Abstract
Introduction
The purpose of the study was to describe demographic and clinical characteristics and outbreak handling of a large methicillin-resistant Staphylococcus aureus (MRSA) outbreak in a neonatal intensive care unit (NICU) in Denmark June 25th–August 8th 2008, and to identify risk factors for MRSA transmission.
Methods
Data were collected retrospectively from medical records and the Danish Neobase database. All MRSA isolates obtained from neonates, relatives and NICU health care workers (HCW) as well as environmental cultures were typed.
Results
During the 46 day outbreak period, 102 neonates were admitted to the two neonatal wards. Ninety-nine neonates were subsequently sampled, and 32 neonates (32%) from 25 families were colonized with MRSA (spa-type t127, SCCmec V, PVL negative). Thirteen family members from 11 of those families (44%) and two of 161 HCWs (1%) were colonized with the same MRSA. No one was infected. Five environmental cultures were MRSA positive. In a multiple logistic regression analysis, nasal Continuous Positive Airway Pressure (nCPAP) treatment (p = 0.006) and Caesarean section (p = 0.016) were independent risk factors for MRSA acquisition, whereas days of exposure to MRSA was a risk factors in the unadjusted analysis (p = 0.04).
Conclusions
MRSA transmission occurs with high frequency in the NICU during hospitalization with unidentified MRSA neonates. Caesarean section and nCPAP treatment were identified as risk factors for MRSA colonization. The MRSA outbreak was controlled through infection control procedures.
Introduction
Infections due to MRSA have become an increasing clinical problem, causing considerable morbidity and mortality worldwide [1]. Interestingly, some MRSA clones have had the capacity for pandemic spread, while others gave predominantly local epidemics [1], [2]. This globally changing epidemiology has, in part, been caused by community-associated MRSA (CA-MRSA) – new MRSA types initially not found in the hospital and characterized by carrying small SCCmec cassette type IV or V and sometimes the PVL gene [2]. This increase of CA-MRSA has also been seen in Denmark, where the annual number of new MRSA cases has increased significantly from 100 in 2002 to 1293 cases in 2011 [3], though the prevalence of MRSA in Staphylococcus aureus bacteraemias still remains low (1.6%–21 of 1293 patients in 2011) [3]. Major health-care associated MRSA outbreaks are rare in Denmark with 22 identified outbreaks in 2011– the largest outbreaks occurred at neonatal departments in the Copenhagen area and Zealand, comprising a total of 26 cases [3]. Furthermore, we have recently shown, in other Copenhagen hospitals, that CA-MRSA has a 9.3 fold lower risk of starting an outbreak than HA-MRSA clones [4]. Interestingly, neonates are highly susceptible for staphylococcal colonization within days of birth, but as they grow older many lose carriage by the age of two [4]–[8]. In some cases colonization will be with MRSA, and MRSA outbreaks in NICU’s have previously been reported in numerous countries (Germany [9], Great Britain [10], [11] Israel [12], [13] Japan [14] Scotland [15], Taiwan [16], USA [17], [18]). Most of these outbreaks have been caused by HA-MRSA, but many of these countries have identified CA-MRSA in the community. Thus the spread of CA-MRSA in their NICU’s may either be introduced by the families or hospital-acquired from HCWs, the NICU environmental, or by inter-hospital transfer of neonates [10], [15], [16], [19], [20]. MRSA outbreaks in NICU’s in countries with low MRSA prevalence are uncommon and have generally been small [21], [22].
When screening for MRSA is used in outbreak control, many neonates are identified as MRSA carriers, but invasive infections have been seen in 14–26% of cases [23], [24].
Attempts have been made to control MRSA in the NICU through treatment of colonized infants, [14], [25], [26]. Poor treatment success has been associated to MRSA colonization of the pharynx [14] and although many NICU use chlorhexidine gluconate body wash, there are safety issues regarding usage in preterm infants and newborns [27].
The purpose of this study was to describe demographic and clinical characteristics and risk factors associated with the first and largest hospital associated (HA)-MRSA outbreak in a NICU in Denmark. This outbreak was caused by introduction of a CA-MRSA.
Materials and Methods
The Neonatal Ward
The Neonatal ward at Glostrup Hospital is a level II care centre that receives neonates from three hospitals. The Neonatal ward has two units: The neonatal intensive care unit (NICU) with 20 beds is located on the 6th floor, while the special care baby unit (SCBU) has 10 beds on the 2nd floor. Staffing of these units is by separate nursing teams; but NIC specialists and obstetricians move between neonates. The NICU has a large room for four neonates, with 3 open incubators, facilities for nasal continuous positive airway pressure (nCPAP) and short-term ventilator treatment. A further eight rooms can hold two neonates and five of these rooms can be used for nCPAP. The eight rooms in the SCBU are designed for 10 infants and their mothers.
Neonates from 28 weeks of gestational age and/or with a birth weight of at least 800 grams are admitted to the units. Neonates requiring mechanical ventilation are transferred to a tertiary care centre in Copenhagen, as are infants needing surgical or more complex interventions. Neonates, who have been transferred to the tertiary care centre, will often return when stabilized. NICU neonates are often transferred to the SCBU before being discharged.
The Neonatal ward is open all hours for parents. In the Neonatal ward, the parents are invited to participate in the care of their child under guidance from the staff. Most of the parents take part in tube feeding, bathing and general care of the child. Mothers are supported in early breastfeeding. The parents share a small kitchen with dining facilities and have access to one toilet in each unit.
The MRSA Outbreak
The outbreak was discovered on July 28th 2008, when MRSA was isolated from a pharyngeal swab from a triplet, who had received prolonged nCPAP. Following MRSA detection, the triplets were isolated and screened for MRSA. The triplets had been transferred from the tertiary care centre NICU in Copenhagen 34 days prior to identification of the outbreak. They were born at the tertiary care centre, and during the first 15 days of hospitalization they were located in a room adjacent to an isolation room, with a neonate isolated due to MRSA spa-type t127. A further 15 neonates were transferred to Glostrup Hospital from the same tertiary care centre NICU during the outbreak period; none of them were MRSA colonized. The triplets were, for this reason, considered to be the index patients at Glostrup Hospital, and their admittance date June 25th marked the beginning of the MRSA exposure period. All 28 neonates hospitalized at the Neonatal ward were isolated and sampled, and the 2 neonatal units were closed for new admittances on August 8th, effectively ending the exposure period.
An outbreak management group was established with representatives from the hospital management, pediatric department and daily managers of the two units, a clinical microbiologist and an infection control nurse. The infection control nurse instructed the staff on infection control measures focusing on hand hygiene and hand disinfection, environmental disinfective cleaning and use of protective equipment; gloves and isolation gowns when caring for the neonates. The staff was tested August 5th-13th or as soon as possible after those dates. The parents received both written and oral information about the outbreak, hand hygiene and disinfection. All neonates discharged during the exposure period were recalled and screened for MRSA. The remaining NICU neonates were transferred to the SCBU on August 25th, and the NICU was temporarily closed, cleaned and disinfected with 3 cycles of vaporised of H2O2 and silver ions using SterinisT, (Gloster Sante Europe) [28]. The last MRSA colonized neonates were discharged from the SCBU (September 19th), 52 days after the index patient was tested positive. The SCBU was then thoroughly cleaned and disinfected with SterinisT. During the outbreak, none of the colonized neonates, family members or HCW developed an infection with MRSA.
Study Design
All neonatal patients, who had been admitted to the Neonatal ward during the outbreak period, were included in the study. Demographic and clinical data were collected from medical records and from the database, Neobase. Neobase was established in 1995 and registers data from all neonatal wards in Denmark on neonates, while they are hospitalized. Data registered were: date of birth, gender, vaginal delivery or caesarean section, singleton or multiple birth, weight and gestational age at birth, Apgar score at 1 and 5 minutes, asphyxia at birth or chronic lung disease (defined as: Neonate with gestational age (GA) <32 weeks and continued need of oxygen treatment at 36 weeks postmenstrual age), duration of nCPAP treatment, peripheral venous catheter (PVC), antibiotic therapy, dates of admission and discharge or transfer to other hospitals. Discrepancies between the two datasets were thoroughly checked and resolved.
When patients were admitted to the Neonatal ward, the Guardians signed an informed consent that their childs data would be stored in the National database Neobase. This database is in accordance with the rules of the Danish Data Protection Agency and the present study was approved by them (GLO-2009-06). As MRSA is a notifiable disease in Denmark, the study did not require approval from the regional ethical committee but adhered to the ethical guidelines of the hospital. After creation of the research database the data were anonymized.
Surveillance Cultures
Surveillance swabs from all neonates were obtained from nose, throat, axilla, perineum, and from possible infection sites (urine, skin, eyes). MRSA screening (nose and throat) was performed on all household relatives of MRSA colonized children and on all healthcare workers (HCW) of the Neonatal ward. Swabs were transported in Stuart’s medium (SSI, Copenhagen, Denmark) to the Department of Clinical Microbiology.
Environmental Cultures
Direct environmental cultures were performed from the room of the last patient with MRSA discharged from the NICU and SBCU, respectively. Cultures were performed before cleaning, after cleaning and disinfection with persulphate (VirkonT), and after the subsequent disinfection with the dry mist generator SterinisT. Follow-up cultures were performed after 6 and 15 weeks.
Cultures were performed from 10 locations: alarm bottom, water tap, water handle, alcohol dispenser, bed railing, chair seat, arm rest, laundry cupboard handle, wall and floor. Culture of inanimate surfaces was performed with a 10 cm2 staphylococci/Enterobacteriaceae dip slide containing a double agar with neutralizer for detergent and disinfectants (PC2TN) and Baird Parker agar (BV) (Biotrace International, Bridgend, UK). Dip slides were pressed firmly onto the surface, incubated at 35°C and inspected for growth after 1, 2 and 5 days.
MRSA Identification
Surveillance swabs (from neonates) or a sweep through the colonies from dip slides (environmental cultures) were cultured in a MRSA selective enrichment broth including cefoxitin (Department of Clinical Microbiology, Herlev, Denmark) overnight at 35°C. The following day, 1 µL of the enrichment broth was cultured on chromogenic agar plates (MRSA-ID agar®, BioMeriux) and blood agar plates. S. aureus isolates were identified by a positive Staphaurex® test (Remel Europe Ltd., Dartford, UK) and a positive coagulase test (Department of Clinical Microbiology, Herlev). All MRSA isolates were mecA positive, Panton-Valentine leukocidin (PVL) gene negative by PCR, spa typed by sequencing the staphylococcal protein A gene and SCCmec typed as previously described [29], [30]. Antimicrobial susceptibility was performed by disc diffusion for penicillin, erythromycin, clindamycin, gentamicin, vancomycin, linezolid, moxifloxacin, rifampicin and fusidic acid according to SRGA recommendations [31]. An isolate from this outbreak (H597) has been whole genome sequenced (Illumina MiSeq). The potential virulence gene content was identified by a BLAST analysis of the assembled genome against a database of 143 unique DNA sequences designed to detect gene (groups) generally believed to be related to virulence in S. aureus with a selected threshold equal to 90.00% identity within at least 60% of a given gene (Table 1).
Table 1. Virulence gene profile of a MRSA isolate from the neonatal outbreak (Mette Theilgaard Christiansen, personal communication).
| Virulence factors | Relatedgenes | Nucleotidehomology | Virulence factors | Relatedgenes | Nucleotidehomology | Virulence factors | Relatedgenes | Nucleotidehomology |
| Adherence | Exoenzyme | Toxin | ||||||
| Autolysin | atl | 100.00% | Cysteine protease | sspB | 100.00% | Beta hemolysin | hlb | 100.00% |
| Cell wall associated fibronectin binding protein | ebh | 93.57% | sspC | 99.70% | Delta hemolysin | hld | 100.00% | |
| Clumping factor B | clfB | 92.61% | Hyaluronate lyase | hysA | 100.00% | Exfoliative toxin type A | eta | 99.47% |
| Collagen adhesion | cna | 98.51% | Lipase | lip | 100.00% | Exotoxin/superantigen-like proteins | set2 | 100.00% |
| Extracellular adherence protein/MHC analogous protein | eap/map | 92.92% | geh | 100.00% | set3 | 100.00% | ||
| Fibronectin binding proteins | fnbA | 97.93% | Serine protease | splA | 100.00% | set4 | 100.00% | |
| fnbB | 100.00% | splB | 100.00% | set16 | 100.00% | |||
| Intercellular adhesin | icaR | 100.00% | splD | 100.00% | set17 | 100.00% | ||
| icaA | 100.00% | splE | 100.00% | set18 | 100.00% | |||
| icaD | 100.00% | splF | 100.00% | set20 | 100.00% | |||
| icaB | 100.00% | Serine V8 protease | sspA | 97.78% | set21 | 100.00% | ||
| icaC | 100.00% | Staphylokinase | sak | 100.00% | set22 | 100.00% | ||
| Ser-Asp rich fibrinogen-binding proteins | sdrC | 92.14% | Thermonuclease | nuc | 100.00% | set23 | 99.86% | |
| sdrD | 96.77% | Secretion system | set24 | 100.00% | ||||
| sdrE | 100.00% | Type VII secretion system | esxA | 100.00% | set25 | 100.00% | ||
| sdrH | 97.11% | esaA | 100.00% | set26 | 100.00% | |||
| Staphylococcal protein A | spa | 99.11% | essA | 100.00% | hlgA | 100.00% | ||
| von Willebrand factor | vwb | 99.93% | esaB | 100.00% | hlgC | 100.00% | ||
| Host Immune evasion | essB | 100.00% | hlgB | 100.00% | ||||
| Exoprotein SCIN | scn | 99.72% | essC | 100.00% | lukD | 100.00% | ||
| Capsule Type 1, 5 and 8 | capA | 100.00% | esaC | 98.99% | lukE | 100.00% | ||
| capB | 100.00% | esxB | 99.37% | |||||
| capC | 100.00% | |||||||
| capD | 100.00% | |||||||
| capP | 99.15% |
Statistical Analysis
All statistical data were generated using SPSS version 16.0 (SPSS Inc. (now IBM)). Data are shown as medians and interquartile ranges. P values for the differences between groups were calculated using Fisher’s Exact Test for categorical variables and Mann-Whitney test for continuous variables. Risk of MRSA colonization were tested in a logistic regression analysis and expressed by odds ratio (OR) estimates. Variables with a P-value of 0.2 or less in the univariate analysis were tested in the multivariate analysis. P<0.05 was considered statistically significant.
Results
Patients
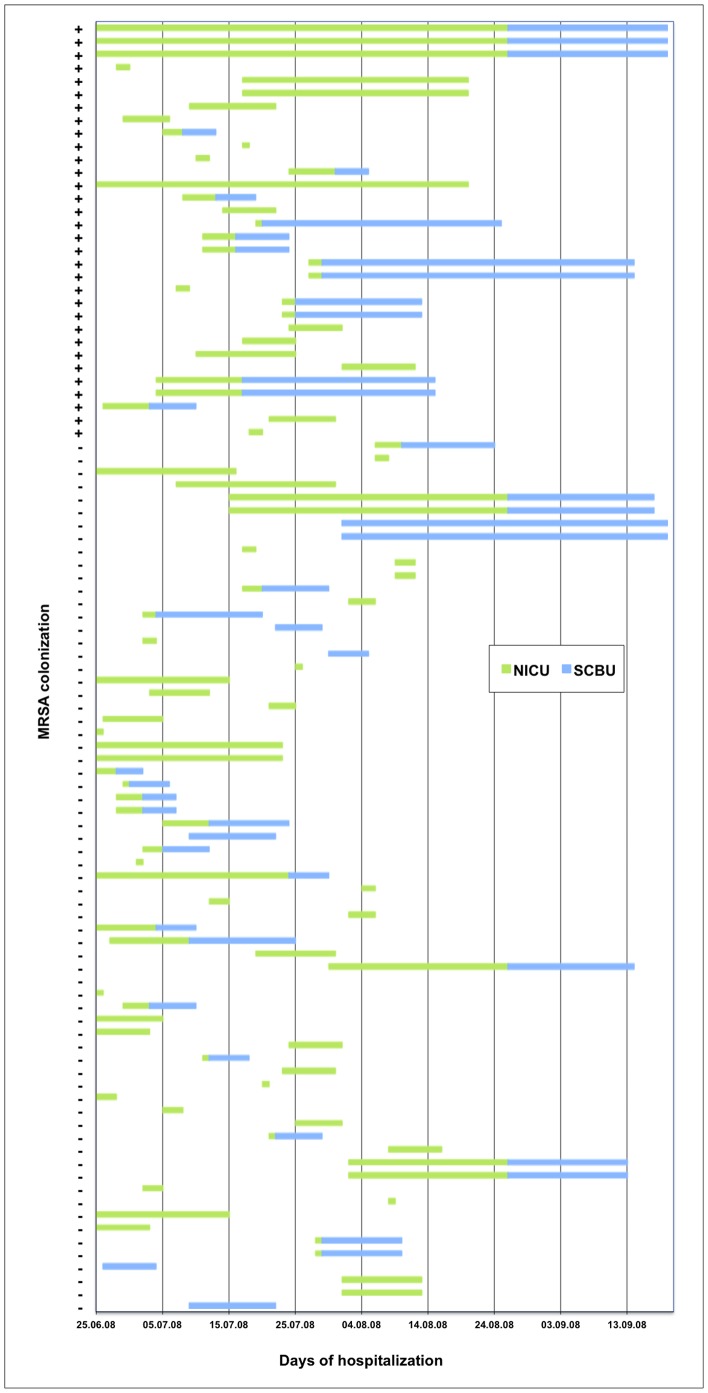
One hundred and two neonates were admitted to the Neonatal ward during the 42-day MRSA exposure period (June 25th–August 8th). One neonate died due to perinatal asphyxia, and two discharged children could not be contacted. Thus, the study comprised 99 children from 84 families. Figure 1 shows the time period of hospitalization at the two neonatal units and the carriage of MRSA for each individual neonate. Ninety-two neonates were initially admitted to the NICU unit, and 52 were discharged to their homes directly from the NICU (16 colonized with MRSA) and 40 neonates were transferred to the SCBU (16 colonized with MRSA). The first case of transferral to the SCBU occurred on July 2nd. Three neonates were readmitted to the NICU from the SCBU. Seven neonates were admitted directly to the SCBU, none of them became colonized with MRSA. Thirty-two percent (32/99) of neonates were colonized with MRSA, comprising 30% of the families (25/84). A higher proportion of colonized neonates were hospitalized at the discovery of the MRSA outbreak (on July 28th), compared to discharged neonates (48% (15/31) vs. 25% (17/68), P = 0.04).
Figure 1. Time course of MRSA outbreak at the 2 neonatal wards.
The figure shows the time period of hospitalization for each individual neonate at NICU (green) and SCBU (blue). The first 32 lines (marked with a+sign) are the MRSA colonized neonates and non-colonized neonates are marked with a - sign.
The MRSA Strain
The MRSA outbreak strain was characterized as mecA positive, spa-type t127, Sequence Type ST 1835, a single locus variant of ST1 and PVL negative. The isolates were SCCmec V and contained SCCfus (Anders Rhod Larsen, personal communication). All isolates were susceptible to erythromycin, clindamycin, gentamicin, vancomycin, linezolid, moxifloxacin, rifampicin, but resistant to fusidic acid. A virulence gene profile is shown in Table 1. The isolate contained Exfoliative toxin type A, however, scalded skin syndrome was not part of the clinical presentation.
Risk Factors for MRSA Acquisition
Caesarean section and nCPAP therapy were more frequent in neonates colonized with MRSA than in non-colonized children (53% vs. 30%, P = 0.03 and 66% vs. 27%, P<0.001, respectively, Table 2). Neonates colonized with MRSA had been exposed to MRSA for a longer period, than those not colonized (12.5 days vs. 8 days, respectively, P = 0.04). The risk of being colonized with MRSA increased with 1% per day of the total hospitalization (before and during the triplets admittance); 2.7% per day when exposed to MRSA; 3% per day when treated with nCPAP during the total hospitalization; and 22% per day when treated with nCPAP during the MRSA exposure period.
Table 2. Risk factors* for MRSA acquisition.
| (No./total) or medians with | Colonized | Not Colonized | Univariate logistic regression | Multivariate logistic regression | ||
| interquartile range | (N = 32) | (N = 67) | OR (95% CI) | p-value | OR (95% CI) | p-value |
| Gender (male) | 53% (17/32) | 52% (35/67) | 1.04 (0.45–2.41) | 1 | ||
| Gestational age (days) | 249 (228–266) | 262 (226–279) | 1.00 (0.98–1.01) | 0.27 | ||
| Birth weight (grams) | 2729 (1742–3519) | 2490 (1800–3280) | 1.00 (1.00–1.00) | 0.69 | ||
| Hospital of birth (Glostrup) | 56% (18/32) | 54% (36/67) | 1.11 (0.47–2.59) | 0.83 | ||
| Caesarean section | 53% (17/32) | 30% (20/67) | 2.66 (1.12–6.35) | 0.03 | 3.74 (1.27–11.0) | 0.016 |
| Twins or triplets | 41% (13/32) | 24% (16/67) | 2.18 (0.89–5.37) | 0.10 | 1.48 (0.37–5.87) | 0.6 |
| nCPAP treatment | 66% (21/32) | 27% (18/67) | 5.20 (2.10–12.88) | <0.001 | 5.88 (1.67–20.7) | 0.006 |
| Transferred from tertiary care centre NICU | 9% (3/32) | 23.9% (16/67) | 0.33 (0.09–1.23) | 0.11 | 0.07 (0.006–1.27) | 0.09 |
| Chronic lung disease | 6% (2/32) | 3% (2/67) | 2.17 (0.29–16.12) | 0.59 | ||
| Intravascular devices | 59% (19/32) | 45% (30/67) | 1.80 (0.77–4.24) | 0.20 | 0.89 (0.20–3.95) | 0.9 |
| Treatment with antibiotics | 44% (14/32) | 30% (20/67) | 1.83 (0.76–4.38) | 0.18 | 1.44 (0.33–6.37) | 0.6 |
| Days hospitalized | 12.5 (7.25–34) | 9 (4–27) | 1.01 (0.99–1.03) | 0.19 | 0.99 (0.93–1.05) | 0.8 |
| MRSA exposure in days | 12.5 (7.25–34) | 8 (4–17) | 1.03 (1.00–1.05) | 0.04 | 3.11 (0.40–24.6)# | 0.28 |
Data with a p-value of 0.2 or less were tested in the multivariate analysis.
Period of MRSA exposure was logarithmically transformed in the multivariate (Logistic Regression) analysis.
There was no association between MRSA acquisition and gender, gestational age, birth weight, presence of chronic lung disease, PVC or previous treatment with antibiotics (Table 2, P>0.05). In the multivariate logistic regression analysis, only treatment with nCPAP and delivery by Caesarean section were independent risk factors for MRSA colonization (P = 0.006 and P = 0.016, respectively).
Family Members and HCW
Among the 25 families with MRSA colonized neonates, two of nine (22%) families with hospitalized neonates had colonized household members; whereas nine out of 16 (56%) families of discharged neonates had colonized household members. In total, 13 family members out of 68 tested (20%) were colonized with MRSA. Two of 161 HCWs (1,2%) were colonized with MRSA.
Environmental Cultures
In the NICU, MRSA was found on two locations: an alarm button and the floor. In the SBCU, MRSA was found on three locations; a chair seat, a laundry cupboard handle and the floor. Environmental cultures after SterinisT disinfection of the NICU and SCBU were all without growth of MRSA.
Discussion
In this large NICU outbreak 32 children from 25 families became colonized with MRSA. The MRSA was introduced into the NICU with the arrival of triplets transferred from the Copenhagen tertiary care NICU. Most likely these triplets have been exposed to the MRSA of a neonate in isolation at this hospital. The MRSA isolates were in all cases the rare spa-type t127, resistant to fucidic acid, described by others as CA-MRSA and previously involved in a CA-MRSA outbreak in the UK [2], [10]. The triplets were in our NICU for 43 days, before MRSA was found in a clinical sample, and during this period 32% of exposed hospitalized infants became colonized, showing the rapid expansion of this clone. Infants still hospitalized at the point of discovery of the MRSA outbreak, compared to infants allready discharged, were more frequently colonized. This was probably due to physical aspects of the ward. The closer to discharge, the further away from the nurses station the infants were placed. In our study the significant risk factors in both univariate and multivariate logistic regression analysis for MRSA colonization were: delivery by caesarean section and nCPAP therapy. We did not find premature birth, low birth weight or multiple gestation as colonization risk factors, although they have been described by others [24], [32], [33]. In MRSA colonized infants, risk factors for MRSA infection have been; gender, gestational age, birth weight, intravascular device, term surgical neonates, multiple gestation, gavage feeding, intubation, [32], [34]–[36]. The MRSA carriage rate in our HCWs was low, 1.2%, but we do not know, if there has been transient carriage. Similar low rates have been found in other countries [12], [19], but MRSA carriage rates of up to 25% have also been described [13], [16]. As found by other investigators environmental contamination with MRSA was an issue, stressing the importance of cleaning [11], [16], [17].
A NICU is a complex hospital ward, regarding infection control procedures, because HCWs and parents are closely involved in the care of the neonates. Although parents are instructed, they are not trained to have good infection control procedures. In our outbreak, Caesarean section and nCPAP were identified as risk factors. Both of these procedures may involve more HCW handling of the neonates at least for the first days after birth. Control of the outbreak was gained through barrier precautions, isolation procedures and intensified disinfective cleaning followed by a final intensive cleaning, when the last MRSA neonate was discharged. The NICU was briefly closed for admittance of new patients. No neonates were treated for MRSA carriage.
Our study was unique in several ways. The MRSA outbreak was very large involving more than one hundred suspected cases, of which 98% were screened. All MRSA belonged to the same CA-MRSA clone, and there is no suspicion of more than one introduction to the NICU. Clinical data collection was complete as all analyzed information could be found in the Neobase and the neonates’ medical records, allowing us to perform multivariate logistic analysis without missing values.
Our study has nevertheless limitations. Only family members with MRSA colonized neonates were screened for MRSA, which could have resulted in unidentified cases.
Also, each individual HCW took their own MRSA surveillance cultures, which could have resulted in sampling failures. However, no swabs were without growth of any microorganisms. Previous studies have shown a greater degree of transmission of MRSA in wards where the nurse staff had an excessive workload [21], [37], but we were not able to include data regarding ratio between staff numbers and neonates in the exposure period due to the retrospective aspect of our investigation.
In conclusion this NICU MRSA outbreak was caused by a CA-MRSA that rapidly spread in a bacterial naive population. Control was established by an outbreak management group that focused on MRSA screening, barrier procedures, isolation, infection control instruction and cleaning. As a consequence of this outbreak all neonates transferred between the four NICUs in Copenhagen are isolated and screened for MRSA on arrival in a new NICU.
Acknowledgments
The authors would like to thank Mette Theilgaard Christiansen for supplementing the virulence gene profile of a MRSA isolate from the neonatal outbreak (Table 1).
Funding Statement
The authors have no support or funding to report.
References
- 1. Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, et al. (2010) Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med 7: e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, et al. (2011) A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6: e17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EPI-NEWS No 47–2012 - Statens Serum Institut. Available: http://www.ssi.dk/English/News/EPI-NEWS/2012/No%2047%20-%202012.aspx. Accessed 13 December 2012.
- 4. Hetem DJ, Westh H, Boye K, Jarløv JO, Bonten MJ, et al. (2012) Nosocomial transmission of community-associated methicillin-resistant Staphylococcus aureus in Danish Hospitals. J Antimicrob Chemother 2012 Jul 67(7): 1775–80. [DOI] [PubMed] [Google Scholar]
- 5. Lebon A, Moll HA, Tavakol M, van Wamel WJ, Jaddoe VW, et al. (2010) Correlation of bacterial colonization status between mother and child: the Generation R Study. J Clin Microbiol 48: 960–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lindberg E, Nowrouzian F, Adlerberth I, Wold AE (2000) Long-time persistence of superantigen-producing Staphylococcus aureus strains in the intestinal microflora of healthy infants. Pediatr Res 48: 741–747. [DOI] [PubMed] [Google Scholar]
- 7. Pinter DM, Mandel J, Hulten KG, Minkoff H, Tosi MF (2009) Maternal-infant perinatal transmission of methicillin-resistant and methicillin-sensitive Staphylococcus aureus. Am J Perinatol 26: 145–151. [DOI] [PubMed] [Google Scholar]
- 8. Peacock SJ, Justice A, Griffiths D, de Silva GDI, Kantzanou MN, et al. (2003) Determinants of Acquisition and Carriage of Staphylococcus aureus in Infancy. J Clin Microbiol 41: 5718–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heinrich N, Mueller A, Bartmann P, Simon A, Bierbaum G, et al. (2011) Successful management of an MRSA outbreak in a neonatal intensive care unit. Eur J Clin Microbiol Infect Dis 30: 909–913. [DOI] [PubMed] [Google Scholar]
- 10. David MD, Kearns AM, Gossain S, Ganner M, Holmes A (2006) Community-associated meticillin-resistant Staphylococcus aureus: nosocomial transmission in a neonatal unit. J Hosp Infect 64: 244–250. [DOI] [PubMed] [Google Scholar]
- 11. Otter JA, Klein JL, Watts TL, Kearns AM, French GL (2007) Identification and control of an outbreak of ciprofloxacin-susceptible EMRSA-15 on a neonatal unit. J Hosp Infect 67: 232–239. [DOI] [PubMed] [Google Scholar]
- 12. Regev-Yochay G, Rubinstein E, Barzilai A, Carmeli Y, Kuint J, et al. (2005) Methicillin-resistant Staphylococcus aureus in neonatal intensive care unit. Emerg Infect Dis 11: 453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stein M, Navon-Venezia S, Chmelnitsky I, Kohelet D, Schwartz O, et al. (2006) An outbreak of new, nonmultidrug-resistant, methicillin-resistant Staphylococcus aureus strain (sccmec type iiia variant-1) in the neonatal intensive care unit transmitted by a staff member. Pediatr Infect Dis J 25: 557–559. [DOI] [PubMed] [Google Scholar]
- 14. Hayakawa T, Hayashidera T, Katsura S, Yoneda K, Kusunoki T (2000) Nasal mupirocin treatment of pharynx-colonized methicillin resistant Staphylococcus aureus: preliminary study with 10 carrier infants. Pediatr Int 42: 67–70. [DOI] [PubMed] [Google Scholar]
- 15. Gould IM, Girvan EK, Browning RA, MacKenzie FM, Edwards GF (2009) Report of a hospital neonatal unit outbreak of community-associated methicillin-resistant Staphylococcus aureus. Epidemiol Infect 137: 1242–1248. [DOI] [PubMed] [Google Scholar]
- 16. Lin YC, Lauderdale TL, Lin HM, Chen PC, Cheng MF, et al. (2007) An outbreak of methicillin-resistant Staphylococcus aureus infection in patients of a pediatric intensive care unit and high carriage rate among health care workers. J Microbiol Immunol Infect 40: 325–334. [PubMed] [Google Scholar]
- 17. Nambiar S, Herwaldt LA, Singh N (2003) Outbreak of invasive disease caused by methicillin-resistant Staphylococcus aureus in neonates and prevalence in the neonatal intensive care unit. Pediatr Crit Care Med 4: 220–226. [DOI] [PubMed] [Google Scholar]
- 18. Gerber SI, Jones RC, Scott MV, Price JS, Dworkin MS, et al. (2006) Management of outbreaks of methicillin-resistant Staphylococcus aureus infection in the neonatal intensive care unit: a consensus statement. Infect Control Hosp Epidemiol 27: 139–145. [DOI] [PubMed] [Google Scholar]
- 19. Bratu S, Eramo A, Kopec R, Coughlin E, Ghitan M, et al. (2005) Community-associated methicillin-resistant Staphylococcus aureus in hospital nursery and maternity units. Emerg Infect Dis 11: 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McDonald JR, Carriker CM, Pien BC, Trinh JV, Engemann JJ, et al. (2007) Methicillin-resistant Staphylococcus aureus outbreak in an intensive care nursery: potential for interinstitutional spread. Pediatr Infect Dis J 26: 678–683. [DOI] [PubMed] [Google Scholar]
- 21. Andersen BM, Lindemann R, Bergh K, Nesheim BI, Syversen G, et al. (2002) Spread of methicillin-resistant Staphylococcus aureus in a neonatal intensive unit associated with understaffing, overcrowding and mixing of patients. J Hosp Infect 50: 18–24. [DOI] [PubMed] [Google Scholar]
- 22. Sax H, Posfay-Barbe K, Harbarth S, Francois P, Touveneau S, et al. (2006) Control of a cluster of community-associated, methicillin-resistant Staphylococcus aureus in neonatology. J Hosp Infect 63: 93–100. [DOI] [PubMed] [Google Scholar]
- 23. Gregory ML, Eichenwald EC, Puopolo KM (2009) Seven-year experience with a surveillance program to reduce methicillin-resistant Staphylococcus aureus colonization in a neonatal intensive care unit. Pediatrics 123: e790–e796. [DOI] [PubMed] [Google Scholar]
- 24. Huang YC, Chou YH, Su LH, Lien RI, Lin TY (2006) Methicillin-resistant Staphylococcus aureus colonization and its association with infection among infants hospitalized in neonatal intensive care units. Pediatrics 118: 469–474. [DOI] [PubMed] [Google Scholar]
- 25. Milstone AM, Budd A, Shepard JW, Ross T, Aucott S, et al. (2010) Role of decolonization in a comprehensive strategy to reduce methicillin-resistant Staphylococcus aureus infections in the neonatal intensive care unit: an observational cohort study. Infect Control Hosp Epidemiol 31: 558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Milstone AM, Song X, Coffin S, Elward A (2010) Society for Healthcare Epidemiology of America's Pediatric Special Interest Group (2010) Identification and eradication of methicillin-resistant Staphylococcus aureus colonization in the neonatal intensive care unit: results of a national survey. Infect Control Hosp Epidemiol 31: 766–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chapman AK, Aucott SW, Milstone AM (2012) Safety of chlorhexidine gluconate used for skin antisepsis in the preterm infant. J Perinatol 32: 4–9. [DOI] [PubMed] [Google Scholar]
- 28. Bartels MD, Kristoffersen K, Slotsbjerg T, Rohde SM, Lundgren B, et al. (2008) Environmental meticillin-resistant Staphylococcus aureus (MRSA) disinfection using dry-mist-generated hydrogen peroxide. J Hosp Infect 70: 35–41. [DOI] [PubMed] [Google Scholar]
- 29. Bartels MD, Boye K, Rhod Larsen A, Skov R, Westh H (2007) Rapid increase of genetically diverse methicillin-resistant Staphylococcus aureus, Copenhagen, Denmark. Emerg Infect Dis 13: 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boye K, Bartels MD, Andersen IS, Møller JA, Westh H (2007) A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin Microbiol Infect 13: 725–727. [DOI] [PubMed] [Google Scholar]
- 31.Disk diffusion - basic SRGA methodology. Disk diffusion - basic SRGA methodology. Available: http://www.srga.org/RAFMETOD/rafmet.htm. Accessed 13 December 2012.
- 32. Khoury J, Jones M, Grim A, Dunne WM, Fraser V (2005) Eradication of methicillin-resistant Staphylococcus aureus from a neonatal intensive care unit by active surveillance and aggressive infection control measures. Infect Control Hosp Epidemiol 26: 616–621. [DOI] [PubMed] [Google Scholar]
- 33. Maraqa NF, Aigbivbalu L, Masnita-Iusan C, Wludyka P, Shareef Z, et al. (2011) Prevalence of and risk factors for methicillin-resistant Staphylococcus aureus colonization and infection among infants at a level III neonatal intensive care unit. Am J Infect Control 39: 35–41. [DOI] [PubMed] [Google Scholar]
- 34. Carey AJ, Duchon J, Della-Latta P, Saiman L (2010) The epidemiology of methicillin-susceptible and methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit, 2000–2007. J Perinatol 30: 135–139. [DOI] [PubMed] [Google Scholar]
- 35. Babazono A, Kitajima H, Nishimaki S, Nakamura T, Shiga S, et al. (2008) Risk factors for nosocomial infection in the neonatal intensive care unit by the Japanese Nosocomial Infection Surveillance (JANIS). Acta Med Okayama 62: 261–268. [DOI] [PubMed] [Google Scholar]
- 36. Sakaki H, Nishioka M, Kanda K, Takahashi Y (2009) An investigation of the risk factors for infection with methicillin-resistant Staphylococcus aureus among patients in a neonatal intensive care unit. Am J Infect Control 37: 580–586. [DOI] [PubMed] [Google Scholar]
- 37. Laing IA, Gibb AP, McCallum A (2009) Controlling an outbreak of MRSA in the neonatal unit: a steep learning curve. Arch Dis Child Fetal Neonatal Ed 94: F307–F310. [DOI] [PubMed] [Google Scholar]