Abstract
Peptide antagonists of the human papillomavirus type 11 (HPV-11) E2-DNA association were identified using a filamentous bacteriophage random peptide library. Synthetic peptides antagonized the E2-DNA interaction, effectively blocked E2-mediated transcriptional activation of a reporter gene in cell culture, and inhibited E1-E2-mediated HPV-11 DNA replication in vitro. These peptides may prove to be useful tools for characterizing E2 function and for exploring the effectiveness of E2-inhibitor-based treatments for HPV-associated diseases.
Present estimates are that 24 million Americans are infected with human papillomavirus (HPV) and that 5 million new cases occur annually. As there are no specific antiviral therapies for HPV infection, there is a clear unmet need for the development of safe and effective therapies for HPV-associated disease.
Papillomaviruses lack the enzymes generally targeted by most currently available antiviral agents (e.g., virus-encoded DNA polymerases, reverse transcriptases, and proteases). Papillomaviruses do encode E1, a helicase-ATPase, but it has so far proven a difficult enzyme to develop for classic drug screening. As such, successful development of anti-HPV therapies may require identification of novel antiviral targets, such as viral transcription factors and replication proteins (2, 21, 25) such as the HPV-encoded E2 protein.
The 50-kDa E2 protein is comprised of three functional domains (19). The amino-terminal domain of E2 is necessary for viral trans-activation and for direct association with E1. The smaller carboxyl-terminal domain (E2C) encodes the DNA binding and dimerization functions. Linking the amino- and carboxyl-terminal domains is a small, poorly conserved hinge region. E2 function requires binding to a 12-bp palindromic nucleotide sequence, ACCN6GGT, that is present at several locations throughout the HPV genome and is repeated several times near the viral origin of replication. Once associated with DNA, E2 interacts with a variety of host cell transcription factors to modulate viral transcription. The papillomavirus E2 protein is also required for origin (ORI)-specific viral DNA replication. Because association of E1 with E2 enhances the affinity of both proteins for the viral ORI, DNA sequence-specific binding by E2 effectively mediates sequence-specific DNA binding by E1 (6).
Since the E2-DNA interaction is central to the coordination of the essential papillomavirus transcription and replication processes, disruption of E2-DNA binding should inhibit HPV replication. To this end, we constructed a filamentous bacteriophage 10-mer random peptide library and screened for sequences that disrupted the E2-DNA association by binding to the E2 protein.
Phage display selection of E2-DNA binding inhibitors.
We used peptide phage display (reviewed in reference 16) to identify peptide binders to bio-E2C, a biotinylated E2 fusion protein consisting of 88 carboxyl-terminal amino acids of the HPV type 11 (HPV-11) E2 DNA binding domain fused to the 99-residue polypeptide substrate of Escherichia coli biotin holoenzyme synthetase (3). Bio-E2C, expressed under the control of a T7 promoter in E. coli, was purified similarly to the method described by Alexander and Phelps (1).
A phage library, fGWX10, displaying 10-mer random peptide sequences was constructed as follows. DNA sequence encoding a Flag tag (AspTyrLysAspAspAspLys) and XhoI and KpnI restriction sites was inserted into the vector fTC (24). Dual BbsI sites were then engineered into the modified fTC vector by using XhoI- and KpnI-digested fTC, and the XhoI site was removed by site-directed mutagenesis, yielding the phage display vector fGW. fGW was digested with BbsI and ligated with a DNA cassette encoding a 30-amino-acid sequence containing a 10-residue stretch of random amino acids (NH2-EDGGSXXXXXXXXXXGGGGSGGGGSGGGGS, where X represents any of the 20 common amino acids). The DNA cassette encoding the 10-mer random peptide sequences was constructed by a synthetic ligase chain reaction procedure (10). Based on transformation efficiency and sequencing of primary transformants, the library was estimated to contain about 1010 different peptide sequences. A total of 2 × 1011 fGWX10 phage CFU in 100 μl of TBSDM buffer (25 mM Tris-HCl [pH 7.0], 150 mM NaCl, 5 mM dithiothreitol [DTT], 2 mM MgCl2) containing 0.5% milk was added to wells containing purified bio-E2C captured on NeutrAvidin-coated 96-well plates (Pierce, Rockford, Ill.). Phage screening was repeated for a total of six rounds essentially as described previously (23). For each round the ability of randomly picked single phage clones to bind to immobilized full-length E2 (FL-E2; purified according to the method described in reference 11) was assessed by phage enzyme-linked immunosorbent assay (ELISA) (4). Sequencing of DNA from positive phage clones for each round revealed two classes of E2C-binding phage clones, encoding CF/LXC and ES/TWXXWWL/A E2-binding motifs (Table 1). Of the phage in the ES/TWXXWWL/A motif class, phage clone 6N30 showed the best specific binding to FL-E2 as determined by phage ELISA (data not shown). However, the strongest binding signal was obtained with the 6N40 phage clone, which contains the CF/LXC motif (data not shown).
TABLE 1.
Peptide sequence groups selected from the 10-mer phage library
| Phage clone | No. of identical sequences in round of biopanning:
|
Peptide sequencec | |||||
|---|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | 4th | 5th | 6th | ||
| Group 1 | |||||||
| 2N18 | 0 | 1 | 0 | 0 | 0 | 0 | S V F Y A CaF A CbF |
| 4N27 | 0 | 0 | 0 | 1 | 0 | 0 | W S E Q C F T C W W |
| 6N40 | 0 | 0 | 1 | 6 | 24 | 9 | F M C L W C G E V H |
| Motif | C F/L X C | ||||||
| Group 2 | |||||||
| 6N30 | 0 | 0 | 1 | 2 | 7 | 11 | W I E S W P E W W L |
| 2N19 | 0 | 2 | 0 | 0 | 0 | 0 | S W R Q W W L A G |
| 6X27 | 0 | 0 | 1 | 1 | 0 | 1 | N A E S W H A W W L |
| 6N21 | 0 | 0 | 0 | 0 | 7 | 27 | M E T W E A W W A G |
| Motif | E S/T W X X W W L/A | ||||||
Position of cysteine numbered as 3.
Position of cysteine numbered as 6.
Residues in boldface are relatively conserved.
Determination of equilibrium inhibition constants (Kis) for peptide antagonism of E2-DNA interaction by surface plasmon resonance (SPR) spectroscopy.
Apparent equilibrium Kis for synthetic peptides (Table 2) were determined using a solution affinity method (14) on a BIAcore 3000 instrument (BIAcore, Inc., Piscataway, N.J.). We captured biotinylated E2BS7592 DNA (see reference 1) on a streptavidin sensor chip to yield ∼600 resonance units of immobilized DNA. Both the E2BS7592 and control flow cells were blocked with biotinylated bovine serum albumin. Samples containing fixed concentrations of E2 proteins (15 nM E2C, purified by a method similar to that described in reference 1, or 50 nM FL-E2, purified according to the method of reference 11) in HEPES-buffered saline (0.01 M HEPES [pH 7.4], 0.15 M NaCl, 3 mM EDTA, 0.005% polysorbate 20, 1 mM DTT) and increasing amounts of peptide (0.5 to 100 μM) were injected over the sensor chip. For each of three independent experiments, a calibration curve consisting of serially diluted E2 protein only (between 0.3 and 100 nM E2C or FL-E2) was performed to estimate the free E2 protein concentration in each of the E2-peptide mixtures. Competition curves in at least two independent experiments were analyzed using the solution affinity analysis in the BIAcore evaluation software.
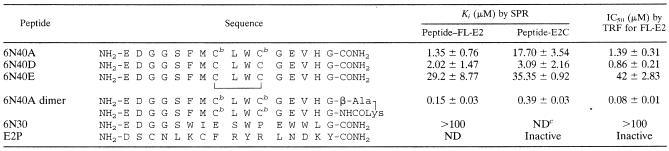
TABLE 2.
Peptide Kis and IC50s for HPV-11 E2-binding peptides for antagonism of E2-DNA interactionsa
Apparent Kis were calculated from two or more independent SPR analyses. IC50s were determined by measuring peptide-mediated inhibition of E2-DNA association as described in the text.
Cysteine protection group Acm, -CH2-NH-COCH3.
ND, not determined.
The Kis for selected peptides are shown in Table 2. 6N40D had the highest affinity of the set of 6N40-based peptides, followed by 6N40A (Acm-protected cysteines), and 6N40E (cyclic disulfide version). Individually, the peptides bound with similar affinities to FL-E2 and E2C, with the exception of 6N40A, which had a higher affinity for FL-E2. The fact that 6N40A, with protected cysteines, possessed FL-E2-binding and DNA-antagonist activities similar to those of native 6N40D (Table 2) indicated that E2-DNA antagonism did not require covalent cross-linking of the peptide with a cysteine in the E2 DNA recognition helix (18). Mass spectroscopy analyses of 6N40D solutions incubated with FL-E2 or E2C provided further evidence against peptide-protein cross-linking (data not shown). The cyclic disulfide version, 6N40E, had a significantly decreased effect, with a 14-fold increase of the Ki for the FL-E2-DNA interaction, but still possessed greater affinity for FL-E2 than did 6N30.
In an effort to increase the affinity of E2-DNA antagonist peptides for E2, a synthetic dimer of peptide 6N40A was synthesized. The dimeric 6N40A peptide showed a 9- to 45-fold-increased affinity compared to the monomeric 6N40A peptide for FL-E2 and E2C, respectively. This increase in apparent peptide affinity is most likely a consequence of the chelate effect (5, 20), wherein linking peptides increases the effective concentration of E2-binding sites.
A peptide derived from the E1-E2-binding domain has been shown to inhibit the E2-E1 interaction (15). We therefore synthesized and tested peptide E2P (Table 2), which is derived from the DNA recognition helix of E2 protein (18); however, E2P did not prevent DNA binding to FL-E2.
It was possible that peptide 6N40D could bind directly to E2BS DNA instead of to HPV E2 protein, thereby inhibiting the HPV E2-DNA interaction. However, SPR-based DNA-peptide binding experiments using 6N40D peptide and immobilized E2BS DNA showed no evidence of 6N40D binding to E2BS DNA (data not shown). Additionally, phage ELISA experiments using immobilized biotinylated E2BS DNA and 6N40 phage gave no evidence of phage-DNA association (data not shown).
Measurement of peptide IC50s for antagonism of FL-E2-DNA association by using TRF.
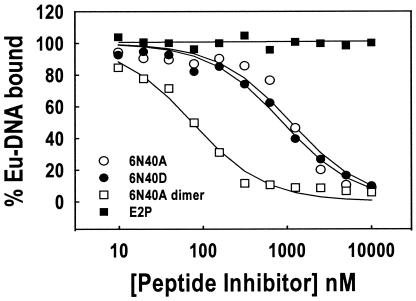
As another measure of E2-DNA binding antagonism, time-resolved fluorescence (TRF) assays were performed with synthetic peptides versus the Eu-DNA (10 nM Kd E2C-binding sequence from reference 1) and FL-E2 interaction. The inhibition curves and resulting 50% inhibitory concentration (IC50) results for peptide-mediated inhibition of the E2-DNA interaction are shown in Fig. 1 and summarized in Table 2, respectively. Consistent with the SPR results, peptides 6N40A and 6N40D were potent antagonists of FL-E2-DNA association, and the 6N40A dimer peptide antagonized E2-DNA association most efficiently (Table 2). Finally, peptide E2P was inactive for antagonism of FL-E2-DNA binding by TRF assay.
FIG. 1.
Comparison of synthetic peptides by TRF. IC50 results are reported in Table 1. One microgram of anti-E2 monoclonal antibody 93.2a12 (anti-E2 antibody generated at GlaxoSmithKline) was added to each well in a 96-well plate and incubated at 4°C overnight. Wells were blocked with 1% milk in Tris-buffered saline and then washed with TBSDM buffer. Peptide samples were serially diluted in TBSDM containing 10 nM FL-E2 and incubated at room temperature for 30 min. Annealed oligonucleotides were end labeled for 30 min at room temperature in the presence of 16 nM (each) dATP, dTTP, and dGTP plus 16 nM Eu-dCTP and 0.017 U of Klenow enzyme/μl. Eu-DNA (100 pM) was then added to each FL-E2 sample and incubated at room temperature for an additional 30 min. The Eu-DNA-FL-E2 reaction mixtures were transferred into the monoclonal antibody-coated wells and incubated at room temperature for 1 h. Each well was then washed, and 100 μl of TRF enhancement solution (EG & G Wallac, Gaithersburg, Md.) was added and incubated for 10 min at room temperature. Bound Eu-DNA fluorescence was then determined on a Victor 1420 multiple label counter (EG & G Wallac). Data from three independent experiments were analyzed with the equation f = Vmax{1 − [x/(K + x)]}, where f is the signal from bound Eu-DNA, Vmax is the maximum signal, x is the peptide concentration, and K is the IC50 of synthetic peptides for inhibition of the FL-E2-Eu-DNA interaction.
Peptide-mediated inhibition of transcriptional activation in cell culture.
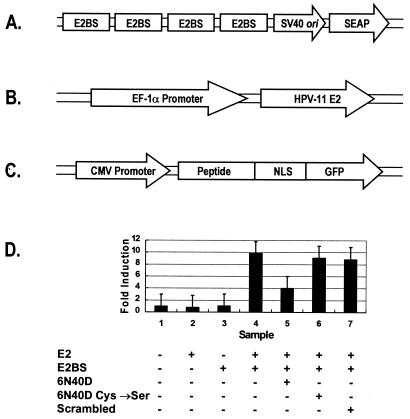
We designed a transient-transcription assay to test if peptides could inhibit E2-DNA binding in cell culture. The reporter plasmid, pSEAP-2 (Fig. 2A), contained four tandem E2-binding sites upstream of a minimal simian virus 40 promoter and a secreted alkaline phosphatase (SEAP) reporter gene. When the E2 expression plasmid pFastBac-E2 (Fig. 2B) was cotransfected into cells with the pSEAP-2 reporter plasmid, a ∼10-fold-increased trans-activation was seen based on SEAP reporter values (Fig. 2D).
FIG. 2.
Inhibition of E2-mediated reporter gene transcription by 6N40D peptide expressed in Vero cells. (A) Four tandem repeats of the E2-binding-site palindromes (nucleotides 28 to 66 within the HPV-11 origin of replication) were inserted upstream of the minimal simian virus 40 (SV40) promoter and sequence encoding a secreted form of human placental alkaline phosphatase to create the reporter plasmid pSEAP-2. (B) An HPV-11 E2 expression plasmid, pFastBac-E2, was generated by inserting the open reading frame sequence for full-length HPV-11 E2 into pFastBac-EF1αP. Expression from this vector is directed from the EF1α promoter. (C) A DNA cassette encoding the 6N40D E2 inhibitor peptide and a nuclear localization signal peptide (NLS) was inserted into the enhanced GFP fusion vector, pEGFP-N1 (Clontech), and then subcloned into the pFastBac-MAM-1 expression vector, yielding the baculovirus expression construct pFastBac-MAM/6N40D. pFastBac-MAM/6N40D derivative vectors were made which contained the NLS-GFP fusion with double Cys→Ser mutated 6N40D peptide sequence (pFastBac-MAM/6N40DCys→Ser) or the NLS-GFP fusion with a scrambled 6N40D peptide sequence (NH2-SCGDEHMGLECGWVGF-CONH2) (pFastBac-MAM/scrambled). CMV, cytomegalovirus. (D) To test the effects of 6N40D peptides on E2-mediated transcription control in vivo, Vero cells were transiently transfected with 0.125 μg of the reporter plasmid pSEAP-2 and/or 0.5 μg of the E2 transcription activator plasmid pFastBac-E2. The transfected cells were inoculated 24 h later with baculoviruses that expressed 6N40D, double Cys→Ser mutated 6N40D, or scrambled 6N40D-NLS-GFP fusion proteins, according to the bottom panel. Medium supernatant (50 μl) was assayed for SEAP activity 48 h after the transductions. Fold induction represents SEAP activity in Vero cells with mock-transfected cells as reference. At least three independent experiments were performed in triplicate, and the average values are shown. The error bars indicate the standard deviations.
Baculovirus expression systems have been used extensively to characterize effects of protein expressed in mammalian cell lines (e.g., see references 8 and 22). To determine the effects of 6N40 inhibitor peptide on E2-mediated trans-activation, a baculovirus construct, pFastBac-MAM/6N40-NLS-GFP, was created that would constitutively express a fusion polypeptide (8, 22). This fusion polypeptide contained the 6N40 amino acid sequence, a nuclear localization signal (NLS), and green fluorescent protein (GFP) as an indicator for baculovirus transduction. When the pFastBac-MAM/6N40-NLS-GFP expression construct (Fig. 2C) was transduced into cells containing the pFastBac-E2 expression plasmid, a 58% ± 9% reduction in E2-mediated trans-activation of the SEAP reporter gene was observed, consistent with peptide-mediated inhibition of trans-activation. Control constructs expressing the double Cys→Ser mutated peptide or a scrambled 6N40D peptide sequence showed no significant reduction in SEAP reporter activity. These data were confirmed in additional transient-expression assays using the E2 inhibitor peptide 6N40-NLS-GFP expressed from a mammalian expression plasmid rather than from a baculovirus (data not shown). The levels of fusion mRNA and fusion proteins for the different experiments were determined to be equivalent by using anti-GFP DNA probes and anti-GFP antibodies (data not shown), though global transcription levels were not measured.
Cell-free DNA replication assays.
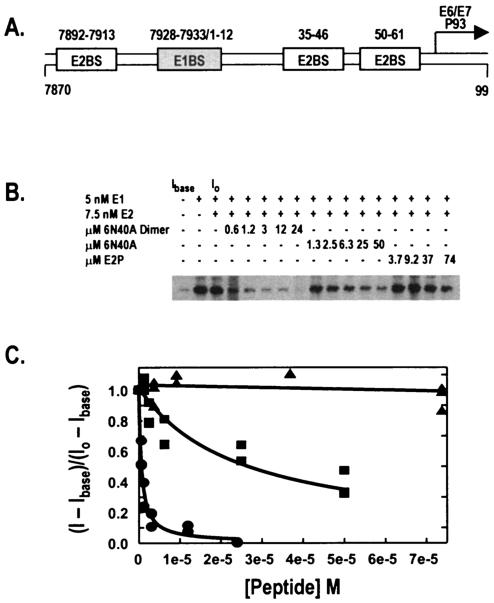
An in vitro HPV-11 DNA replication assay was used as described previously (6, 13) to more directly measure the effects of E2-DNA antagonist peptides on E2-stimulated DNA replication. Purified HPV-11 E1 helicase and E2 protein were added to a cytoplasmic preparation of 293 cells and a DNA replication template (pUC18/7870-99; Fig. 3A) containing the HPV-11 origin of replication. Assay conditions were optimized to maximize the dose response to increasing E2 protein. HPV-11 E1 and E2 proteins were homogeneously purified as previously described (11). Viral proteins E1 and E2 and test peptides were added as indicated (Fig. 3B). Addition of E2 caused a two- to threefold stimulation of origin-dependent viral replication (data not shown). Digested replication products were run on a 1% agarose gel, and the gel was dried and exposed to RXB film (Denville Scientific). An image of a representative gel is shown in Fig. 3B, and plotted data points are shown in Fig. 3C. Results were quantified with a PhosphorImager (Molecular Dynamics).
FIG. 3.
Inhibition of HPV-11 in vitro DNA replication by peptides 6N40A and 6N40A dimer. (A) pUC18/7870-99, an in vitro DNA replication template that contains 182 bp (nucleotides 7870 to 0099) from the HPV-11 locus control region inserted into pUC18. (B) NdeI-digested replication products from cell-free replication assays using HPV-11 ORI-containing plasmid (pUC18/7870-99) and increasing concentrations of 6N40A, 6N40A dimer, and E2P. E1, E2, and peptide were added as noted. Lanes corresponding to initial and baseline intensities of counts (I0 and Ibase, respectively) are indicated. (C) Replication assays with increasing peptide concentrations were quantified by both PhosphorImager and filter-binding techniques, and the resulting data for both were fitted to two-parameter hyperbolic decay curves. 6N40A dimer inhibited replication (circles) with an IC50 of 631 nM, while monomeric 6N40A inhibited replication (squares) less efficiently (IC50 = 27.3 μM). Peptide E2P (triangles) did not inhibit in vitro DNA replication at the concentrations tested.
Addition of peptide 6N40A to the replication mixture suppressed origin-dependent DNA replication with an IC50 of 27 μM. Consistent with other results described above, dimeric 6N40A peptide was predictably an even more potent antagonist of E1-E2-dependent DNA replication, with an IC50 of 670 nM, while peptide E2P was inactive for suppression of in vitro DNA replication.
HPV E2-DNA binding as an antiviral target.
The binding of the E2 protein to viral origin DNA is central to the coordination of viral transcription and replication. Several investigators have attempted to block E2 expression or to antagonize E2 functions. Cowsert et al. used phosphorothioate antisense oligonucleotides to block E2 mRNA translation and E2-mediated transcriptional trans-activation in cultured cells (7, 9). More recently, a peptidic nucleic acid was designed to bind to a double-stranded DNA bovine papillomavirus E2-binding site, thereby antagonizing E2-DNA binding by peptide-DNA association. This peptidic nucleic acid had some activity in a cell culture-based assay (17). A 15-mer peptide derived from the N terminus of the E2 protein was shown to inhibit the protein-protein interaction between E1 and E2 and also diminished HPV DNA replication in vitro (15). Finally, using a structure and activity relationship by nuclear magnetic resonance approach, a small molecule was generated that inhibited E2 binding to E2BS DNA in vitro with an IC50 of 10 μM (12). Despite these efforts, no effective E2-DNA binding antagonists have been developed that can be readily delivered into HPV-infected cells.
Our cell culture-based transcriptional activation experiments demonstrated that 6N40D-NLS-GFP inhibited E2-stimulated transcription, presumably due to the peptide component of the fusion polypeptide antagonizing the E2-ORI association. Theoretically, a higher-affinity antagonist peptide would more fully inhibit E2-DNA binding in cell culture; however, it is also possible that the NLS and GFP regions of the fusion peptide caused some steric hindrance of 6N40D-E2 association. Note that transcription inhibition studies with phosphorothioate oligonucleotide E2-DNA binding antagonists (7) indicated that an oligonucleotide of lower E2-binding affinity caused only a 50% decrease in trans-activation activity, while an oligonucleotide of higher affinity decreased trans-activation 10-fold. Thus, the incomplete suppression of E2-mediated trans-activation by the 6N40D-NLS-GFP fusion protein might simply be a reflection of suboptimal E2-DNA binding antagonism.
Taken together, our results indicate that the E2-DNA interaction can be antagonized successfully both in vitro and in cultured cells. We believe that this is the first example of the successful use of filamentous phage biopanning for the identification of protein-DNA binding antagonists that function specifically through direct binding to protein rather than to DNA. The ability of these peptides to inhibit the interaction between E2C and DNA suggests that the protein-peptide interaction site is within the carboxyl-terminal DNA binding domain of E2.
Finally, our results indicate that the E2-ORI interaction, which is required for the trans-activation and replication functions of E2, is a potential target for antiviral drug development. The placement of the E2 protein as a central regulator of viral transcription and replication allows antagonism of a single molecular interaction, E2 with its 12-bp palindromic binding site, to suppress two primary processes required for development and propagation of papillomavirus infection.
Acknowledgments
We thank Kathy Lindley for generating anti-E2 monoclonal antibody 93.2a12 and Anne Hassell for providing E2C. K.A.A. acknowledges the support of the Duke University Cancer Center Tissue Culture Facility.
K.A.A. and K.A.H. were supported by National Institutes of Health grant R01 CA81214.
REFERENCES
- 1.Alexander, K. A., and W. C. Phelps. 1996. A fluorescence anisotropy study of DNA binding by HPV-11 E2C protein: a hierarchy of E2-binding sites. Biochemistry 35:9864-9872. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, K. A., and W. C. Phelps. 2000. Recent advances in diagnosis and therapy of human papillomaviruses. Expert Opin. Investig. Drugs 9:1753-1765. [DOI] [PubMed] [Google Scholar]
- 3.Barker, D. F., and A. M. Campbell. 1981. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J. Mol. Biol. 146:451-467. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj, D., S. S. Singh, S. Abrol, and V. K. Chaudhary. 1995. Monoclonal antibodies against a minor and the major coat proteins of filamentous phage M13: their application in phage display. J. Immunol. Methods 179:165-175. [DOI] [PubMed] [Google Scholar]
- 5.Brown, B. M., and R. T. Sauer. 1993. Assembly of the Arc repressor-operator complex: cooperative interactions between DNA-bound dimers. Biochemistry 32:1354-1363. [DOI] [PubMed] [Google Scholar]
- 6.Chao, S.-F., W. J. Rocque, S. Daniel, L. E. Czyzyk, W. C. Phelps, and K. A. Alexander. 1999. Subunit affinities and stoichiometries of the human papillomavirus type 11 E1:E2:DNA complex. Biochemistry 38:4586-4594. [DOI] [PubMed] [Google Scholar]
- 7.Clark, P. R., M. L. Roberts, and L. M. Cowsert. 1998. A novel drug screening assay for papillomavirus specific antiviral activity. Antivir. Res. 37:97-106. [DOI] [PubMed] [Google Scholar]
- 8.Condreay, J. P., S. M. Witherspoon, W. C. Clay, and T. A. Kost. 1999. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. USA 96:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowsert, L. M., M. C. Fox, G. Zon, and C. K. Mirabelli. 1993. In vitro evaluation of phosphorothioate oligonucleotides targeted to the E2 mRNA of papillomavirus: potential treatment for genital warts. Antimicrob. Agents Chemother. 37:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, S. J., C. R. MacKenzie, and S. A. Narang. 1993. Simultaneous randomization of antibody CDRs by a synthetic ligase chain reaction strategy. Nucleic Acids Res. 21:4418-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon, E. P., G. L. Pahel, W. J. Rocque, J. A. Barnes, D. C. Lobe, M. H. Hanlon, K. A. Alexander, S.-F. Chao, K. Lindley, and W. C. Phelps. 2000. The E1 helicase of human papillomavirus type 11 binds to the origin of replication with low sequence specificity. Virology 270:345-357. [DOI] [PubMed] [Google Scholar]
- 12.Hajduk, P. J., J. Dinges, G. F. Miknis, M. Merlock, T. Middleton, D. J. Kempf, D. A. Egan, K. A. Walter, T. S. Robins, S. B. Shuker, T. F. Holzman, and S. W. Fesik. 1997. NMR-based discovery of lead inhibitors that block DNA binding of the human papillomavirus E2 protein. J. Med. Chem. 40:3144-3150. [DOI] [PubMed] [Google Scholar]
- 13.Hartley, K. A., and K. A. Alexander. 2002. Human TATA binding protein inhibits human papillomavirus type 11 DNA replication by antagonizing E1-E2 protein complex formation on the viral origin of replication. J. Virol. 76:5014-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson, R., A. Michaelsson, and L. Mattsson. 1991. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor-based analytical system. J. Immunol. Methods 145:229-240. [DOI] [PubMed] [Google Scholar]
- 15.Kasukawa, H., P. M. Howley, and J. D. Benson. 1998. A fifteen-amino-acid peptide inhibits human papillomavirus E1-E2 interaction and human papillomavirus DNA replication in vitro. J. Virol. 72:8166-8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koivunen, E., W. Arap, D. Rajotte, J. Lahdenranta, and R. Pasqualini. 1999. Identification of receptor ligands with phage display peptide libraries. J. Nucl. Med. 40:883-888. [PubMed] [Google Scholar]
- 17.Kurg, R., U. Langel, and M. Ustav. 2000. Inhibition of the bovine papillomavirus E2 protein activity by peptide nucleic acid. Virus Res. 66:39-50. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto, T., N. Nakashima, K. Takase, H. Hirochika, and H. Mizuno. 1997. A mutation study of the DNA binding domain of human papillomavirus type 11 E2 protein. J. Biochem. 121:138-144. [DOI] [PubMed] [Google Scholar]
- 19.McBride, A. A., H. Romanczuk, and P. M. Howley. 1991. The papillomavirus E2 regulatory proteins. J. Biol. Chem. 266:18411-18414. [PubMed] [Google Scholar]
- 20.Page, M. I., and W. P. Jencks. 1971. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc. Natl. Acad. Sci. USA 68:1678-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelps, W. C., J. A. Barnes, and D. C. Lobe. 1998. Molecular targets for human papillomaviruses: prospects for antiviral therapy. Antivir. Chem. Chemother. 9:359-377. [DOI] [PubMed] [Google Scholar]
- 22.Sarkis, C., C. Serguera, S. Petres, D. Buchet, J. L. Ridet, L. Edelman, and J. Mallet. 2000. Efficient transduction of neural cells in vitro and in vivo by a baculovirus-derived vector. Proc. Natl. Acad. Sci. USA 97:14638-14643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott, J. K., and G. P. Smith. 1990. Searching for peptide ligands with an epitope library. Science 249:386-390. [DOI] [PubMed] [Google Scholar]
- 24.Smith, M. M., L. Shi, and M. Navre. 1995. Rapid identification of highly active and selective substrates for stromelysin and matrilysin using bacteriophage peptide display libraries. J. Biol. Chem. 270:6440-6449. [DOI] [PubMed] [Google Scholar]
- 25.Underwood, M. R., L. M. Shewchuk, A. M. Hassell, and W. C. Phelps. 2000. Searching for antiviral drugs for human papillomaviruses. Antivir. Ther. 5:229-242. [PubMed] [Google Scholar]