers H, Taghian AG, Luo CM, Treszezamsky A, Sgroi DC, et al. (2009) Utility of DNA repair protein foci for the detection of putative BRCA1 pathway defects in breast cancer biopsies. Mol Cancer Res 7: 1304–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gossage L, Madhusudan S (2007) Current status of excision repair cross complementing-group 1 (ERCC1) in cancer. Cancer Treat Rev 33: 565–577. [DOI] [PubMed] [Google Scholar]
- 46. Latimer JJ, Johnson JM, Kelly CM, Miles TD, Beaudry-Rodgers KA, et al. (2010) Nucleotide excision repair deficiency is intrinsic in sporadic stage I breast cancer. Proc Natl Acad Sci U S A 107: 21725–21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alexander BM, Sprott K, Farrow DA, Wang X, D'Andrea AD, et al. (2010) DNA repair protein biomarkers associated with time to recurrence in triple-negative breast cancer. Clin Cancer Res 16: 5796–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deans AJ, West SC (2011) DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11: 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kee Y, D'Andrea AD (2010) Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24: 1680–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, et al. (2006) DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 355: 983–991. [DOI] [PubMed] [Google Scholar]
- 51. Simon GR, Sharma S, Cantor A, Smith P, Bepler G (2005) ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest 127: 978–983. [DOI] [PubMed] [Google Scholar]
- 52. Friboulet L, Barrios-Gonzales D, Commo F, Olaussen KA, Vagner S, et al. (2011) Molecular Characteristics of ERCC1-Negative versus ERCC1-Positive Tumors in Resected NSCLC. Clin Cancer Res 17: 5562–5572. [DOI] [PubMed] [Google Scholar]
- 53. Carrassa L, Damia G (2011) Unleashing Chk1 in cancer therapy. Cell Cycle 10: 2121–2128. [DOI] [PubMed] [Google Scholar]
- 54. Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, et al. (2007) The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor/progesterone receptor/HER-2 breast carcinomas. Cancer Res 67: 6574–6581. [DOI] [PubMed] [Google Scholar]
- 55. Domagala P, Huzarski T, Lubinski J, Gugala K, Domagala W (2011) PARP-1 expression in breast cancer including BRCA1-associated, triple negative and basal-like tumors: possible implications for PARP-1 inhibitor therapy. Breast Cancer Res Treat 127: 861–869. [DOI] [PubMed] [Google Scholar]
- 56. Ossovskaya V, Koo IC, Kaldjian EP, Alvares C, Sherman BM (2010) Upregulation of Poly (ADP-Ribose) Polymerase-1 (PARP1) in Triple-Negative Breast Cancer and Other Primary Human Tumor Types. Genes Cancer 1: 812–821. [DOI] [PMC free article] [PubMed] [Google Scholar]