Abstract
Drosophila olfactory sensory neurons express either odorant receptors or ionotropic glutamate receptors (IRs). The sensory neurons that express IR64a, a member of the IR family, send axonal projections to either the DC4 or DP1m glomeruli in the antennal lobe. DC4 neurons respond specifically to acids/protons, whereas DP1m neurons respond to a broad spectrum of odorants. The molecular composition of IR64a-containing receptor complexes in either DC4 or DP1m neurons is not known, however. Here, we immunoprecipitated the IR64a protein from lysates of fly antennal tissue and identified IR8a as a receptor subunit physically associated with IR64a by mass spectrometry. IR8a mutants and flies in which IR8a was knocked down by RNAi in IR64a+ neurons exhibited defects in acid-evoked physiological and behavioral responses. Furthermore, we found that the loss of IR8a caused a significant reduction in IR64a protein levels. When expressed in Xenopus oocytes, IR64a and IR8a formed a functional ion channel that allowed ligand-evoked cation currents. These findings provide direct evidence that IR8a is a subunit that forms a functional olfactory receptor with IR64a in vivo to mediate odor detection.
Introduction
The olfactory system detects odors through a large repertoire of diverse sensory receptors expressed on the surface of olfactory sensory neurons (OSNs). In mammals, binding of odorant ligands to odorant receptors (ORs) leads to a cascade of events including the activation of G-proteins and adenylyl cyclases, the elevation of cyclic AMP level, and the opening of cyclic nucleotide-gated channels (Ronnett and Moon, 2002; Touhara and Vosshall, 2009). The signaling pathway in the insect olfactory system appears to be more complex. It has been proposed, for example, that ORs function as both G-protein-coupled receptors (Wicher et al., 2008) and as ligand-gated ion channels (Sato et al., 2008; Wicher et al., 2008). There may also be a greater variety of chemosensory receptors in insects. For example, the Drosophila olfactory organ expresses two gustatory receptors (Suh et al., 2004; Jones et al., 2007) that use the Gαq signaling pathway to detect carbon dioxide (Yao and Carlson, 2010). Additionally, a family of chemosensory ionotropic glutamate receptors (IRs) was identified previously that is expressed in both the olfactory and gustatory systems of the Protostomia clade, which encompasses nematodes, insects, and mollusks and crustaceans (Benton et al., 2009; Croset et al., 2010). These IRs are members of the ionotropic glutamate receptor family (iGluRs), which includes the AMPA, NMDA, and kainate receptors. The chemosensory IRs share conserved transmembrane domains with classic iGluRs, but contain large variations within their putative amino terminal domains, ligand binding domains, and C termini.
In Drosophila melanogaster, IRs and ORs are expressed largely by nonoverlapping populations of OSNs located inside sensilla—specialized sensory hairs located on the surface of the insect olfactory organs, antenna, and maxillary palp. IRs are expressed by OSNs that innervate the coeloconic sensilla, whereas ORs are expressed by OSNs that innervate the basiconic and trichoid sensilla with the exception of OR35a, which is expressed in OSNs innervating coeloconic sensilla (Vosshall and Stocker, 2007; Benton et al., 2009). The axons of OSNs that express the same type of ORs or IRs converge their axons onto a single pair of glomeruli in the antennal lobe (AL). IR- and OR-expressing OSNs project to complementary sets of glomeruli in the AL that appear restricted in the posterior and anterior zones of the AL, respectively (Couto et al., 2005; Fishilevich and Vosshall, 2005; Silbering et al., 2011). In contrast, the axon terminals of the projection neurons (PNs) postsynaptic to IR or OR OSNs are interdigitated within higher brain centers—the mushroom body (MB) and the lateral horn (LH) (Silbering et al., 2011).
Approximately 60 IRs have been identified in the Drosophila genome, 17 of which are expressed in the antenna (Benton et al., 2009; Croset et al., 2010). Each IR+ neuron expresses two to four IRs, which are thought to form a functional receptor complex (Abuin et al., 2011; Silbering et al., 2011), although the in vivo evidence supporting physical interactions among these IRs is missing. In contrast to ORs, which are broadly tuned to alcohols, ketones, and esters, IRs are tuned primarily to acids and amines (Yao et al., 2005; Hallem and Carlson, 2006; Silbering et al., 2011). A growing body of evidence suggests that some IRs respond specifically to a single class of odorants and that the OSNs that express these IRs mediate hardwired innate behaviors (Ai et al., 2010; Grosjean et al., 2011). In a previous study, we found that IR64a is required in the Drosophila olfactory system for acid detection (Ai et al., 2010). Specifically, inactivation of IR64a or IR64a+ neurons led to impaired physiological and behavioral responses to acids/protons, while stimulation of IR64a+ neurons was sufficient to elicit an aversive behavioral response (Ai et al., 2010). IR64a+ OSNs send their dendrites to coeloconic sensilla located in the third chamber of a specialized internal structure in the antenna called the sacculus and project their axons to either the DC4 or DP1m glomerulus in the antennal lobe. Interestingly, the DC4 glomerulus responds specifically to acids, whereas DP1m is activated by a wide variety of odorants. The molecular mechanism that distinguishes the function of DC4 from that of DP1m remains unclear. Here we report that IR8a physically associates with IR64a, and that IR8a is required for trafficking and stability of IR64a protein. Furthermore, we found that IR64a and IR8a form a functional ligand-gated cation channel.
Materials and Methods
Transgenic flies and fly stocks.
The IR64a-HA genomic rescue construct (Ai et al., 2010) to produce a transgenic fly line was engineered in pCaSpeR4 using 8 kb of the 5′-UTR of the IR64a genomic sequence, 4 kb of the IR64a genomic coding sequence (including introns), and an in-frame HA coding sequence followed by 1.4 kb of the 3′-UTR of the IR64a genomic sequence. PromoterIR64a-mCherry was made in pCaSpeR4 by cloning 8 kb of the 5′-UTR of IR64a fused with a DNA sequence encoding mCherry. IR8a-GAL4 was generated by cloning a DNA sequence upstream of IR8a start codon (676 bp) fused directly to the sequences from IR8a intron 1 (59 bp), intron 2 (131 bp), and intron 3 (789 bp) into pCaSpeR-AUG-GAL4. Transgenic lines expressing UAS-IR8a RNAi [Vienna Drosophila RNAi Center (VDRC) IDs 51142 and 29917] were obtained from the VDRC Stock Center (Vienna, Austria). nSyb-GAL4 was a gift from J. Simpson (Janelia Farm Research Campus, Ashburn, VA). Flies carrying RNAi lines also bear UAS-Dicer2. Other flies were described previously: IR64ami (Ai et al., 2010); ORCO−/− (Larsson et al., 2004) and IR8a1 (Abuin et al., 2011); UAS-GCaMP3.0 (Tian et al., 2009); and UAS-C3PA and UAS-SPA (Ruta et al., 2010).
Fly rearing.
We raised ∼200,000 control (Canton S) and experimental (IR64ami-expressing IR64a-HA genomic rescue transgene) flies for each round of the coimmunoprecipitation (co-IP) experiment in custom-built population cages at room temperature. Each population cage was made of a plastic cylinder (30 cm in diameter) with a cloth cap at one end and a spandex bundle forming an entrance at the other end. The cages were placed horizontally and the fly food was provided in a pan placed inside the cage. A breeding cage contained ∼500 adult male and female flies. New fly food pans were put in the breeding cage twice a week. The old food pans containing eggs and larvae were placed in new population cages (rearing cages) to allow flies to grow. Each rearing cage produced ∼10,000 adult flies on average. Four- to seven-day-old adult flies from rearing cages were harvested by CO2 anesthesia, weighed, and stored at −80°C until needed. One adult fly weighs ∼1 mg.
Coimmunoprecipitation.
Four- to seven-day-old flies were frozen in liquid nitrogen and passed through a set of cold fine metal sieves (10 cm diameter) of different mesh size (710, 250, and 180 μm) by vigorous shaking. Upon sieving, dismantled body parts were sorted according to their size: bodies were retained in the 710 μm sieve, heads and wings were retained in the 250 μm sieve, and legs were retained in the 180 μm sieve, while antennae and halteres passed through all sieves and thus became enriched in the bottom vessel. By visual inspection, the bottom fraction contained ∼50% antennae, ∼25% broken legs, ∼20% halteres, and ∼5% other tissues. For each round of sieving, 30∼35 g of frozen flies (one fly weighs ∼1 mg on average) were used. The sieving procedure was repeated six times to sieve ∼200,000 flies (∼200 g).
The antennal tissues were collected and homogenized using a mortar and pestle in the presence of liquid nitrogen. Proteins were extracted by incubating the homogenized tissues in lysis buffer (25 mm HEPES, pH 7.25, 150 mm NaCl, 2 mm EDTA, 0.5% n-dodecyl-β-maltoside, 0.15% CHAPS, 1× protease inhibitor cocktail) at 4°C for 3 h. A preabsorption step was performed in which the supernatant was incubated with rat-IgG agarose beads (Santa Cruz Biotechnology; catalog #sc-2344) at 4°C for 2 h. The preabsorbed supernatant was then incubated with rat anti-HA agarose beads (Roche; catalog #1815016) at 4°C overnight. The beads were washed three times for 10 min each in wash buffer (25 mm HEPES, pH 7.25, 500 mm NaCl, 0.1 mm EDTA, 0.5% n-dodecyl-β-maltoside, 0.15% CHAPS, 1× protease inhibitor cocktail) at room temperature. Immunoprecipitated proteins were eluted in 0.1 m glycine solution, pH 2.0, neutralized immediately by 10N NaOH, mixed with SDS-PAGE loading buffer, and stored at −20°C before further analysis.
Western blotting.
Protein samples were subjected to SDS-PAGE gel electrophoresis. Proteins were then transferred to a PVDF membrane, which was blocked with 5% milk and subsequently blotted by primary antibodies at 4°C overnight. Primary antibodies used for Western blots were as follows: rat anti-HA (Roche; catalog #11867423001), 1:1000; guinea pig anti-IR8a (Abuin et al., 2011), 1:1000; mouse anti-tubulin (Covance; catalog #MMS-410P), 1:40,000. The PVDF membrane was incubated with horseradish peroxide-conjugated secondary antibodies at room temperature for 1 h. Protein bands were visualized by using an ECL SuperSignal chemiluminescent substrate (Thermo Scientific; catalog #34094).
Mass spectrometry.
About 90% of the eluted fraction from anti-HA beads was concentrated and loaded onto an SDS-PAGE gel. After electrophoresis, the gel was stained using the Invitrogen Silver Quest silver stain (catalog #LC6070). Both HA and control lanes were each cut into seven equal individual slices without regard to the gel staining pattern. The samples were destained, reduced with 20 mm DTT, and alkylated with 50 mm iodoacetamide. The samples were then digested overnight with 0.1 μg trypsin per gel slice. Tryptic peptides were extracted, dried under a vacuum, and then resuspended in 12 μl 0.1% formic acid. Eight microliters of each sample were loaded onto a 75 μm × 12 cm column self-packed with 3 μm ReproSil-Pur C18-AQ beads (Dr. Maisch), eluted with a gradient of 2–40% acetonitrile in 0.1% formic acid over 50 min at 300 nl/min, and analyzed using an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). Proteins were identified using the Andromeda search engine (MaxQuant version 1.2.2.5) with cysteine carbamidomethylation specified as a fixed modification and methionine oxidation as a variable modification to search the UniProt Drome.Fasta database (downloaded October 30, 2011). Relative quantities of the proteins were determined using the iBAQ feature of MaxQuant.
Immunohistochemistry.
Antibodies used for immunostaining were as follows: rat anti-HA (Roche; catalog #11867423001), 1:1000; guinea pig anti-IR8a (Abuin et al., 2011), 1:1000; rabbit anti-IR64a (Ai et al., 2010), 1:1000; monoclonal 21A6 (Developmental Studies Hybridoma Bank), 1:200; rabbit anti-DsRed (Clontech; catalog #632496), 1:1000; chicken anti-green fluorescent protein (GFP; Abcam; catalog #ab13970), 1:1000.
Immunostaining of whole-mount brain and cryosectioned antenna were performed as described previously (Ai et al., 2010). Whole-mount antennae staining by anti-HA as seen in Figure 1A was done similarly to the immunostaining of whole-mount brains. Because the antibody cannot penetrate the cuticle of the antennae, only the sensillar tips, but not the cell bodies, of IR64a+ neurons were stained.
Figure 1.
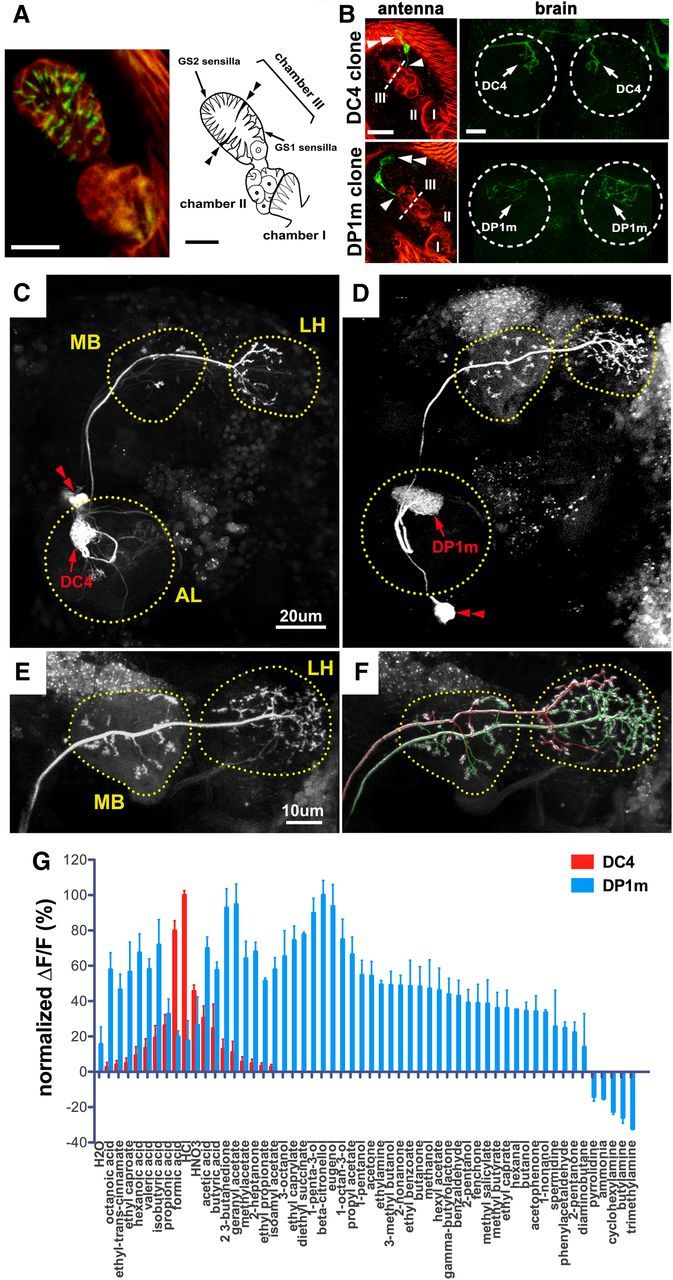
Anatomic separation and functional distinction of neurons innervating DC4 and DP1m. A, Left, Anti-HA (green) immunostaining of a whole-mount antenna from an IR64a-HA transgenic fly reveals the sensilla tips (green) of IR64a+ OSNs. Red autofluorescence depicts the outline of the sacculus. Right, Drawing of the sacculus. Double arrowheads point to a pair of thick cuticular flaps that separate dorsal and ventral compartments of Chamber III of the sacculus. B, Flip-out labeling of single cell clones (green) in flies carrying IR64a-GAL4, UAS-frt-Stop-frt-CD8GFP, and hs-Flip. Examples of single cells projecting bilaterally to either DC4 (top row) or DP1m (bottom row) are shown with images of the antennae on the left and the corresponding antennal lobes on the right. Arrowheads show sites of dendritic innervation in the sacculus; double arrowheads show cell bodies. The dotted line indicates cuticular flaps that separate dorsal and ventral compartments of Chamber III. The dotted circle indicates antennal lobe. Scale bars: A, B, 10 μm. C, D, PA-GFP labeling of a DC-PN (C) and a DP1m-PN (D) in the brains of nSyb-GAL4; UAS-C3PA flies. The outline of each neuropil was determined by the background fluorescence of PA-GFP. Red double arrowheads indicate PN cell bodies. E, F, PA-GFP labeling of a single DP1m-PN (E) followed by labeling of a single DC4-PN (F) in the same brain of a fly carrying nSyb-GAL4; UAS-C3PA. The axonal projections of the DP1m PN and DC4 PN were mapped in 3D space and labeled in green and red, respectively, by using the Vaa3D software (Peng et al., 2010). Note that axonal termini of the DP1m-PN (green) and DC4-PN (red) occupy largely nonoverlapping space within the LH. The double labeling was repeated three times with similar results. G, Odor tuning properties of DC4 (red) and DP1m (blue) glomeruli in flies carrying IR64a-GAL4; UAS-GCaMP3.0 were measured by calcium imaging. For each glomerulus, the GCaMP fluorescence intensity changes (ΔF/F) were normalized to the maximal ΔF/F response, which was defined as 100%.
In vivo calcium imaging.
Live fly preparation and in vivo calcium imaging experiments were performed as described previously (Ai et al., 2010). Flies described in Figure 4 were 8–10 d old, and their genotypes were as follows: wild-type (UAS-GCaMP3; IR64a-GAL4), IR8a1 (IR8a1; UAS-GCaMP3; IR64a-GAL4), IR8a-RNAi (IR64a-GAL4, UAS-dcr2; UAS-GCaMP3.0, UAS-IR8a-RNAi), IR25a-RNAi (IR64a-GAL4, UAS-dcr2; UAS-GCaMP3.0, UAS-IR25a-RNAi), and IR64ami (UAS-GCaMP3; IR64a-GAL4, IR64ami).
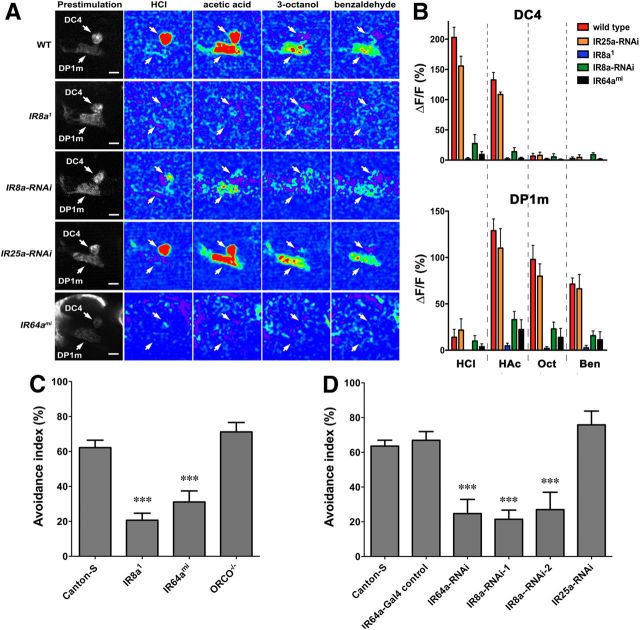
Figure 4.
IR8a is required specifically in IR64a+ neurons for the physiological and behavioral responses to odorants. A, Calcium imaging of flies carrying IR64a-GAL4; UAS-GCaMP3.0 in wild-type (WT), IR8a mutant (IR8a1), IR8a-RNAi (IR64a-GAL4, UAS-dcr2, UAS-IR8aRNAi), IR25a-RNAi (IR64a-GAL4, UAS-dcr2, UAS-IR25aRNAi), or IR64a mutant (IR64ami) backgrounds. Arrows indicate DC4 and DP1m glomeruli. Scale bars, 10 μm. B, Fluorescence intensity changes (ΔF/F) of DC4 (top) and DP1m (bottom) in response to odorants were quantified. N = 5. HCl, Hydrochloric acid (3.6%); HAc, acetic acid (1%); Oct, 1-octanol (1%); Ben, benzaldehyde (1%). C, Avoidance to acetic acid in wild-type and different mutant flies in a T maze. N = 10∼16. D, Avoidance to acetic acid in flies carrying different UAS-RNAi transgenes driven by IR64a-GAL4. N = 8∼16. ***p < 0.01 (ANOVA with Tukey's test).
Behavioral analysis.
For acid avoidance assays using a T maze, 10 μl of acetic acid solution (10% v/v diluted in distilled water) or water as a control was dispensed onto a piece of filter paper (5 × 5 mm) placed in a 14 ml tube (Thermo Fisher Scientific; catalog #149598). Tubes were sealed with Parafilm and allowed to stand for at least 10 min at room temperature before experiments. Flies (8–10 d old) were introduced into the elevator of a T maze by gentle tapping. A tube containing acetic acid was placed onto one side of the T maze, and a control tube was placed onto the other side. Flies in the elevator were then given a choice between the “acid side” and the “control side” for 30 s. The avoidance index was calculated as follows: (the number of flies in control tube − the number of flies in acid tube)/number of flies in both tubes. In Figure 4, flies carrying UAS-RNAi transgenes also carried UAS-Dicer2.
Xenopus oocyte expression and electrophysiological recordings.
cDNA sequences encoding full-length IR64a and IR8a were cloned into oocyte expression vector pGEM-HE (Liman et al., 1992). cRNA were synthesized using a standard protocol. Twenty to 30 ng of cRNA were injected into healthy Stage V and VI oocytes. Electrophysiological measurements were performed 2–3 d after injection. Currents were recorded under voltage clamp at a holding potential of −50 mV or as indicated. At each holding potential, an odor-evoked current (Iodor) was calculated by subtracting the current measured in control buffer (pH 7.3) from the current measured in response to stimuli (1 mm acetate or pH 5.5 buffer).
Single-cell labeling by photoactivatable GFP.
The brain from a fly carrying nSyb-GAL4; UAS-C3PA was used for the labeling of single cells. Brains from <1-d-old flies were dissected in a buffer [containing the following (in mm): 108 NaCl, 5 KCl, 2 CaCl2, 8.2 MgCl2, 4 NaHCO3, 1 NaH2PO4, 5 trehalose dihydrate, 10 sucrose, 5 HEPES, pH 7.5] and immobilized by pinning down onto silicone gel. Before photoconversion, the low-intensity fluorescence of photoactivatable (PA)-GFP protein was visualized by two-photon illumination at a 925 nm wavelength. Both DC4 and DP1m glomeruli were easily identifiable according to their position within the antennal lobe. The three-dimensional structure of the glomerulus was stimulated using weak photoconverting light at a 715 nm wavelength for five cycles with 30 s intervals in between. After the photoconversion, PA-GFP proteins within the glomerulus enhanced fluorescent intensity and diffused and labeled the PN cell bodies. Then stronger photoconverting two-photon light was applied to a single labeled PN cell body for 60 cycles with 30 s intervals, which led to robust labeling of the entire structure of the cell including axonal and dendritic terminals. Sequential labeling of DP1m- and DC4-PNs in the same brain was done similarly. The axonal projections of the PNs were traced in three-dimensional space by using the autotracing function of the Vaa3d software (Peng et al., 2010) with the assistance of a manual tracing function.
Results
Two populations of OSNs that express IR64a
IR64a is expressed by ∼15 OSNs that innervate their dendritic terminals into the sacculus (Shanbhag et al., 1995), an internal pit-like, tripartite structure in the antenna (Fig. 1A). The sacculus is lined with two types of sensilla: basiconic sensilla, which line the entrance to the sacculus (Chamber I, found on the posterior surface of the antenna), and coeloconic sensilla, which line Chambers II (the middle chamber) and III. Chamber III is divided further by a thick cuticular flap (Fig. 1A, double arrowheads) into ventral and dorsal compartments containing thick type 1 grooved (GS1) sensilla and slender GS2 sensilla, respectively. Odorants presumably diffuse through these chambers and activate coeloconic sensilla in Chambers II and III. To determine the exact location of IR64a+ dendrites within the sacculus, we immunostained the whole-mount antennae of IR64a-HA genomic rescue transgenic flies with anti-HA antibody. The IR64a-HA fusion protein confers a functional IR64a receptor as it rescued IR64a mutant phenotypes (Ai et al., 2010). Notably, we found that the stain was taken up at the tip of each sensillum in Chamber III (Fig. 1A, green), but not by its proximal dendrite or the cell body. This suggests that the IR64a receptors at the dendritic tips are accessible to the antibody and thus to odorants in the external environment. Furthermore, this result shows that IR64a+ dendrites innervate both the dorsal and ventral compartments of Chamber III.
IR64a+ neurons project their axons to two adjacent glomeruli in the antennal lobe: DC4 and DP1m. It is not known whether a single IR64a+ neuron innervates both glomeruli or whether one IR64a+ subpopulation targets DC4 while another targets DP1m. To determine the pattern of innervation in this region, we labeled individual IR64a+ cells using the flip-out technique by driving the expression of UAS-frt-Stop-frt-mCD8GFP (Basler and Struhl, 1994) under the control of IR64a-GAL4. After inducing heat shock briefly in third-instar larvae to express flipase (hs-Flp), we obtained adult flies in which the stop cassette was excised randomly in a single IR64a+ cell, which as a result expresses GFP. Indeed, of the 87 flip-out flies analyzed, we found seven flies with a single GFP+ cell in their antennae. We also found that each of these IR64a+ neurons innervated either the DC4 glomerulus (four cells) or the DP1m glomerulus (three cells), but not both (Fig. 1B; Table 1).
Table 1.
Summary of flip-out experiments
| DC4 |
DP1m |
|||
|---|---|---|---|---|
| Dorsal sacculus | Ventral sacculus | Dorsal sacculus | Ventral sacculus | |
| Number of single-cell flip-out flies | 0 | 4 | 3 | 0 |
| Number of multiple-cell flip-out flies | 0 | 1 fly (4 cells) | 3 flies (∼2–4 cells each) | 0 |
Flies with single or multiple flip out cells that project exclusively to either DC4 or DP1m were categorized according to their dendritic innervation to either the dorsal or ventral compartment of sacculus Chamber III.
Interestingly, we found evidence of a correlation between the anatomic location of IR64a+ dendrites in the sacculus and their glomerular target in the antennal lobe. The dendrites of DC4-targeting neurons innervated the ventral compartment of Chamber III, while the dendrites of DP1m-targeting cells innervated the dorsal compartment (Fig. 1B). In further support of this, we found one additional flip-out fly that labeled four DC4 neurons, all of which extended their dendrites to the ventral compartment of Chamber III, and four additional flip-out flies that labeled two to four DP1m neurons, all of which innervate the dorsal compartment of Chamber III (Table 1). We did not find a correlation between the anatomic location of the IR64a+ cell body and the target of their axon or dendrite. Together, our findings suggest that DC4 neurons innervate the thick GS1 type coeloconic sensilla in the ventral compartment of Chamber III, and DP1m neurons innervate the thin GS2 coeloconic sensilla in the dorsal compartment.
DC4 and DP1m projection neurons at higher brain centers
We next asked how olfactory information downstream of DC4 and DP1m glomeruli is represented anatomically in higher brain centers. We used PA-GFP (Patterson and Lippincott-Schwartz, 2002; Datta et al., 2008; Ruta et al., 2010) to label individual PNs postsynaptic to DC4 and DP1m OSNs. Specifically, we expressed improved versions of PA-GFP, UAS-C3PA (Ruta et al., 2010) driven by a pan-neuronal driver nSyb-GAL4, and used a two-photon laser to illuminate the DC4 or DP1m glomerulus and subsequently the PN cell bodies (for details, see Materials and Methods). By doing so, we labeled single DC4- and DP1m-PNs with strong GFP fluorescence and found that they project their axons along the inner antennocerebral tract (iACT) to innervate both the MB and the LH (Fig. 1C,D). The DC4-PN axonal terminal bifurcates to form stereotypical dorsal and ventral branches in the anterior medial region of the LH (Fig. 1C), which was not described previously because available PN drivers such as GH146-GAL4 and acj-GAL4 do not label DC4-PN. In contrast to the DC4-PN, we found that the DP1m-PN had extensive axonal branches that occupy large regions of the LH (Fig. 1D), which is consistent with previous reports (Marin et al., 2002; Jefferis et al., 2007). While we found two to three DC4-PNs cell bodies in each brain by PA-GFP labeling, there is apparently only one DP1m-PN. We have successfully labeled a total of 10 individual DC4-PNs and 8 DP1m-PNs in different animals, all of which project to the MB and the LH via iACT with the stereotypic branching patterns.
To further compare the axonal branching of DC4-PN and DP1m-PN axons in the LH, we labeled a single DP1m-PN followed by the labeling of a single DC4-PN in the same brain. As shown in Figure 1F, the DC4-PN (labeled in red) and DP1m-PN (labeled in green) occupy largely nonoverlapping space within the LH: the dorsal axonal branch of the DC4-PN innervates regions adjacent to that occupied by DP1m-PN axons, while the ventral branch of the DC4-PN occupies distinct regions in the LH. This finding suggests that the olfactory information detected by the functionally and anatomically distinct DC4 and DP1m OSNs is presented in spatially distinct loci within the LH. We did not systematically compare the innervation of DC4 versus DP1m PNs in the MB since innervation of PNs in the MB is less stereotyped.
Odorant tuning of DC4 and DP1m OSNs
Having determined the anatomical characteristics of DC4 and DP1m cells, we sought to characterize the odorant response profiles for DC4 and DP1m OSNs by monitoring their responses to 56 different odorants. Using in vivo calcium imaging, we found that DC4 and DP1m have different odor tuning properties: DC4 responds specifically to acidic odorants, and DP1m is broadly tuned to many odorants (Fig. 1G), which is consistent with previous results (Ai et al., 2010). The distinct odor response profile of these two populations of IR64a+ neurons supports a hypothesis that additional factors in DC4 and DP1m neurons exist and function with IR64a to confer odor sensitivity.
IR8a physically interacts with IR64a in vivo
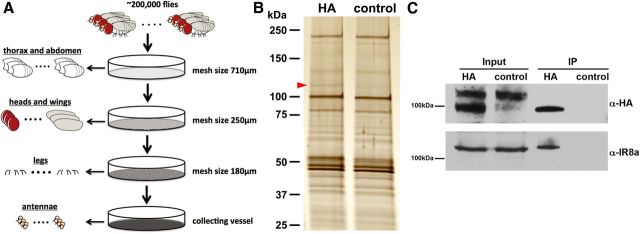
To gain insights into the molecular composition of the IR64a-containing receptor complex that is likely to be responsible for distinct odor tuning profiles in DC4 and DP1m, we performed co-IP coupled with mass spectrometry (MS) analysis to identify IR64a-associated proteins. We began the immunoprecipitation process by inducing the expression of the IR64-HA genomic rescue transgene in both DC4 and DP1m neurons in an IR64a mutant background. This allowed us to tag all functional IR64a proteins in flies with HA and thus made sure that we accumulated enough material to carry out the co-IP experiments, a crucial step considering the limited number of neurons (∼15) in each antenna that express IR64a. Once ∼200,000 flies were available, we isolated their antennae (Fig. 2A) (see Materials and Methods) and then performed co-IP of IR64a-HA proteins using anti-HA affinity resin. For control, the antennae from ∼200,000 wild-type flies without the IR64a-HA transgene was subjected to the same co-IP procedure. Silver staining of the co-IP product reproducibly showed a band of ∼120 kDa in the experimental group, but not in control group (Fig. 2B, arrowhead). Using MS, we found that the main component of the 120 kDa band is IR8a, another member of the Drosophila IR family. These large-scale co-IP experiments were performed three times, and IR8a was identified by MS in an amount roughly similar to that of IR64a-HA (ratio IR64a-HA/IR8a, 1.3 ± 0.7, average ± SD) in immunoprecipitates of IR64a-HA antennae lysates, but not from wild-type antennae lysates in all the trials. We performed additional small-scale co-IP experiments using ∼10,000 pairs of independently prepared antennae and performed Western blot analysis using anti-IR8a antibody (Abuin et al., 2011). We consistently detected endogenous IR8a protein in IR64a-HA, but not control co-IP lysates (Fig. 2C). Together, these results strongly suggest that IR8a and IR64a are present in the same complex and likely interact with each other directly in vivo.
Figure 2.
IR8a binds to IR64a in vivo. A, Schematic drawing of large-scale isolation of antennal tissue for co-IP experiments. B, Silver stain of co-IP proteins from IR64a-HA transgenic flies (HA) and wild-type flies (control). The red arrowhead points to a silver stain positive band that is present in the HA sample but absent in control sample. C, Western blot analysis of independently prepared antennal tissue showing that IR8a is coimmunoprecipitated with IR64a-HA.
IR8a is expressed in IR64a+ neurons
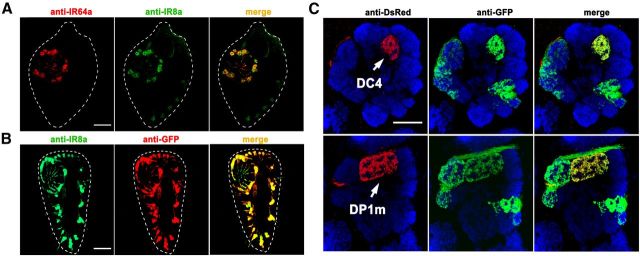
IR8a was shown previously to be broadly expressed along with other IR receptors in many coeloconic neurons (Abuin et al., 2011). To determine whether IR8a is expressed in IR64a+ neurons, we performed immunohistochemistry on antennae sections using anti-IR64a and anti-IR8a antibodies. As shown in Figure 3A, all of the IR64a+ neurons expressed IR8a. To determine whether these IR8a-expressing neurons innervate the DC4 or DP1m glomerulus, we generated an IR8a-GAL4 transgene to drive the expression of a reporter transgene, UAS-mCD8GFP. The resulting IR8a-GAL4, which faithfully recapitulated endogenous IR8a expression (Fig. 3B), had a pattern of expression similar to that induced by a previously reported IR8a-GAL4 driver (Silbering et al., 2011). Immunostaining of the antennal lobes from flies carrying IR8a-GAL4, UAS-mCD8GFP, and a mCherry transgene fused directly downstream from the IR64a promoter (PromoterIR64a-mCherry) revealed that IR8a-GAL4 is expressed in ∼10 glomeruli, including both DC4 and DP1m, and that its expression overlaps with the mCherry expression in both of these glomeruli (Fig. 3C). These findings indicate that IR8a and IR64a are coexpressed in sensory neurons that innervate the DC4 and DP1m glomeruli.
Figure 3.
IR8a is expressed in IR64a+ neurons. A, Fluorescence micrographs of a cryosectioned wild-type antenna immunostained by anti-IR64a (red) and anti-IR8a (green) polyclonal antibodies. The dotted line outlines the antenna. B, A section of an antenna from IR8a-GAL4; UAS-GFP flies immunostained by anti-IR8a (green) and anti-GFP (red) showing that IR8a-GAL4 faithfully recapitulated endogenous IR8a expression. C, An antennal lobe from a fly carrying IR8a-GAL4; UAS-CD8GFP and PromoterIR64a-mCherry immunostained by anti-dsRed/mCherry (red; corresponding to IR64a promoter expression), anti-GFP (green; corresponding to IR8a promoter expression), and nc82 (blue). Note that red and green fluorescence represent the glomeruli labeled by IR64a and IR8a promoters, respectively. Top row, Focal plane showing the DC4 glomerulus. Bottom row: Focal plane showing the DP1m glomerulus of the same antennal lobe. Scale bars: 20 μm.
IR8a is required for physiological and behavioral responses to odorants
To determine whether IR8a is required for the response of DC4 and DP1m glomeruli to odorants, we compared the glomerular responses to odorants in wild-type and IR8a1 (Abuin et al., 2011) and IR64ami (Ai et al., 2010) mutant flies using in vivo calcium imaging. As shown in Figure 4, A and B, the responses of both DC4 and DP1m glomeruli were significantly reduced in IR64a and IR8a mutants. The effect of the IR8a mutation on the response of DC4 and DP1m neurons appeared to be specific since RNAi knockdown of IR8a expression in IR64a+ neurons using an IR64a-GAL4 driver also resulted in a significant reduction in odor-evoked responses in the DC4 and DP1m glomeruli (Fig. 4A,B). As a control, knockdown of IR25a, another broadly expressed IR family member, using the IR64a-GAL4 driver had no effect on DC4 or DP1m glomerular response to odorants (Fig. 4A,B).
We showed previously that IR64a is required for acid avoidance behavior. To determine whether IR8a is functionally involved in behavioral responses to acid, we performed an acid avoidance assay using a T maze. We found that, similar to IR64a mutant flies, IR8a mutant flies demonstrated a significantly reduced avoidance to acetic acid (Fig. 4C). Furthermore, RNAi knockdown of IR8a, but not IR25a, in IR64a+ neurons resulted in a significant reduction in acid avoidance (Fig. 4D). Together, these results demonstrated that IR8a functions in IR64a+ neurons to mediate acid-evoked physiological and behavioral responses.
IR8a regulates IR64a protein stability
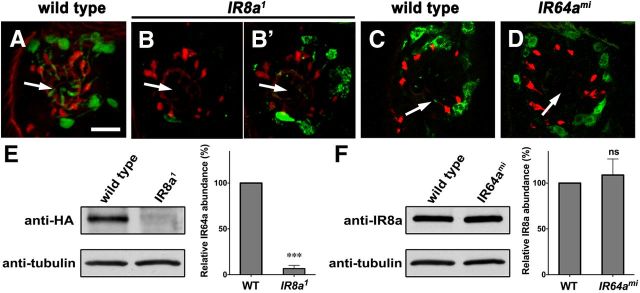
Having found evidence that IR8a and IR64a coexist in a receptor complex and that mutations in each receptor cause similar defects, we attempted to determine whether the loss of either IR64a or IR8a affects the expression and trafficking of the other protein. To determine whether the loss of IR8a affects the production of IR64a, we immunostained antennae sections using anti-IR64a antibody. In so doing, we found that the IR64a protein levels decreased dramatically to below detection levels in most IR8a-null mutant antennae (Fig. 5, compare A, B). Occasionally, we detected a weak immunopositive anti-IR64a signal in IR8a mutant antennae. By increasing the laser power and detector gain, we were able to visualize the residual IR64a protein in some antennal tissues and found that it had failed to localize to the dendritic terminals (Fig. 5B′). These results suggest that IR8a is required for IR64a protein trafficking and stability. To determine whether the loss of IR64a affects IR8a, we stained antennae sections with anti-IR8a antibody. We found that IR64a mutation did not affect IR8a protein levels; however, IR8a failed to correctly localize to the dendritic terminals in IR64a mutant antennae (Fig. 5, compare C, D). This observation is consistent with previous findings regarding the trafficking of coexpressed IR receptors; i.e., the trafficking of one receptor depends on the trafficking of the other (Abuin et al., 2011).
Figure 5.
IR8a affects IR64a protein abundance. A–D, Cryosectioned antennae from wild-type, IR8a mutant, or IR64a mutant flies immunostained by anti-IR64a (A–B′, green) or anti-IR8a (C, D, green) and monoclonal antibody 21A6 (red). B′, An image of the same sacculus region as in B taken with increased laser power and more sensitive detector gain to overexpose the green fluorescence. Arrows point to the sensilla within the sacculus. Scale bar, 10 μm. E, F, Left, Western blot of dissected antennae (35 pairs of antennae per lane). Right, Relative protein abundance from Western blot was quantified by the gel analysis function of the ImageJ software. N = 3. ***p = 0.0015. ns, Not significantly different by Student's t test.
To quantify the relative amounts of protein in mutant versus wild-type antennae, we measured the IR64a and IR8a band intensity by performing Western blot analysis. Because anti-IR64a antibody failed to cleanly detect IR64a in Western blot, we monitored IR64a-HA protein levels in flies carrying the genomic rescue transgene IR64a-HA and found that IR64a-HA protein levels in IR8a-null antennae were reduced to 6.6 ± 3.6% (mean ± SEM) of the protein levels observed in wild-type controls (Fig. 5E). In contrast, IR8a protein levels did not change significantly in IR64a-null mutants compared to wild-type controls (Fig. 5F).
Reconstitution of a functional IR64a and IR8a receptor complex
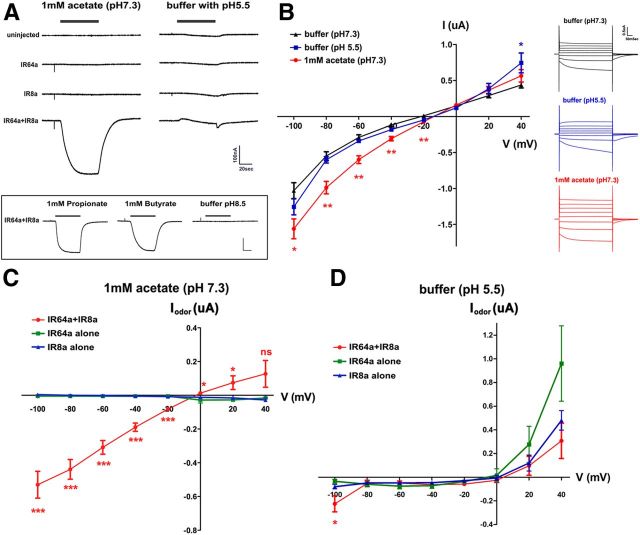
Few Drosophila IRs were shown to form ligand-gated cation channels (Abuin et al., 2011). To determine whether IR64a and IR8a form a functional ion channel, we expressed these two proteins in Xenopus oocytes and characterized their electrophysiological properties. Oocytes expressing IR8a and IR64a had a depolarized resting membrane potential of −23.8 ± 1.3 mV (mean ± SEM) compared to noninjected control oocytes (−55.9 ± 2.3 mV) or IR64a-injected oocytes (−53.3 ± 0.9 mV). Expression of IR8a alone also led to a slightly depolarized resting potential (−45.9 ± 1.2 mV).
To characterize the current through this channel, we voltage clamped the oocytes at −50 mV and measured currents in IR64a- and IR8a-expressing oocytes in response to acetate and low pH, which activate DP1m and DC4 in vivo, respectively (Fig. 1G). Oocytes coexpressing IR64a and IR8a (IR64a+IR8a) exhibited large inward currents in response to acetate (Fig. 6A), but oocytes expressing either protein alone did not respond. IR64a+IR8a-expressing oocytes also showed similar responses to 1 mm propionate and butyrate (Fig. 6A, bottom), two of the ligands that activate the DP1m glomerulus in vivo. Exposure to an acidic buffer (pH 5.5) did not induce an inward current in oocytes expressing both IR64 and IR8a (Fig. 6A). Nor did they respond to basic buffer (pH 8.5) (Fig. 6A, bottom).
Figure 6.
IR64a and IR8a form functional ion channels in Xenopus oocytes. A, Top left, Acetate (1 mM) induced an inward current in oocyte expressing both IR64a and IR8a. Top right, Acidic buffer induced a small and transient outward current and a small off response. Bottom, Responses to propionate, butyrate, and pH 8.5 buffer. Calibration: 100 nA, 20 s. B, I/V relationship in IR64a+IR8a-expressing oocytes in response to different stimuli. Insets, right, Examples of currents from an IR64a+IR8a-expressing oocyte clamped from −100 to 40 mV with 20 mV steps. *p < 0.05; **p < 0.01 [unpaired Student's t test compared to buffer (pH 7.3) control]. C, D, Relationship of odor-evoked currents (Iodor) and holding potentials. In each oocyte, evoked currents measured at each holding potential were normalized to the currents at the same holding potential in control buffer (pH 7.3) (see Materials and Methods). *p < 0.05; ***p < 0.01 (ANOVA with Tukey's test).
We further measured current/voltage (I/V) relationship. We found that under basal conditions (pH 7.3), IR64a+IR8a-expressing oocytes showed measurable currents at different holding potentials (Fig. 6B, black trace). Lowering the pH to pH 5.5 did not cause significantly larger currents except when holding at +40 mV (Fig. 6B, blue trace). In contrast, the currents evoked by 1 mm acetate, pH 7.3, were significantly larger than those evoked by acidic (or control neutral) buffers when the membrane was held at negative potentials (Fig. 6B, red trace). We further calculated the odor-evoked current (Iodor) by subtracting the baseline current measured in pH 7.3 buffer from the current measured in the presence of odor, and we compared Iodor in oocytes expressing IR64a, IR8a, and IR64a+IR8a. As shown in Figure 6C, acetate induced significant Iodor at all negative holding potentials in IR64a+IR8a-expressing oocytes, but not in oocytes expressing either receptor alone. Exposure to acidic solution, pH 5.5, however, did not lead to significant Iodor (Fig. 6D).
Discussion
In this study, we identified IR8a as a physically associated receptor subunit of IR64a in vivo. IR8a functions to regulate both IR64a protein abundance and trafficking. Like IR64a, IR8a is required for acid-evoked physiological and behavioral responses in flies. Furthermore, IR8a and IR64a proteins form a functional ligand-gated ion channel in a heterologous system (i.e., Xenopus oocytes). Our findings lay a foundation for structure and function analyses of these IR receptor channels and could serve as a precursor for future crystallography efforts to characterize these receptors. Similar crystallography studies have been highly informative in revealing the properties of other members of the ionotropic glutamate receptor family (Sobolevsky et al., 2009). Moreover, it is intriguing that both IR64a and IR8a are highly conserved in several mosquito species (Croset et al., 2010) (unpublished data). As the fly IR64a is involved in detecting CO2 (Ai et al., 2010), which is emitted by human hosts and often serves as important cues for mosquito host-seeking behavior, our findings will shed light on the function of the mosquito IR64a+IR8a receptor complex and facilitate future studies in understanding mosquito chemotaxis behavior and preventing mosquito-borne infectious diseases.
We found that IR8a is not only required for the trafficking of IR64a, but also regulates the abundance of IR64a protein (Fig. 5). The latter function was not observed in a previous study (Abuin et al., 2011). A major difference between our study and that of Abuin et al. (2011) is that they overexpressed GFP-fused IR64a or IR84a under the control of IR8a-GAL4, whereas we monitored endogenous IR64a protein levels either by immunohistochemistry using anti-IR64a antibody or Western blot analysis using the genomic rescue IR64a-HA transgene, which is under the control of its endogenous promoter. Thus, our analyses should more closely reflect the endogenous conditions. However, neither approach has revealed the mechanism responsible for the reduction in the IR64a protein levels in IR8a mutants. It is possible that IR8a functions as a chaperone to facilitate IR64a protein folding. In the absence of IR8a, IR64a might become misfolded and undergo misfolding-mediated proteasome degradation.
OSNs innervating the DC4 and DP1m glomeruli express both IR64a and IR8a, but they have distinct odor response profiles. The molecular mechanism underlying this difference is not yet understood. It is possible that additional yet-to-be identified factors are present specifically in DC4 or DP1m neurons to modulate the ligand-binding specificity of the IR64a+IR8a receptor complex in vivo. Such factors may not be restricted to cell-autonomous proteins such as coreceptors and intracellular proteins. Nonautonomous factors secreted by adjacent support cells (Shanbhag et al., 1995, 2000) could modulate the IR64a+IR8a receptor complex. This possibility is supported by our finding that DC4- and DP1m-OSN dendrites encounter different local microenvironments as they innervate the morphologically distinct sensilla GS1 and GS2 in the ventral and dorsal compartments of Chamber III of the sacculus (Fig. 1A,B). An alternative explanation for the distinct odor profiles of DC4 and DP1m may lie in their expression of different IR64a or IR8a splice isoforms, which could lead to distinct receptor subunits in these neurons. To date, no splice isoforms have been identified for either IR64a or IR8a, however. The expression of antenna-specific isoforms is likely limited, and thus they could have been missed by the expressed sequence tag or RNAseq (Roy et al., 2010). The identification of additional factors or isoforms required for IR64a and IR8a to function may require the development of more sensitive genomic or proteomic methods.
Many canonical glutamate receptors such as AMPAR and NMDA receptors bind to PDZ domain-containing intracellular scaffolding proteins through their C termini (Leonard et al., 1998; Sheng and Sala, 2001; Kim and Sheng, 2004; Funke et al., 2005). We did not identify such scaffolding proteins in our study. This may not be surprising, given that IR64a is predicted to have a very short intracellular C terminus (19 amino acid residues) that lacks any recognizable protein binding motifs. Additionally, there is no evidence of direct binding between a major Drosophila PDZ domain protein, Disc-large, and glutamate receptors (Qin et al., 2005). On the other hand, IR8a is predicted to have a longer intracellular tail (109 aa), which contains no identifiable conserved domain motifs. Future experiments using co-IP procedures to precipitate the IR8a intracellular tail might provide some insights into the nature of the intracellular pathways downstream of these olfactory IRs.
IR64a and IR8a formed functional ion channels in the Xenopus oocytes. We observed leaky inward currents in oocytes that express both IR64a and IR8a. This may reflect a unique property of the IR64a-containing receptor complex since no leak currents were reported from IR84a+IR8a or IR75a+IR8a receptor complexes (Abuin et al., 2011). Interestingly, insect odorant receptors expressed in heterologous systems also appear to be partially active in the absence of their ligands: OR22a+ORCO produced basal currents in HEK293 cells that could be diminished by the application of OR22a antagonist (Wicher et al., 2008); OR47a+ORCO expression in HeLa cells caused an elevated resting intracellular Ca2+ that could be reduced by extracellular application of EGTA (Sato et al., 2008). Together with the basal activities of these insect ORs, the leaky currents we observed in IR64a+IR8a-expressing Xenopus oocytes may reflect the underlying mechanism of odorant receptor-dependent spontaneous electric activities in the sensilla (Hallem et al., 2004; Larsson et al., 2004; Abuin et al., 2011).
IR64a+IR8a-expressing oocytes exhibited specific inward currents in response to acetate, propionate, and butyrate, but not to acidic pH. This response is similar to the odor response of the DP1m glomerulus in vivo, but differs from that of DC4. However, the expression of R64a+IR8a in Xenopus oocytes did not completely recapitulate the full spectrum of the odor response profile of the endogenous DP1m glomerulus. For example, β-citronellol, benzaldehyde, and 3-octanol, all of which strongly activated the DP1m glomerulus in vivo, failed to elicit inward currents in IR64a+IR8a-expressing oocytes (data not shown). These findings suggest that additional factors may exist in DP1m neurons that modulate the function of the IR64a+IR8a receptor complex. Consistent with this hypothesis, ectopic expression of IR64a in some (but not all) IR8a+ neurons conveyed odor responsiveness (Ai et al., 2010).
We also found that DC4 and DP1m pathways are anatomically segregated from each other: (1) In the antennae, DC4 and DP1m OSNs innervate their dendrites to ventral and dorsal compartments of Chamber III in the sacculus, respectively (Fig. 1B). (2) In the LH, the axonal projections of DC4- and DP1m-PNs occupy nonoverlapping space (Fig. 1C–F). These results suggest that sensory information encoded by DC4 and DP1m OSNs are represented in distinct areas within the LH, a brain structure that is proposed to encode sensory valence and direct innate behaviors. In fact, an example of the topographic segregation of distinct sensory information within the LH was reported previously (Jefferis et al., 2007). The DC4 pathway detects acids and mediates innate avoidance behavior, whereas the DP1m pathway responds to a wide variety of odorants, but its biological function is not clear. Inferred from these findings, the spatial segregation of DC4 and DP1m pathways within the LH would encode accurate distinction of these two different sensory inputs in the brain.
Footnotes
This work was supported by the National Research Service Award Fellowship to M.A., NIH NINDS Grant P30 NS050276 to T.A.N., and the Whitehall Foundation, the Irma T. Hirschl/Weill Caulier Trust Award, and NIH RO1 grants (National Institute of General Medical Sciences and NIDCD) to G.S.B.S. We thank Ruth Lehmann for sharing her two-photon microscope, Niels Ringstad for assistance with Xenopus oocyte electrophysiological recordings, Guoan Zhang for discussion and technical assistance with MS analysis, and Richard Benton for providing IR8a antibody and fly stocks.
References
- Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, Suh GS. Acid sensing by the Drosophila olfactory system. Nature. 2010;468:691–695. doi: 10.1038/nature09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Vasconcelos ML, Ruta V, Luo S, Wong A, Demir E, Flores J, Balonze K, Dickson BJ, Axel R. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452:473–477. doi: 10.1038/nature06808. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- Grosjean Y, Rytz R, Farine JP, Abuin L, Cortot J, Jefferis GS, Benton R. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011;478:236–240. doi: 10.1038/nature10428. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr, Luo L. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Leonard AS, Davare MA, Horne MC, Garner CC, Hell JW. SAP97 is associated with the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem. 1998;273:19518–19524. doi: 10.1074/jbc.273.31.19518. [DOI] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-A. [DOI] [PubMed] [Google Scholar]
- Marin EC, Jefferis GS, Komiyama T, Zhu H, Luo L. Representation of the glomerular olfactory map in the Drosophila brain. Cell. 2002;109:243–255. doi: 10.1016/S0092-8674(02)00700-6. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat Biotechnol. 2010;28:348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Schwarz T, Kittel RJ, Schmid A, Rasse TM, Kappei D, Ponimaskin E, Heckmann M, Sigrist SJ. Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila. J Neurosci. 2005;25:3209–3218. doi: 10.1523/JNEUROSCI.4194-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnett GV, Moon C. G proteins and olfactory signal transduction. Annu Rev Physiol. 2002;64:189–222. doi: 10.1146/annurev.physiol.64.082701.102219. [DOI] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, Axel R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Shanbhag SR, Singh K, Singh RN. Fine structure and primary sensory projections of sensilla located in the sacculus of the antenna of Drosophila melanogaster. Cell Tissue Res. 1995;282:237–249. doi: 10.1007/BF00319115. [DOI] [PubMed] [Google Scholar]
- Shanbhag SR, Müller B, Steinbrecht RA. Atlas of olfactory organs of Drosophila melanogaster 2. Internal organization and cellular architecture of olfactory sensilla. Arthropod Struct Dev. 2000;29:211–229. doi: 10.1016/S1467-8039(00)00028-1. [DOI] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GS, Benton R. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 2011;31:13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh GS, Wong AM, Hergarden AC, Wang JW, Simon AF, Benzer S, Axel R, Anderson DJ. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431:854–859. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol. 2009;71:307–332. doi: 10.1146/annurev.physiol.010908.163209. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Yao CA, Carlson JR. Role of G-proteins in odor-sensing and CO2-sensing neurons in Drosophila. J Neurosci. 2010;30:4562–4572. doi: 10.1523/JNEUROSCI.6357-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]