Abstract
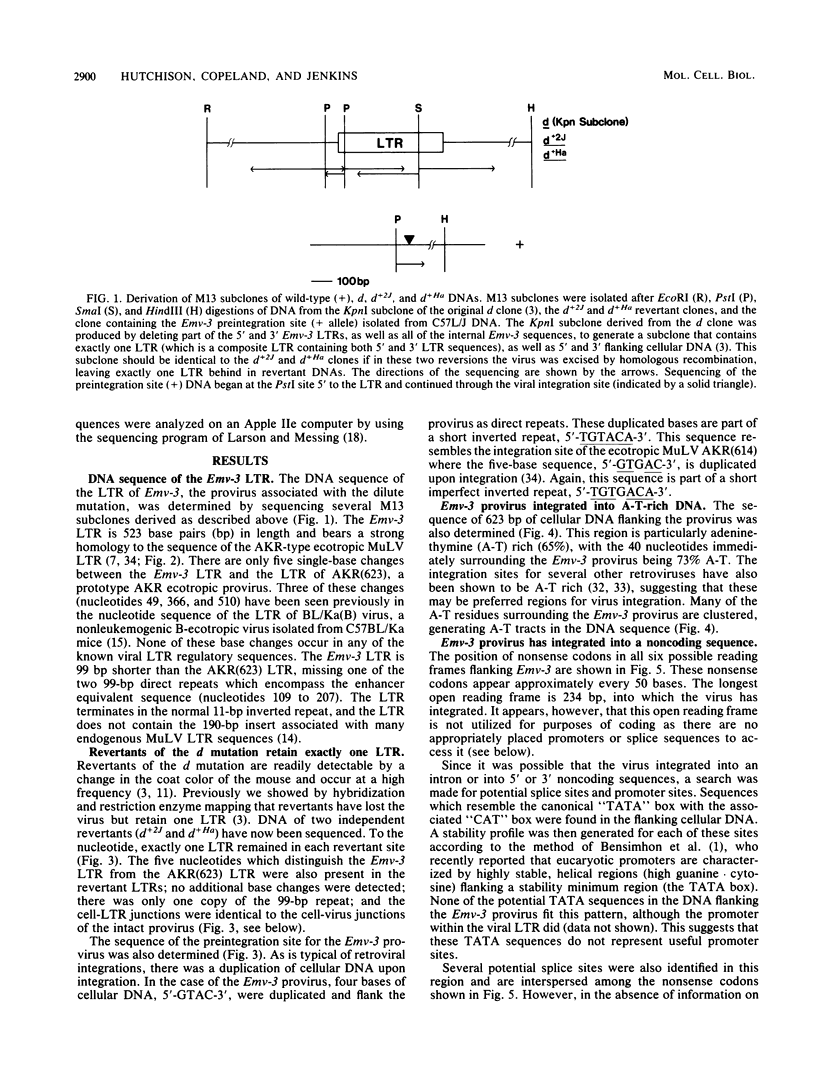
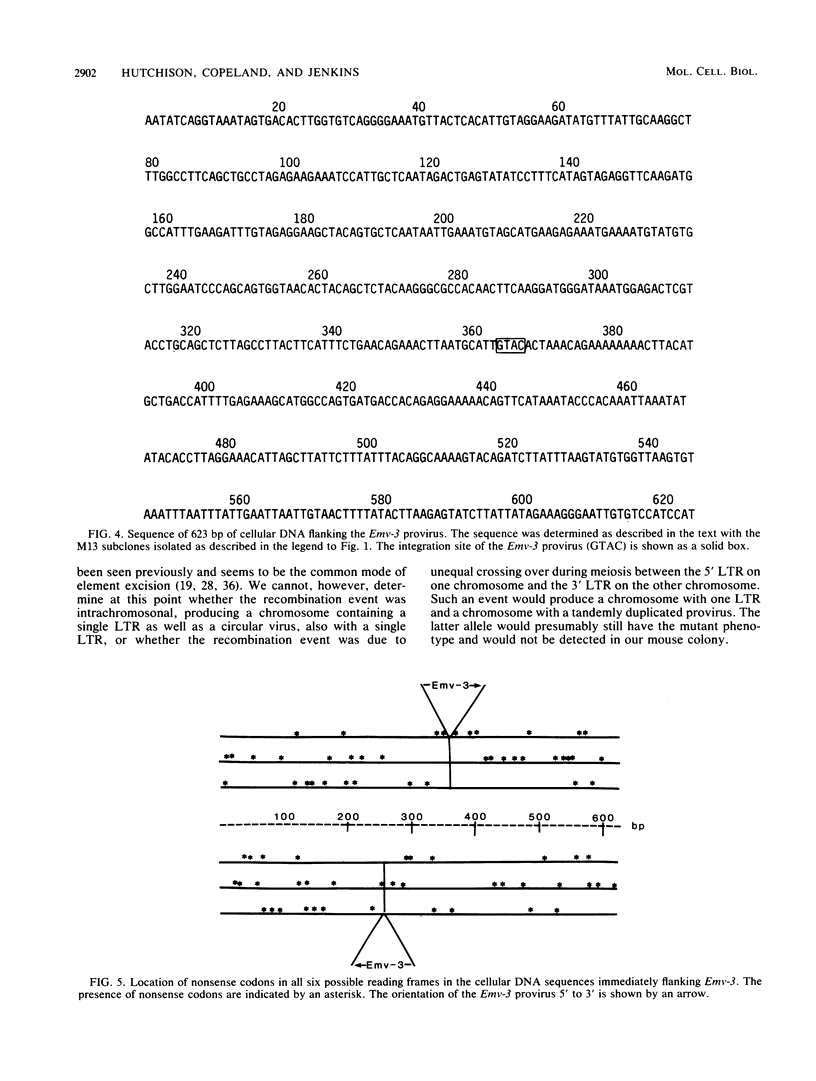
The unstable dilute-coat-color mutation (d) of DBA/2J mice has been shown to be the result of integration of an ecotropic murine leukemia virus within the mouse genome. Molecular cloning and restriction enzyme analysis of the dilute allele and the viral preintegration site (+ allele), as well as two independent dilute revertants (d+2J and d+Ha), suggested that reversion is due to virus excision occurring by homologous recombination involving the viral long terminal repeats. The DNA sequence has now been determined for the cell-virus junctions of the provirus associated with the d mutation, for the viral preintegration site, and for the two revertant sites. These data (i) indicate that the d mutation was caused by a normal virus integration, (ii) confirm that virus excision occurs by precise homologous recombination, as exactly one long terminal repeat is present in each revertant site, and (iii) suggest that the virus induced the d mutation by integration into a noncoding sequence.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bensimhon M., Gabarro-Arpa J., Ehrlich R., Reiss C. Physical characteristics in eucaryotic promoters. Nucleic Acids Res. 1983 Jul 11;11(13):4521–4540. doi: 10.1093/nar/11.13.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L., Vargas C., Spear B. B., Cox E. C. Transposable elements as mutator genes in evolution. Nature. 1983 Jun 16;303(5918):633–635. doi: 10.1038/303633a0. [DOI] [PubMed] [Google Scholar]
- Copeland N. G., Hutchison K. W., Jenkins N. A. Excision of the DBA ecotropic provirus in dilute coat-color revertants of mice occurs by homologous recombination involving the viral LTRs. Cell. 1983 Jun;33(2):379–387. doi: 10.1016/0092-8674(83)90419-1. [DOI] [PubMed] [Google Scholar]
- Harbers K., Kuehn M., Delius H., Jaenisch R. Insertion of retrovirus into the first intron of alpha 1(I) collagen gene to embryonic lethal mutation in mice. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1504–1508. doi: 10.1073/pnas.81.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R. G., Shulman M. J., Murialdo H., Gibson D. M., Hozumi N. Mutant immunoglobulin genes have repetitive DNA elements inserted into their intervening sequences. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7425–7429. doi: 10.1073/pnas.79.23.7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Herr W. Nucleotide sequence of AKV murine leukemia virus. J Virol. 1984 Feb;49(2):471–478. doi: 10.1128/jvi.49.2.471-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenaga H., Saigo K. Insertion of a movable genetic element, 297, into the T-A-T-A box for the H3 histone gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4143–4147. doi: 10.1073/pnas.79.13.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg R. R., Syvanen M. DNA gyrase is a host factor required for transposition of Tn5. Cell. 1982 Aug;30(1):9–18. doi: 10.1016/0092-8674(82)90006-x. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Harbers K., Schnieke A., Löhler J., Chumakov I., Jähner D., Grotkopp D., Hoffmann E. Germline integration of moloney murine leukemia virus at the Mov13 locus leads to recessive lethal mutation and early embryonic death. Cell. 1983 Jan;32(1):209–216. doi: 10.1016/0092-8674(83)90511-1. [DOI] [PubMed] [Google Scholar]
- Jenkins N. A., Copeland N. G., Taylor B. A., Lee B. K. Dilute (d) coat colour mutation of DBA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature. 1981 Oct 1;293(5831):370–374. doi: 10.1038/293370a0. [DOI] [PubMed] [Google Scholar]
- Jenkins N. A., Copeland N. G., Taylor B. A., Lee B. K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982 Jul;43(1):26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsrud L., Calos M. P., Miller J. H. The transposon Tn9 generates a 9 bp repeated sequence during integration. Cell. 1978 Dec;15(4):1209–1219. doi: 10.1016/0092-8674(78)90047-8. [DOI] [PubMed] [Google Scholar]
- Khan A. S., Martin M. A. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci U S A. 1983 May;80(9):2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. P., Kaplan H. S., Fry K. E. Characterization of an infective molecular clone of the B-tropic, ecotropic BL/Ka(B) murine retrovirus genome. J Virol. 1982 Oct;44(1):217–225. doi: 10.1128/jvi.44.1.217-225.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuff E. L., Feenstra A., Lueders K., Rechavi G., Givol D., Canaani E. Homology between an endogenous viral LTR and sequences inserted in an activated cellular oncogene. Nature. 1983 Apr 7;302(5908):547–548. doi: 10.1038/302547a0. [DOI] [PubMed] [Google Scholar]
- Kühn S., Fritz H. J., Starlinger P. Close vicinity of IS1 integration sites in the leader sequence of the gal operon of E. coli. Mol Gen Genet. 1979 Jan 2;167(3):235–241. doi: 10.1007/BF00267414. [DOI] [PubMed] [Google Scholar]
- Larson R., Messing J. Apple II computer software for DNA and protein sequence data. DNA. 1983;2(1):31–35. doi: 10.1089/dna.1.1983.2.31. [DOI] [PubMed] [Google Scholar]
- Levis R., Rubin G. M. The unstable wDZL mutation of Drosophila is caused by a 13 kilobase insertion that is imprecisely excised in phenotypic revertants. Cell. 1982 Sep;30(2):543–550. doi: 10.1016/0092-8674(82)90251-3. [DOI] [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel B. G., Hayward W. S., Robinson H. L., Fang J., Astrin S. M. Avian leukosis virus-induced tumors have common proviral integration sites and synthesize discrete new RNAs: oncogenesis by promoter insertion. Cell. 1981 Feb;23(2):323–334. doi: 10.1016/0092-8674(81)90128-8. [DOI] [PubMed] [Google Scholar]
- Noori-Daloii M. R., Swift R. A., Kung H. J., Crittenden L. B., Witter R. L. Specific integration of REV proviruses in avian bursal lymphomas. Nature. 1981 Dec 10;294(5841):574–576. doi: 10.1038/294574a0. [DOI] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Nusse R., van Ooyen A., Cox D., Fung Y. K., Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984 Jan 12;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Bishop J. M., Varmus H. E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982 Jan 21;295(5846):209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Courtneidge S. A., Crittenden L. B., Fadly A. M., Bishop J. M., Varmus H. E. Analysis of avian leukosis virus DNA and RNA in bursal tumours: viral gene expression is not required for maintenance of the tumor state. Cell. 1981 Feb;23(2):311–322. doi: 10.1016/0092-8674(81)90127-6. [DOI] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Farabaugh P. J., Chaleff D. T., Fink G. R. The origins of gene instability in yeast. Science. 1980 Sep 19;209(4463):1375–1380. doi: 10.1126/science.6251544. [DOI] [PubMed] [Google Scholar]
- Russell L. B. Definition of functional units in a small chromosomal segment of the mouse and its use in interpreting the nature of radiation-induced mutations. Mutat Res. 1971 Jan;11(1):107–123. doi: 10.1016/0027-5107(71)90036-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller H. The intergenic region and the origins for filamentous phage DNA replication. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):401–408. doi: 10.1101/sqb.1979.043.01.046. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Tamura T., Takano T. Long terminal repeat (LTR)-derived recombination of retroviral DNA: sequence analyses of an aberrant clone of baboon endogenous virus DNA which carries an inversion from the LTR to the gag region. Nucleic Acids Res. 1982 Sep 11;10(17):5333–5343. doi: 10.1093/nar/10.17.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Quintrell N., Ortiz S. Retroviruses as mutagens: insertion and excision of a nontransforming provirus alter expression of a resident transforming provirus. Cell. 1981 Jul;25(1):23–36. doi: 10.1016/0092-8674(81)90228-2. [DOI] [PubMed] [Google Scholar]
- Wolf D., Rotter V. Inactivation of p53 gene expression by an insertion of Moloney murine leukemia virus-like DNA sequences. Mol Cell Biol. 1984 Jul;4(7):1402–1410. doi: 10.1128/mcb.4.7.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]


