Abstract
The pregenomic RNA (pgRNA) of hepadnaviruses is packaged into capsids where it is reverse transcribed to yield mature DNA genomes. This report describes differences between the 3′ region and other regions of the pgRNA isolated from capsids. Analysis of capsid pgRNA isolated by using an established method involving micrococcal nuclease treatment demonstrated reduced levels of the 3′ region of the pgRNA compared to the 5′ region. This underrepresentation of the 3′ region was partly a result of microccocal nuclease digestion of the 3′ region because isolation of capsid pgRNA by an alternative method that did not involve nuclease treatment led to a greater, but not complete, recovery of the 3′ region. These results indicate that the 3′ region of the capsid pgRNA is susceptible to micrococcal nuclease digestion during its isolation and that the 3′ region can still be underrepresented when capsid pgRNA is isolated without nuclease digestion. Additional experiments show that the 3′ ends of capsid pgRNA isolated by micrococcal nuclease treatment are heterogeneously dispersed from nucleotide 2577 to the poly(A) tail. These data provide evidence that the 3′ region of the capsid pgRNA has biochemical properties different from those of its 5′ region. Possibly, the 3′ region of the pgRNA is not packaged into the interior of the capsid but rather is associated with a part of the capsid where it is susceptible to microccocal nuclease digestion.
Hepadnaviruses, also known as hepatitis B viruses, have a DNA genome and replicate via reverse transcription of a pregenomic RNA (pgRNA) (6, 17). Hepadnaviruses infect the livers of their hosts, where they can cause diseases such as liver cirrhosis and hepatocellular carcinoma. One important member of the hepadnavirus family is the duck hepatitis B virus (DHBV). DHBV has been a useful model for understanding the molecular biology of hepadnaviruses.
The pgRNA is a multifunctional transcript. It is the template for reverse transcription (18) and the mRNA for the production of the viral polymerase and the capsid protein subunit, called P and C, respectively (4, 16). The pgRNA is transcribed from the covalently closed circular DNA episome present in the nucleus of infected cells (2). Transcription of the DHBV pgRNA results in a 3.3-kb polyadenylated RNA that is terminally redundant by approximately 270 nucleotides (nt). In addition, several subgenomic transcripts (sgRNAs) are synthesized that are mRNAs for the viral surface proteins (2, 14). All DHBV RNA transcripts are 3′ colinear because of a single polyadenylation signal on the covalently closed circular DNA.
Within the cytoplasm of the infected cell, the pgRNA and P protein are packaged into capsids in a process known as RNA encapsidation (1, 7). The P protein is required for encapsidation of the pgRNA; however, the reverse transcriptase activity of P protein is not required for this process (21, 23). The cis-acting elements for DHBV encapsidation are well defined (3, 8, 15); however, little is known about how the pgRNA is associated with its capsid. RNA encapsidation initiates a series of events that lead to virion production. After RNA packaging, reverse transcription of the pgRNA can occur within the capsid (18). Upon synthesis of the mature viral DNA genome, capsids can be secreted from the cell (22). Therefore, pgRNA encapsidation is essential for all subsequent events in the production of infectious virus.
Historically, pgRNA that is associated with capsids, called capsid pgRNA, has been distinguished from cytosolic, nonencapsidated pgRNA because of its resistance to nuclease digestion (3, 8, 9). Presumably, resistance of the capsid pgRNA to nuclease digestion is attributable to the capsid shell that surrounds the pgRNA. However, we report here that although the 5′ region of the capsid pgRNA is protected from nuclease, the 3′ region, including the 3′-terminal redundancy and the poly(A), of the capsid pgRNA is susceptible to micrococcal nuclease digestion. Furthermore, isolation of capsid pgRNA by a method that does not rely on the nuclease resistance of the RNA leads to a greater, but not complete, recovery of the 3′ region of the capsid pgRNA. These different properties between the 5′ region and the 3′ region of the capsid pgRNA give insights into the process and consequences of RNA encapsidation.
MATERIALS AND METHODS
Plasmids.
All plasmids that express DHBV pgRNA contain 1.5 copies of the DHBV3 genome and are derived from the wild-type construct, pD1.5G, or wild-type DHBV (8). DHBV PY96F contains two nucleotide substitutions, C454G and A456T, which change the amino acid residue used for priming DNA synthesis on the P protein from a tyrosine to a phenylalanine. DHBV PYMHA contains two nucleotide substitutions, G1706C and A1710C, which change the putative catalytic site of the reverse transcriptase activity of the P protein from the amino acid sequence YMDD to YMHA. Expression of DHBV PY96F or DHBV PYMHA leads to pgRNA encapsidation, but these variants do not synthesize DNA due to the respective mutations in the P protein. ΔSpacer contains a deletion of nt 1094 to 1243 in the background of DHBV PYMHA and maintains the open reading frame of the P protein. 3′LACZ is a substitution in the background of DHBV PYMHA and contains 137 nt of LacZ (nt 2663 to 2799 of the LacZ open reading frame) fused to 226 nt of the simian virus 40 (SV40) polyadenylation signal replacing the 3′ copy of DHBV nt 2531 to 3021. 3′LACZ expresses a pgRNA with the normal 3′-terminal redundancy replaced with 284 nt of the LacZ/SV40 fusion. pT7PG was used as a template for the generation of an in vitro-transcribed pgRNA (IVT). pT7PG contains DHBV nt 2529 to 3021, followed by nt 1 to 2850 downstream of a T7 promoter. In vitro transcription with T7 RNA polymerase yields an RNA that is terminally redundant by approximately 320 nt. Details of the molecular cloning of any of these plasmids is available upon request.
Cell cultures and transfection.
The chicken hepatoma cell line, LMH, was cultured as described previously (12). DNA transfections were performed by the calcium phosphate precipitation method. For each transfection, 9 μg of DHBV expression plasmid was transfected with 1 μg of a green fluorescent protein expression plasmid.
Preparation of cytoplasmic lysates.
At 3 days posttransfection LMH cells were washed with HBS+EGTA (2 mM HEPES [pH 7.45], 150 mM NaCl, and 0.5 mM EGTA), and cytoplasmic lysates were prepared. To generate a cytoplasmic lysate by using nonionic detergent, cells were treated with Nonidet P-40 (NP-40) lysis buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 0.2% NP-40), and nuclei were pelleted. To generate a cytoplasmic lysate by Dounce homogenization, cells were resuspended in a solution of 10 mM Tris (pH 7.5), 1 mM EDTA, and 20 μg of aprotinin/ml, and cells were disrupted on ice by 60 strokes in the Dounce homogenizer (13). The cytoplasmic lysates were subsequently used for the isolation of viral RNA.
Isolation of viral RNA.
The isolation of cytoplasmic poly(A) RNA (A RNA) was performed as previously described (15). For the isolation of micrococcal nuclease-treated capsid RNA (M RNA), the cytoplasmic lysate was adjusted to 5 mM CaCl2, and 22 U of micrococcal nuclease (Worthington) was added. The samples were incubated at 37°C for 60 min. The solution was adjusted to 10 mM EDTA-0.2% sodium dodecyl sulfate-50 mM NaCl-0.2 mg of pronase/ml to inactivate the micrococcal nuclease and digest proteins. The capsid RNA was extracted once with phenol-chloroform and once with chloroform and then stored in ethanol.
Immunoprecipitated capsid RNA (I RNA) was isolated by incubating the cytoplasmic lysate with 5 μl of αDHBc polyclonal antibody (a generous gift from Jesse Summers, University of New Mexico-Albuquerque) prebound to 5 mg of protein A-Sepharose (Amersham Biosciences). The immunoprecipitate was washed three times with 1 ml of 1× phosphate-buffered saline. Next, the capsids were resuspended in 10 mM EDTA-0.2% sodium dodecyl sulfate -50 mM NaCl-0.2 mg of pronase/ml to digest the proteins. Finally, the capsid RNA was extracted once with phenol-chloroform and once with chloroform and then stored in ethanol.
RNase protection analysis.
Riboprobes were transcribed in vitro from linearized DNA templates by using T7, SP6, or T3 RNA polymerase and labeled with [α-32P]UTP. Riboprobes were gel purified and RNase protection analysis was carried out as described previously (15). RNA from 1/10 of a transfected plate was used in each analysis.
RNase H mapping and Northern blotting analysis.
RNA from 1/10 of a transfected plate was analyzed by RNase H mapping. RNA was precipitated with ethanol, washed in 70% ethanol, and resuspended in 20 μl of annealing buffer (1 mM EDTA [pH 7.5], 0.2 M KCl, and 25 pmol each of oligonucleotides D2347− alone or D2347− and T21−). D2347− contains DHBV minus-sense sequence from nt 2347 to 2327. T21− is an oligonucleotide with a tract of 21 thymine bases. Samples were heated at 95°C for 3 min and incubated at room temperature for 20 min. Next, 20 μl of RNase H digestion buffer (40 mM Tris-HCl, 28 mM MgCl2, and 1 U of RNase H [Amersham]) was added. The samples were incubated at 37°C for 45 min. The reaction was adjusted to 10 mM EDTA, 0.2% sodium dodecyl sulfate, 50 mM NaCl, and 0.2 mg of pronase/ml and then incubated at 37°C for 20 min. The reaction was phenol-chloroform extracted and ethanol precipitated. RNA was dissolved in formaldehyde RNA loading buffer, and Northern blot analysis was performed as previously described (11). The Northern blot was probed with a 32P end-labeled oligonucleotide, D2445− (DHBV minus-sense nt 2445 to 2428), and analyzed by using Molecular Dynamics Storm and ImageQuant software.
RESULTS
The 3′ region of the DHBV pgRNA is underrepresented in RNA isolated from capsids.
Studies of DHBV pgRNA encapsidation lead us to examine the 5′ and 3′ regions of the capsid pgRNA. RNA encapsidation was analyzed by isolating RNA from LMH cells transfected with DHBV expression plasmids. A cytoplasmic lysate was prepared from transfected cells, and two fractions of RNA were isolated from the lysate: cytoplasmic polyadenylated RNA (A) and micrococcal nuclease-treated capsid RNA (M). The A fraction was isolated by oligo(dT) selection of all cytoplasmic RNAs, and it served as a control for RNA accumulation. DHBV pgRNAs and sgRNAs were present in the A fraction. The M fraction was isolated by treating the cytoplasmic lysate with microccocal nuclease, inactivating the nuclease, and then extracting RNA. The M fraction represents capsid pgRNA because of its resistance to micrococcal nuclease digestion.
The A and M fractions were analyzed by using an RNase protection assay (RPA) with a probe (nt 2529 to 3021) that annealed to regions of the pgRNA that contained the terminal redundancies (nt 2529 to 2800). Probe nt 2529 to 3021 was expected to detect the 5′ region of the pgRNA from nt 2529 to 3021 and the 3′ region of the pgRNA and sgRNAs from nt 2529 to 2800, as shown in Fig. 1A. The analysis in Fig. 1B shows that the 5′ region of the pgRNA and the 3′ region of the pgRNA and sgRNAs were detected in the A fraction from LMH transfected with wild-type DHBV (Fig. 1B, lane 4). Surprisingly, only the 5′ region of the pgRNA was clearly detected in the M fraction; the 3′ region from nt 2529 to 2800 was not abundantly detected in the M fraction (Fig. 1B, lane 5). One possibility was that DNA synthesis occurred within capsids, and the 3′ region of the pgRNA was lost as a result of RNase H activity of the P protein. To examine this possibility, A and M RNA fractions were isolated from LMH transfected with the variants, DHBV PY96F or DHBV PYMHA. DHBV PY96F and DHBV PYMHA are functional for RNA encapsidation but deficient in DNA synthesis because of a mutation in the priming moiety of the polymerase or a mutation within the putative reverse transcriptase catalytic site of the polymerase, respectively. Without DNA synthesis in capsids, there is no substrate for RNase H activity and no degradation of the capsid pgRNA. We observed the same result for these DNA synthesis-deficient variants: the 3′ region from nt 2529 to 2800 of the capsid pgRNA was barely detectable, whereas the 5′ region of capsid pgRNA was highly abundant (Fig. 1B, lanes 7 and 9). DHBV PY96F and PYMHA were studied in all subsequent experiments.
FIG. 1.
The 3′ region of DHBV capsid RNA is underrepresented. (A) Nucleotide coordinates of the C and P open reading frames and the positions of features on the pgRNA. The 5′ end of the pgRNA is at nt 2529, and terminal redundancies (designated R) are approximately 270 nt. ɛ is an encapsidation signal located near the 5′ end of the pregenomic RNA. The positions on the pgRNA and sgRNAs detected by RPA probe nt 2529 to 3021 are indicated. For pgRNA, the 5′ fragment from nt 2529 to 3021 and the 3′ fragment from nt 2529 to about nt 2800 are protected. For the sgRNAs, only the 3′ fragment from nt 2529 to 2800 is protected. (B) RPA of cytoplasmic poly(A) RNA (A lanes) and micrococcal nuclease-treated capsid RNA (M lanes) isolated from LMH transfected with DHBV wild type (WT), DHBV PY96F, or DHBV PYMHA. The bands that represent the 5′ region of pgRNA and 3′ region of pgRNA and sgRNAs are indicated. Lane 1, DNA molecular weight marker; lane 2, undigested probe which represents 1/10 of the amount used in the RPA; lane 3, probe digested; lanes 4, 6, and 8, cytoplasmic poly(A) RNA from the indicated DHBV variant; lanes 5, 7, and 9, micrococcal nuclease-treated capsid RNA from the indicated DHBV variant.
We compared the level of the 3′ region from nt 2529 to 2800 that was detected in the M fraction to the level of 3′ region that was predicted to be present based on the level of 5′ region from nt 2529 to 3021, as described in Table 1. The level of the 3′ region from nt 2529 to 2800 in the M fraction for DHBV PYMHA was only 17% of the predicted level. Therefore, the 3′ region from nt 2529 to 2800 was underrepresented compared to the 5′ region in a capsid fraction of RNA isolated by using micrococcal nuclease treatment.
TABLE 1.
Representation of the 3′ region from nt 2529 to 2800 for DHBV PYMHA capsid RNAa
| Cell disruption method | Mean level of region 3′ representation ± SD (n)
|
||
|---|---|---|---|
| Nuclease-treated capsid RNA (M) | Immunoprecipitated capsid RNA (I) | Immunoprecipitated capsids treated with MN (I-M) | |
| NP-40 | 17 ± 11 (7) | 51 ± 13 (4) | 26 ± 11 (2) |
| Dounce homog- enization | 14 ± 9 (6) | 49 ± 8 (6) | 21 ± 4 (4) |
Representation of the 3′ region was determined as the level of the 3′ region from nt 2529 to 2800 that was detected divided by the level of the 3′ region from nt 2529 to 2800 that was predicted multiplied by 100. Based on the expectation that the level of the 5′ and 3′ regions of capsid pgRNA should be equimolar, the level of predicted 3′ region was calculated as the level of the 5′ region that was detected multiplied by the ratio of uridine residues in the 3′ RPA fragment to uridine residues in the 5′ RPA fragment. The values in Table 1 were calculated from RPA by using DHBV probe nt 2529 to 3021. n represents the number of independent analyses for each sample.
Isolation of the capsid pgRNA by immunoprecipitation of capsids without nuclease treatment leads to a greater, but not complete, recovery of the 3′ region.
The inability to abundantly detect the 3′ region of the capsid pgRNA could be the result of processes occurring within the cell or a result of the RNA isolation procedure. To distinguish between these possibilities, alternative methods for RNA isolation were investigated. First, the 3′ region of the capsid pgRNA could be susceptible to micrococcal nuclease digestion, so it was isolated by another method which did not use nuclease treatment of the cytoplasmic lysate. This capsid RNA fraction (I) was generated by immunoprecipitation of DHBV capsids. Another possibility was that the NP-40 treatment was leading to the underrepresentation of the 3′ region. Studies have indicated that NP-40 may disrupt the structural integrity of other viral capsids (5). Therefore, cells were disrupted by Dounce homogenization in a hypotonic buffer in the absence of detergent.
The A, M, and I fractions of RNA were isolated from NP-40-disrupted cells or Dounce-homogenized cells that had been transfected with the variants DHBV PY96F or DHBV PYMHA (Fig. 2). Using a second RPA probe (nt 2410 to 2850) that annealed to a region on the pgRNA containing the terminal redundancies, it was observed that isolation of capsid pgRNA by immunoprecipitation of capsids led to a greater recovery of the 3′ region from nt 2410 to 2800 than that found in the M fraction (Fig. 2B, lane 5 versus lane 6 and lane 8 versus lane 9). On the other hand, disruption of the cells by Dounce homogenization produced the same result as NP-40 lysis of the cells (Fig. 2B, lanes 5 and lane 6 versus lanes 8 and 9), ruling out the idea that NP-40 treatment was leading to the loss of the 3′ region. Comparison of the level of the 3′ region from nt 2529 to 2800 detected by using the different methods for capsid isolation demonstrated that the 3′ region for the I RNA was represented at a level threefold higher than in the M RNA (Table 1). However, 100% recovery of the 3′ region of the I RNA was not seen; only ca. 50% of the predicted 3′ region from nt 2529 to 2800 was recovered.
FIG. 2.
Isolation of capsid RNA by immunoprecipitation leads to a greater recovery of the 3′ region. (A) The fragments of pgRNA protected by RPA probes nt 2410 to 2850 and nt 2529 to 3021 are indicated. (B) RPA of A RNA, M RNA, and immunoprecipitated capsid RNA (I lanes) isolated from LMH transfected with DHBV PY96F. Lane 1, marker; lane 2, undigested probe nt 2410 to 2850; lane 3, digested probe; lanes 4, 5, and 6, the indicated fraction of RNA isolated by NP-40 disruption of the cells; lanes 7, 8, and 9, the indicated fraction of RNA isolated by Dounce homogenization of the cells. (C) RPA of A RNA, M RNA, I RNA, and immunoprecipitated and then micrococcal nuclease-treated capsid RNA (I-M) isolated from LMH transfected with DHBV PYMHA. Lanes 1 to 3, same as in panel B, except probe is nt 2529 to 3021; lanes 4 to 7, the indicated fraction of RNA isolated by Dounce homogenization of the cells.
To further demonstrate that underrepresentation of the 3′ region was a result of micrococcal nuclease digestion, the immunoprecipitated capsids were treated with micrococcal nuclease, the nuclease was inactivated, and the capsid RNA was extracted (I-M RNA fraction). As shown in Fig. 2C and Table 1, treating the immunoprecipitated capsids with micrococcal nuclease caused a reduction in the representation of the 3′ region from nt 2529 to 2800 compared to the I fraction. This result indicated that the underrepresentation of the 3′ region of capsid pgRNA was due in part to micrococcal nuclease susceptibility of the 3′ region. However, the inability to completely recover the 3′ region in the absence of nuclease treatment suggested that other events or processes may also contribute to the underrepresentation of the 3′ region of the capsid pgRNA.
The 3′ region of capsid RNA isolated with micrococcal nuclease treatment is abundantly detected approximately 225 nt upstream of the poly(A) tail.
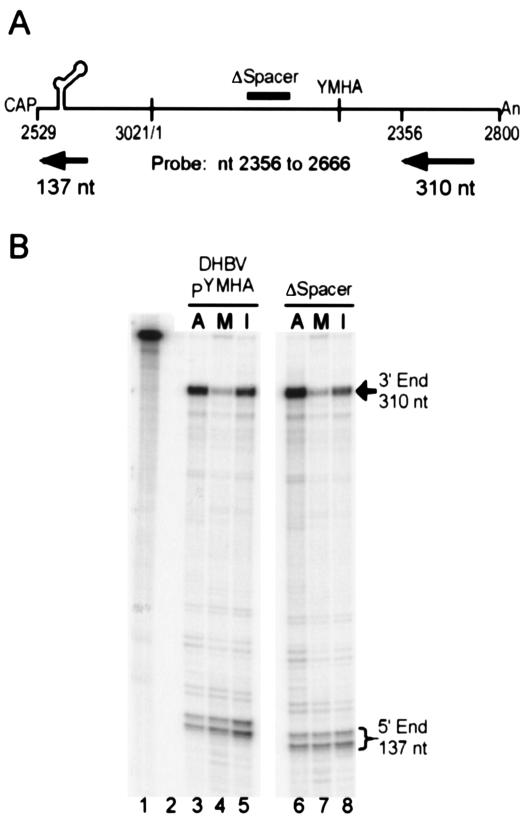
Next, we determined the position in the 3′ region where the capsid pgRNA was not susceptible to micrococcal nuclease digestion. Various RPA probes were used to learn the location of the 3′ end for the M RNA isolated from LMH transfected with DHBV PY96F (as shown in Fig. 3A). The probes—A, B, and C—annealed to sequence that contained different portions of the terminal redundancies of the pgRNA, and therefore, the 3′ region and the 5′ region could be detected by using the same probe. Probe A could detect the 3′ region from nt 2410 to 2800, probe B could detect the 3′ region from nt 2356 to 2755, and probe C could detect the 3′ region from nt 2356 to 2666. For probes A (Fig. 2B), B (data not shown), and C (see Fig. 6B), the representations of the 3′ region for the M RNA were 13, 23, and 24% of the predicted level, respectively. This low level was not because of an inability to simultaneously detect the terminal redundancies by using a single probe. RPA of an in vitro-transcribed pgRNA with probe nt 2529 to 3021 resulted in detection of 100% of the 3′ region (data not shown). In addition to the inability to abundantly detect the 3′ region with probes A, B, and C, we did not detect discrete, smaller bands that would indicate one or more new, distinct positions for the 3′ ends of M RNA (data not shown). RPA with probes that annealed mostly upstream of the 3′-terminal redundancy required a second probe to measure the level of the 5′ region (Fig. 3A, probe sets D, E, and F). When a probe that detected the 3′ region from nt 2356 to 2577 was used (probe set D), the 3′ region of the M RNA was represented at 80% of the level of the 5′ region (Fig. 3A and B). nt 2577 is about 225 nt upstream of the poly(A) tail of pgRNA. Other probes that annealed to the 3′ region upstream of nt 2577 (probe sets E and F; data not shown) detected abundant levels for the 3′ region of the M RNA.
FIG. 3.
Mapping the 3′ region of capsid RNA protected from microccocal nuclease digestion. (A) Probes or probe sets A through F were used for RPA of micrococcal nuclease-treated capsid (M) RNA from DHBV PY96F-transfected cells. The fragments of pgRNA protected by the RPA probes are indicated. The value at the right is the representation of the 3′ region for the given probe set (as described in Table 1). The results represent the mean and standard deviations of three independent analyses for probes A and B and two independent analyses for probes C, D, E, and F. (B) RPA of A RNA and M RNA isolated from LMH transfected with DHBV PY96F using probe set D. Lane 1, marker; lane 2, undigested probes nt 2810 to 84 and nt 2356 to 2577; lane 3, digested probes; lane 4, A RNA; lane 5, M RNA.
FIG. 6.
The 3′ region of a DHBV variant with an internal deletion is still susceptible to nuclease digestion. (A) The fragments of pgRNA protected by RPA probe nt 2356 to 2666 are indicated. The position of the ΔSpacer deletion is also marked. (B) RPA of A, M, and I RNA isolated from LMH transfected with DHBV PYMHA or ΔSpacer. Lane 1, undigested probe nt 2356 to 2666; lane 2, digested probe; lanes 3 to 5, A, M, and I RNA, respectively, isolated from LMH transfected with DHBV PYMHA; and lanes 6 to 8, A, M, and I RNA, respectively, isolated from LMH transfected with ΔSpacer.
Only probes that annealed entirely upstream of nt 2577 abundantly detected the 3′ region of M RNA. On the other hand, probes A, B, and C all annealed to sequence upstream of nt 2577; however, no discrete, smaller bands derived from the 3′ region of capsid RNA were detected. These results indicate that there was not a unique location for the 3′ end for the capsid pgRNA isolated by micrococcal nuclease treatment. These results are consistent with the M RNA fraction containing a heterogeneous population of capsid RNAs with different 3′ ends between nt 2577 and the poly(A) tail. The inability to detect the 3′ region when a probe that anneals 3′ of nt 2577 was used could be because there are many RNA species with different 3′ ends, and any single species may not be abundant enough to be distinguished in the RPA.
Northern blot analysis demonstrates that the 3′ ends of microccocal nuclease-treated capsid RNA are heterogeneous.
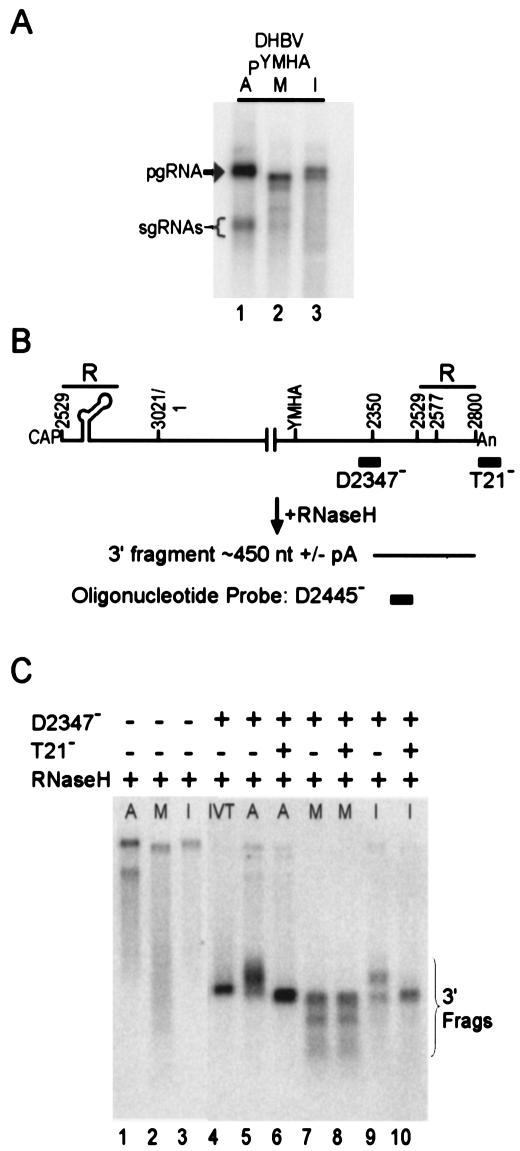
If the 3′ end of capsid pgRNA is susceptible to micrococcal nuclease digestion and heterogeneous over nt 2577 to the poly(A) tail, then capsid RNA isolated by using micrococcal nuclease treatment (M RNA) should be shorter than cytoplasmic polyadenylated pgRNA (A pgRNA). Accordingly, full-length M RNA migrated faster than the A pgRNA when examined by Northern blotting (Fig. 4A). The I RNA consistently contained a doublet that migrated at the position of the pgRNA (Fig. 4A, lane 3). In order to directly examine the 3′ region of A, M, and I fractions of RNA, a Northern blotting protocol that provided better resolution was used (Fig. 4B). This method, called RNase H mapping, involved cleaving the RNA at a position close to the 3′ end and then examining the RNA fragments by Northern blotting. First, a DNA oligonucleotide (D2347−) was annealed close to the 3′ end of the RNA. In addition, an oligonucleotide containing a tract of T's (T21−) could be annealed to the RNA. Next, the RNA-DNA hybrids were cleaved with RNase H, and the 3′ fragments were examined by Northern blotting and probed by using an end-labeled oligonucleotide (D2445−).
FIG. 4.
Northern blot analysis demonstrates that the 3′ end of capsid RNA isolated by using micrococcal nuclease is heterogeneous. (A) Northern blot analysis of A, M, and I RNA (lanes 1, 2, and 3, respectively) isolated from LMH transfected with DHBV PYMHA. The probe in this Northern blot was a positive-strand-specific riboprobe that detects DHBV nt 988 to 1665. Bands representing the pgRNA and sgRNAs are indicated. (B) RNase H mapping strategy to detect the 3′ ends of the pgRNA. Positions where DNA oligonucleotides D2347− and T21− anneal are indicated. RNase H cleavage generated a 3′ fragment of about 450 nt with or without a poly(A) tail. Also indicated is the position where the oligonucleotide probe, D2445−, will detect the 3′ fragments in the Northern analysis. (C) RNase H mapping result. The top of the blot indicates which oligonucleotides were included in the RNase H digestion and whether or not RNase H was included. The bands representing the 3′ fragments are indicated. Lanes 1 to 3, RNase H cleavage in the presence of no oligonucleotide for the A, M, and I RNA; lane 4, IVT, representing 500 nt; lanes 5, 7, and 9, RNase H mapping of A, M, and I RNA, respectively, with D2347−; lanes 6, 8, and 10, RNase H mapping of A, M, and I RNA with D2347− and T21−.
RNase H mapping of an IVT served as a molecular mass marker of 500 nt (Fig. 4C, lane 4, IVT). RNase H digestion of the A fraction by using oligonucleotide D2347− produced a heterogeneous 3′ fragment (Fig. 4C, lane 5). Addition of T21− to the RNase H digestion removed the poly(A) tails and shifted the 3′ fragments in the A fraction to a position of about 450 nt (Fig. 4C, lane 6). RNase H mapping of the 3′ region of M RNA produced a band that migrated at the 450 nt position, along with a smear of faster-migrating, heterogeneous species. The addition of T21− did not change the mobility of the fragments in the M RNA (Fig. 4C, lanes 7 and 8), indicating that the poly(A) tail of M RNA was not present. The heterogeneous species that represent the 3′ fragments of M RNA are consistent with the RPA results described in Fig. 3 that suggested the 3′ ends of capsid RNA isolated by micrococcal nuclease treatment were heterogeneously dispersed. Finally, RNase H mapping of the I RNA generated two populations of 3′ fragments: one population that migrated at 450 nt and one population of a higher molecular weight. The addition of T21− to the I RNA shifted all the 3′ fragments to about 450 nt, indicating that the population of 3′ fragments with the higher molecular weight was polyadenylated. The results of the RNase H mapping corroborate the results from the RPA that indicate the 3′ region of capsid pgRNA is susceptible to micrococcal nuclease digestion and that the isolation of capsid RNA by micrococcal nuclease treatment produces a population of capsid RNAs with heterogeneous 3′ ends.
Substitution of the 3′ terminal redundancy of the pgRNA does not affect the susceptibility of the 3′ region of capsid pgRNA to micrococcal nuclease digestion.
It was possible that there was a cis-acting sequence within the 3′ region that led to its underrepresentation and susceptibility to micrococcal nuclease digestion. In order to examine this idea, the 3′-terminal redundancy of the pgRNA was substituted with heterologous sequence (3′LACZ, Fig. 5A). In 3′LACZ, the approximately 270 nt 3′-terminal redundancy was replaced with 284 nt of LacZ sequence and an SV40 polyadenylation signal. RPA was performed to detect the 5′ and 3′ regions of A, M, and I RNA isolated from LMH transfected with 3′LACZ. The analysis shown in Fig. 5B demonstrates that the 3′ region of the M RNA of 3′LACZ was underrepresented similarly as that of DHBV PYMHA (Fig. 5B, lane 8, and Tables 1 and 2). Also, immunoprecipitation of 3′LACZ capsids lead to a greater, but not complete, recovery of the 3′ region. These results demonstrate that encapsidation of the 3′ region of 3′LACZ pgRNA has characteristics similar to that of encapsidation of DHBV PYMHA. Also, there are no cis-acting sequences in the 3′-terminal redundancy that contribute to its micrococcal nuclease sensitivity and the reduced representation of the 3′ end.
FIG. 5.
The 3′ region of a DHBV variant with a substitution of the 3′ terminal redundancy is also underrepresented and susceptible to nuclease digestion. (A) The fragments of 3′LacZ pgRNA protected by dual RPA probes nt 2529 to 3021 and the 3′LacZ probe. The 3′ substitution of pgRNA with LacZ and the SV40 polyadenylation signal is indicated. (B) RPA of A, M, and I RNA from LMH transfected with DHBV PYMHA or 3′LacZ. Lane 1, probes DHBV nt 2529 to 3021 and 3′LacZ undigested; lane 2, digested probes; lanes 3 to 6, A RNA controls to demonstrate specificity of the probes; lane 3, DHBV PYMHA RNA with dual probes; lane 4, 3′LacZ RNA with dual probes; lane 5, 3′LacZ with only DHBV nt 2529 to 3021 probe; lane 6, 3′LacZ RNA with only 3′LacZ probe; lanes 7 to 9, A, M, and I RNA, respectively, isolated from LMH transfected with 3′LacZ.
TABLE 2.
Representation of the 3′ region of the capsid pgRNA of DHBV variants
| DHBV variant | Mean level of region 3′ representation ± SD (n)
|
|
|---|---|---|
| Micrococcal nuclease capsid RNA (M) | Immunoprecipitated capsid RNA (I) | |
| 3′LACZa | 27 ± 3 (4) | 50 ± 11 (4) |
| ΔSpacerb | 27 ± 1 (2) | 47 ± 1 (2) |
The 5′ and 3′ regions of 3′LACZ were detected by using dual RPA probes, nt 2529 to 3021, for the detection of the 5′ region and 3′LACZ probe for detection of the 3′ region. Representation of the 3′ region was calculated as described in Table 1. n represents the number of independent analyses for each sample.
The representation of the 3′ region for ΔSpacer was calculated as described in Table 1, except values were calculated from RPA by using DHBV probe nt 2356 to 2666.
An internal deletion within the pgRNA does not change the susceptibility of the 3′ region of capsid pgRNA to micrococcal nuclease digestion.
Another possibility was that the 3′ region of the capsid pgRNA was susceptible to micrococcal nuclease digestion because of an RNA packaging limitation of the DHBV capsid that excluded the 3′ region from the interior of the capsid. For example, if the packaging limit was less than the length of the pgRNA and packaging occurred directionally from the 5′ end to 3′ end, then the remainder of the pgRNA in the 3′ region would be not be within the capsid and susceptible to micrococcal nuclease digestion. A prediction based on this hypothesis is that reducing the number of nucleotides of the pgRNA with an internal deletion would permit more of the 3′ region to be packaged into the interior of the capsid and therefore resistant to micrococcal nuclease. To test this idea, encapsidation of a DHBV variant with a 150-nt internal deletion (Fig. 6A, ΔSpacer) was examined. ΔSpacer contains an in-frame deletion within the nonessential spacer region of P. ΔSpacer is functional for encapsidation but cannot undergo reverse transcription as a result of the PYMHA mutation. According to this hypothesis, the internal 150-nt deletion of ΔSpacer would allow more sequence at the 3′ region to be protected from micrococcal nuclease digestion.
However, the 3′ region of ΔSpacer capsid RNA was still underrepresented and susceptible to nuclease digestion (Fig. 6B). RPA was performed with a probe (nt 2356 to 2666) that annealed only 90 nt downstream of the position in the 3′ region that can be detected for the M RNA of DHBV PY96F (nt 2577). The 3′ region of the M RNA of ΔSpacer was underrepresented at a similar level as the M RNA of DHBV PYMHA (Fig. 6B, compare lane 4 with lane 7, and Table 2). Also, immunoprecipitation of ΔSpacer capsids lead to a greater recovery of the 3′ region; however, there was still a reduction in the representation of 3′ region (Fig. 6B, lane 8, and Table 2). These results indicate that encapsidation of the 3′ region of ΔSpacer pgRNA has similar characteristics as encapsidation of DHBV PYMHA and that a 150-nt internal deletion did not affect the susceptibility of the 3′ region of the capsid pgRNA to micrococcal nuclease digestion.
DISCUSSION
These studies reveal that the 3′ region of the DHBV capsid pgRNA has biochemical characteristics different from those of other regions of the pgRNA. The 3′ region of the capsid pgRNA, including sequences within the 3′-terminal redundancy and the poly(A) tail, is susceptible to micrococcal nuclease digestion and underrepresented compared to the 5′ region of the capsid pgRNA. RPA and RNase H mapping of capsid pgRNA isolated by using micrococcal nuclease demonstrated that the 3′ end is variable from nt 2577 to the poly(A) tail. These data indicate there does not seem to be a distinct nucleotide position on the pgRNA where the 3′ end becomes susceptible to micrococcal nuclease. Isolation of capsid pgRNA by immunoprecipitation in the absence of nuclease treatment led to a greater recovery of the 3′ region, but the recovery accounted for only 50% of the molecules of capsid pgRNA. Therefore, the underrepresentation of the 3′ region of the capsid pgRNA was not solely due to micrococcal nuclease treatment. Finally, replacement of the 3′ region with heterologous sequence or introducing an internal deletion within the pgRNA did not change the micrococcal nuclease sensitivity of the 3′ region of the capsid pgRNA.
It is unclear why isolation of capsid RNA by immunoprecipitation of capsids in the absence of nuclease does not lead to complete recovery of the 3′ region of the capsid pgRNA. Our efforts to isolate capsid RNA in the presence of nuclease inhibitors consistently led to about 50% representation of the 3′ region when capsid RNA was isolated by immunoprecipitation in the absence of nuclease (data not shown). The 3′ region could still be degraded during the isolation of capsid pgRNA, or it could be degraded in vivo. Possibly, the immunoprecipitated fraction of capsid pgRNA represents two pools of RNA-containing capsids in cells: one with the 3′ region degraded and one with the 3′ region intact. Also, the 3′ region could be lost before the pgRNA is packaged into capsids. It would be interesting to examine the pgRNA that is not associated with capsids to understand the nature of the 3′ region prior to encapsidation.
Observations from previous reports are consistent with our findings. Büscher et al. (2) described DHBV transcripts isolated from the liver of infected Pekin ducks and identified a 3.0-kb RNA present in a poly(A)− fraction of RNA. This 3.0-kb, nonpolyadenylated RNA was less abundant than the ∼3.5-kb, polyadenylated message that was predicted to be the template for reverse transcription. The 3.0-kb RNA annealed to all positive-strand specific DHBV probes used to characterize the transcripts, indicating that 3.0-kb RNA contained most, if not all, sequences in the DHBV genome. The nature or the function of the 3.0-kb, nonpolyadenylated RNA is not known, but it is possible that this RNA represents capsid pgRNAs that have lost sequence at the 3′ region. Büscher et al. (2) did not use nuclease resistance to isolate RNA, but possibly, the 3.0-kb RNA lost sequences at the 3′ region in vivo or during the RNA isolation. In addition, Hirsch et al. (8) used an RPA probe that detected the 5′ and 3′ regions of the DHBV messages while studying the cis-acting sequences required for encapsidation. Similar to our work, they learned that they could abundantly detect the 5′ region of pgRNA in a capsid fraction of RNA, but they could not efficiently detect the 3′ region of capsid pgRNA. However, their system examined encapsidation of wild-type DHBV that was competent for reverse transcription. They attributed the inability to detect the 3′ region of capsid pgRNA as a consequence of RNase H activity after negative-strand DNA synthesis.
It is not known why the 3′ region of the capsid pgRNA is underrepresented and susceptible to micrococcal nuclease digestion. This characteristic of the capsid pgRNA may give us insight into the conformation of the pgRNA within its capsid. For example, the 3′ region of the pgRNA could be packaged into a part of the capsid that is more porous and leaves the 3′ region accessible to micrococcal nuclease. Another possibility is that the 3′ region of the pgRNA is not packaged into the interior of the capsid. This possibility would indicate that the 3′ region of the capsid pgRNA is positioned outside of the capsid, possibly through a pore in the capsid. The structure of the DHBV capsid, determined by cryoelectron microscopy, demonstrated that there are likely holes within the capsid that are ∼3 nm in diameter (10). The 3′ region of the pgRNA may be associated with these pores such that it is accessible for digestion by micrococcal nuclease.
It is interesting to consider the consequence of the 3′ region of capsid pgRNA being underrepresented and susceptible to micrococcal nuclease digestion. Most of the 3′-terminal redundancy is not used as a template in the synthesis of the mature viral DNA. Negative-strand DNA synthesis initiates within ɛ near the 5′ end of pgRNA and then switches templates to a position close to the 3′ end at nt 2537 (19, 20). Therefore, only sequence upstream of position nt 2538 is used as a template for reverse transcription. This could mean that the pgRNA has an excess of about 260 nt at the 3′ end that do not serve as a template during DNA synthesis. It is possible that the micrococcal nuclease sensitivity of the 3′ region of the pgRNA is related to the fact that this sequence of the pgRNA is not used as a template in the synthesis of mature viral DNA. Perhaps the exclusion of these 3′ sequences from the interior of the capsid is a mechanism to direct the negative-strand template switch or to prevent the nascent negative strand of DNA from switching templates to the incorrect acceptor site. Regardless of the reason for why the 3′ region of the capsid pgRNA is selectively susceptible to nuclease digestion, this new finding provides information about the nature of the capsid pgRNA and how it may have implications on the mechanisms of hepadnaviral replication.
Acknowledgments
We thank Jeff Habig, Ning Liu, and Jesse Summers for helpful discussions and critical review of the manuscript.
This study was supported by NIH grants PO1 CA22443 and P30 CA07175. K.M.O. was supported by a Mary Engsberg Fellowship.
REFERENCES
- 1.Bartenschlager, R., M. Junker-Niepmann, and H. Schaller. 1990. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 64:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buscher, M., W. Reiser, H. Will, and H. Schaller. 1985. Transcripts and the putative RNA pregenome of duck hepatitis B virus: implications for reverse transcription. Cell 40:717-724. [DOI] [PubMed] [Google Scholar]
- 3.Calvert, J., and J. Summers. 1994. Two regions of an avian hepadnavirus RNA pregenome are required in cis for encapsidation. J. Virol. 68:2084-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, L. J., P. Pryciak, D. Ganem, and H. E. Varmus. 1989. Biosynthesis of the reverse transcriptase of hepatitis B viruses involves de novo translational initiation not ribosomal frameshifting. Nature 337:364-368. [DOI] [PubMed] [Google Scholar]
- 5.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 7.Hirsch, R. C., J. E. Lavine, L. J. Chang, H. E. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature 344:552-555. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch, R. C., D. D. Loeb, J. R. Pollack, and D. Ganem. 1991. cis-Acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J. Virol. 65:3309-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenney, J. M., C. H. von Bonsdorff, M. Nassal, and S. D. Fuller. 1995. Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure 3:1009-1019. [DOI] [PubMed] [Google Scholar]
- 11.Loeb, D. D., A. A. Mack, and R. Tian. 2002. A secondary structure that contains the 5′ and 3′ splice sites suppresses splicing of duck hepatitis B virus pregenomic RNA. J. Virol. 76:10195-10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loeb, D. D., and R. Tian. 1995. Transfer of the minus strand of DNA during hepadnavirus replication is not invariable but prefers a specific location. J. Virol. 69:6886-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mabit, H., and H. Schaller. 2000. Intracellular hepadnavirus nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J. Virol. 74:11472-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obert, S., B. Zachmann-Brand, E. Deindl, W. Tucker, R. Bartenschlager, and H. Schaller. 1996. A splice hepadnavirus RNA that is essential for virus replication. EMBO J. 15:2565-2574. [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrow, K. M., and D. D. Loeb. 2002. Characterization of the cis-acting contributions to avian hepadnavirus RNA encapsidation. J. Virol. 76:9087-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlicht, H. J., G. Radziwill, and H. Schaller. 1989. Synthesis and encapsidation of duck hepatitis B virus reverse transcriptase do not require formation of core-polymerase fusion proteins. Cell 56:85-92. [DOI] [PubMed] [Google Scholar]
- 17.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 19.Tavis, J. E., S. Perri, and D. Ganem. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, G. H., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber, M., V. Bronsema, H. Bartos, A. Bosserhoff, R. Bartenschlager, and H. Schaller. 1994. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J. Virol. 68:2994-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei, Y., J. E. Tavis, and D. Ganem. 1996. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J. Virol. 70:6455-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoulim, F., and C. Seeger. 1994. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 68:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]