Abstract
Alzheimer’s disease (AD) is the most common type of dementia in the elderly. It is characterized by the deposition of two forms of aggregates within the brain, the amyloid β plaques and tau neurofibrillary tangles. Currently, no disease-modifying agent is approved for the treatment of AD. Approved pharmacotherapies target the peripheral symptoms but they do not prevent or slow down the progression of the disease. Although several disease-modifying immunotherapeutic agents are in clinical development, many have failed due to lack of efficacy or serious adverse events. Epigenetic changes including DNA methylation and histone modifications are involved in learning and memory and have been recently highlighted for holding promise as potential targets for AD therapeutics. Dynamic and latent epigenetic alterations are incorporated in AD pathological pathways and present valuable reversible targets for AD and other neurological disorders. The approval of epigenetic drugs for cancer treatment has opened the door for the development of epigenetic drugs for other disorders including neurodegenerative diseases. In particular, methyl donors and histone deacetylase inhibitors are being investigated for possible therapeutic effects to rescue memory and cognitive decline found in such disorders. This review explores the area of epigenetics for potential AD interventions and presents the most recent findings in this field.
Keywords: Alzheimer’s disease, DNA methylation, Epigenetics, Histone modification, Memory, Therapy
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder, with over 35 million cases worldwide (Selkoe, 2012). The four acetylcholinesterase inhibitors, donepezil rivastigmine, galantamine and tacrine, along with the NMDA receptor antagonist memantine are the only FDA-approved drugs for AD; however, they merely target the symptoms and do not prevent the progressive loss of memory, cognitive and executive functions in AD patients. With the increase in life expectancy and the absence of disease-modifying agents, the number of people with AD is expected to triple within the upcoming 40 years (Barnes & Yaffe, 2011; Huang & Mucke, 2012; Tricco et al., 2012). The annual costs for AD and other dementias in the US in 2013 are estimated to be over $200 billion and are expected to reach $1.2 trillion in 2050 (Alzheimer’s Association, 2013). With such a heavy socioeconomic burden, there is an urgent need to find novel and improved treatments for AD. Throughout the last century, the advances acquired in health related fields were able to increase the lifespan of AD patients, yet this needs to be matched with discoveries that improve the quality of life of people with this debilitating disorder. New targets should be identified and investigated for possible AD therapeutics that tackle its core pathophysiology as well as prevent the decline in memory and cognitive functions associated with the disease.
AD is characterized by progressive loss of memory and other cognitive and executive functions, with two types of pathological deposits found in the brain, the extracellular amyloid β (Aβ) plaques and the intracellular tau neurofibrillary tangles (NFTs). Senile plaques are mainly composed of Aβ which is cleaved off the larger amyloid β precursor protein (APP) by β-site APP cleaving enzyme (BACE), also known as β-secretase, and γ-secretase (Citron et al., 1995; Shoji et al., 1992). According to the amyloid hypothesis, Aβ and its aggregates are responsible for the neurodegeneration and dementia in AD through mechanisms that involve disturbances in calcium homeostasis which make cells more vulnerable to toxicants that can cause further damage and NFTs (Hardy & Higgins, 1992; Mattson et al., 1992; Selkoe, 1993). The hypothesis was supported by the fact that mutations on APP are connected to hereditary types of AD (Hardy & Higgins, 1992). Early onset familial AD (FAD) could also be due to mutations on genes encoding the presenilin (PS) membrane proteins PS1 and PS2 (Czech et al., 2000; Tanzi et al., 1996). PS mutations increase the production of the more aggregative 42 amino acid-long Aβ (Aβ42) from APP and elevated Aβ42 levels were observed in the blood and brains of FAD patients with PS abnormalities (Czech et al., 2000). In addition, neurons lacking the PS1 gene fail to produce Aβ peptides (De Strooper et al., 1998; Naruse et al., 1998). PS1 was found to be related to the enzyme γ-secretase (De Strooper et al., 1998; Shimojo et al., 2007). These findings suggest that Aβ and its aggregates are involved in the pathology of AD as proposed in the amyloid hypothesis.
NFTs are composed of tau protein which belongs to a family of microtubule-associated proteins that normally promote microtubule assembly (Weingarten et al., 1975). When hyperphosphorylated, tau loses its normal function and becomes prone to form pathological aggregates causing disorders known as tauopathies of which AD is the most common (Alonso et al., 1997; Lee et al., 2001). Both Aβ and tau have been associated with neurodegeneration and memory decline and have been extensively targeted for AD interventions such as immunotherapeutics, enzyme modulators, and aggregation inhibitors (Hardy & Higgins, 1992; Hardy & Selkoe, 2002; Hutton et al., 1998; Iqbal et al., 2009; Lee et al., 2001; Selkoe, 1993).
Epigenetics deals with acquired and heritable modifications on DNA that regulate the expression and functions of genes without affecting the DNA nucleotide sequence. These include DNA methylation and hydroxymethylation, histone modifications and non-coding RNA regulation. Histone modifications consist of acetylation, methylation, crotonylation, ubiquitination, sumoylation, phosphorylation, hydroxylation and proline isomerization (Davie & Spencer, 1999; Houston et al., 2013; Kouzarides, 2007; Peterson & Laniel, 2004). All these pathways act as mediators between the environment and the genome, these epigenetic changes are activated by various conditions such as stress or exposure to environmental toxicants and in turn they result in a variety of responses including gene transcription or silencing. Epigenetic changes are dynamic and unlike genetic mutations, they can be reversed for therapeutic purposes by targeting enzymes or other factors that control or maintain them (Caraci et al., 2012; Feinberg, 2008; Henikoff & Matzke, 1997; Liu et al., 2008; Mill, 2011). As changes within the genetic makeup itself are limited and the environment cannot freely amend the DNA sequence, epigenetics is the mechanism through which the environment can affect gene expression and function which can be employed as a medical intervention for diseases where epigenetics play a pathological role (Jaenisch & Bird, 2003; Mill, 2011). Furthermore, some age related changes are also mediated through epigenetics (Feinberg, 2008).
The majority of AD cases are sporadic or late onset AD (LOAD). Only about 5% of cases are familial or early onset AD which is associated with rare mutations on the APP, PS1 and PS2 genes (Goate et al., 1991; Sherrington et al., 1995; Tanzi, 2012). The sporadic nature of AD suggests that epigenetics plays an important role in the pathology of the disease; a hypothesis that is supported by recent findings from our laboratory and others (Lahiri et al., 2009; Lahiri et al., 2008; Mastroeni et al., 2011; Mill, 2011; Wang et al., 2008a; Wu et al., 2008b; Zawia & Basha, 2005; Zawia et al., 2009). This review explores epigenetic mechanisms as possible targets for AD therapeutics and highlights the current status of epigenetics in AD pathology and drug discovery.
1.1. Epigenetics of the brain and memory formation
Epigenetic dynamics within cells play a major role in their differentiation and in determining their functional type as hepatocytes in the liver, neurons in the brain, skin cells, or other cells, as well as becoming cancerous or not (Chadwick, 2012; Feinberg, 2008). Epigenetics is involved in various brain related disorders and physiologic responses that genetics alone does not completely explain including AD, depression, schizophrenia, glioma, addiction, Rett syndrome, alcohol dependence, autism, epilepsy, multiple sclerosis and stress (Heim & Binder, 2012; Inkster et al., 2013; Jaenisch & Bird, 2003; Kreth et al., 2012; Maric & Svrakic, 2012; Maze & Nestler, 2011; Mifsud et al., 2011; Nguyen et al., 2010; Orr et al., 2012; Qureshi & Mehler, 2010; Shahbazian & Zoghbi, 2002; Taqi et al., 2011; Zawia et al., 2009). As neurons do not divide and cannot be replaced after degeneration, epigenetic changes resulting in neuronal dysfunction need to be targeted and modified to prevent neurodegeneration (Bird, 2007; Selvi et al., 2010).
Recent studies have pointed out the importance of epigenetics in brain development and functions including learning and memory (Feng et al., 2007; Miller & Sweatt, 2007; Molfese, 2011; Sultan & Day, 2011). In particular, DNA methylation and histone acetylation both play an important role in memory formation (Levenson et al., 2006; Miller et al., 2008). Other histone modifications involved in memory are methylation and phosphorylation (Chwang et al., 2006; Gupta et al., 2010; Molfese, 2011).
DNA methylation is catalyzed by DNA methyltransferases (DNMTs) in the presence of the methyl donor S-adenosyl methionine (SAM) (Yen et al., 1992). The DNMT family of enzymes includes DNMT1, DNMT2, DNMT3a, and DNMT3b (Okano et al., 1999). It is found that DNMT3a and DNMT3b are responsible for de novo methylation and establish DNA methylation patterns while DNMT1 has preference for hemi-methylated DNA (Chen et al., 2003; Hsieh, 1999; Okano et al., 1999). DNA methylation occurs on the 5′ position of cytosine in CpG rich regions (Bird, 1986). This epigenetic mechanism regulates gene transcription and plays a particular role in memory functions (Day & Sweatt, 2010; Korzus, 2010; Liu et al., 2009). Memory and learning abilities decline with age which correlates with an overall reduction in DNA methylation (Liu et al., 2009). Furthermore, methylation on certain locations of the APP promoter in the human cortex is reduced with age (Tohgi et al., 1999).
DNMTs are considered crucial for memory functions (Miller & Sweatt, 2007). DNMTs regulate methylation within the promoter of reelin, an extracellular glycoprotein that is involved in memory formation in the adult brain (Levenson et al., 2006; Weeber et al., 2002). Protein levels of DNMT1 and DNMT3a are reduced in the cortex of aged monkeys compared to early time points (Bihaqi et al., 2011). Moreover, conditional knockout mice lacking the expression of Dnmt1 and Dnmt3a genes in forebrain neurons perform worse on the Morris water maze hippocampus related memory task than wild-type littermates or knockout mice lacking the expression of only one of the genes (Feng et al., 2010). The gene expression of Dnmt3a2 decreases with age in mouse cortex and hippocampus (Oliveira et al., 2012). This age-related decline in Dnmt3a2 gene expression is linked to memory decline that can be recovered by restoring DNMT3a2 levels (Oliveira et al., 2012).
Hydroxylation of 5-methylcytosine to 5-hydroxymethylcytosine by ten-eleven translocation (TET) enzymes is an important regulatory pathway involved in brain development, aging and disease (Szulwach et al., 2011). Levels of 5-hydroxymethylcytosine are significantly higher in neurons than in cells of other tissues (Globisch et al., 2010; Szulwach et al., 2011). DNA hydroxymethylation, levels of 5-hydroxymethylcytosine and 5-methylcytidine increase with age, and alterations in DNMT3a have also been reported with aging in mouse hippocampus (Chouliaras et al., 2011; Chouliaras et al., 2012a; Chouliaras et al., 2012b). Such epigenetic changes could be prevented by 50% caloric restriction diet throughout the mice lifetime after weaning (Chouliaras et al., 2011; Chouliaras et al., 2012a; Chouliaras et al., 2012b).
Moreover, mutations on methyl CpG binding protein 2 (MeCP2) may contribute to the development of Rett syndrome; a life-long neurodevelopmental disorder with marked learning disabilities (Amir et al., 1999). Other neurological abnormalities such as autism and infantile encephalopathy have been associated with disturbances in MeCP2 (Chahrour et al., 2008; Moretti & Zoghbi, 2006). MeCP2 binds to methylated cytosine in CpG dinucleotides and inhibits or promotes gene expression by recruiting transcription repressors or activators like cAMP response element-binding protein 1 (CREB1) (Chahrour et al., 2008; Jones et al., 1998; Nan et al., 1998). This also involves MeCP2 binding to histone deacetylase complex (Jones et al., 1998; Nan et al., 1998). MeCP2 levels are found to decrease with age in primates (Bihaqi et al., 2011).
Histone acetylation is also involved in the regulation of learning and memory (Levenson et al., 2004; Martin & Sun, 2004). The most widely studied histone modification is regulated by two groups of enzymes, histone acetyltransferases (HATs) and histone deacetylases (HDACs). The HDACs family of enzymes has been studied extensively for implications in cancer. Aberrant overexpression of various HDACs has been reported in different cancer types including gastric, pancreatic, breast, lung and colon cancer (Barneda-Zahonero & Parra, 2012; Johnstone, 2002; New et al., 2012). Many HDAC inhibitors are in clinical trials for cancer therapy, vorinostat and romidepsin have been approved by the FDA for cutaneous T-cell lymphoma (Kim et al., 2012; Mann et al., 2007; Nebbioso et al., 2012). There are 18 HDACs identified that belong to four classes I, II, III and IV according to their sequence homology (Xu et al., 2007). Class I HDAC2, class IIb HDAC6 and class III sirtuins (SIRTs) 1 and 2 are linked to AD pathology (de Oliveira et al., 2012; Karagiannis & Ververis, 2012). Acetylation of histone H3 in the hippocampus accompanies long-term memory formation in rats as determined in hippocampal tissues collected 1 hour after fear conditioning experiments (Levenson et al., 2004). Increasing histone acetylation by the administration of the HDAC inhibitor sodium butyrate to rats prior to contextual fear conditioning improves memory formation (Levenson et al., 2004). Class I HDAC inhibitors sodium butyrate, sodium valproate and vorinostat enhance cognition in APP/PS1 double transgenic AD mouse model as evaluated by contextual fear conditioning tests (Kilgore et al., 2010). Overexpression of HDAC2 and not HDAC1, both Class I HDACs, in mice impairs memory and chronic administration of the HDAC inhibitors vorinostat or sodium butyrate enhances cognition (Guan et al., 2009). HDAC2 knockout mice display improved memory in fear conditioning experiments over wild-type mice (Guan et al., 2009). Consequently, downregulation or inhibition of HDACs 2 and 6 constitute important therapeutic targets for memory related disorders.
1.2. Epigenetic changes in AD
1.2.1. Histone modifications in AD
Increased levels of HDAC2 have been associated with cognitive impairment in CK-p25 AD mouse model which seems to be mediated through glucocorticoid receptor induced HDAC2 transcription (Graff et al., 2012). Postmortem studies reported that HDAC2 and not HDAC1 or HDAC3 is increased within the hippocampus of AD patients (Graff et al., 2012). Class II HDAC6 levels are elevated in AD cortex and hippocampus by 52% and 91% respectively (Ding et al., 2008). Tau co-localizes with HDAC6 in AD hippocampus and in vitro, and downregulation of HDAC6 decreases tau phosphorylation at Thr231 (Ding et al., 2008). Hyperphosphorylation of tau inhibits its normal functions and promotes its aggregation (Alonso et al., 1997). In particular tau phosphorylation at Thr231 restrains its normal function of binding to microtubules (Sengupta et al., 1998). Class III HDACs or sirtuins is a family of enzymes that includes seven members named SIRT1-7 (Gray & Ekstrom, 2001). In addition to histones, SIRTs are responsible for deacetylation of other molecules like some proteins involved in AD pathology. For example SIRT1 accounts for tau deacetylation which is considered neuroprotective while tau acetylation contributes to tau dysfunction and aggregation (Cohen et al., 2011; Min et al., 2010; Stunkel & Campbell, 2011). SIRT1 is lower in the AD cortex which correlates with presence of tau pathology and memory impairment (Julien et al., 2009). There is evidence that SIRT1 also stimulates α-secretase which cleaves APP within the Aβ sequence and protects against Aβ accumulation (Donmez et al., 2010; Raghavan & Shah, 2012; Wang et al., 2010). SIRT1 is the most studied sirtuin, other sirtuins are also expressed in the brain and SIRT2 has been presented as a drug target for neurodegenerative diseases such as Parkinson’s and Huntington’s diseases (de Oliveira et al., 2012).
1.2.2. DNA methylation in AD
DNA methylation and factors such as DNMT1 are significantly reduced in neurons of entorhinal cortex layer II in AD patients (Mastroeni et al., 2010). Reductions in methylation are particularly localized in tangles containing neurons (Mastroeni et al., 2010). Other studies have demonstrated that there is abnormal methylation in AD patients (Bakulski et al., 2012; Wang et al., 2008a). When studying DNA methylation within the cerebral cortex of AD and control subjects, two out of the fifty loci examined were differentially methylated in AD which represent an acceleration of aging-linked alterations (Siegmund et al., 2007). In an AD discordant pair of monozygotic twins, extensive plaques and NFTs were present and less methylation was found in the cortex of the AD twin compared to the non-AD twin (Mastroeni et al., 2009). However, some studies found no differences in the methylation patterns of AD related genes (Barrachina & Ferrer, 2009). The difficulties in obtaining postmortem AD brain tissues for such studies and the variability among the available tissues as well as the different end points of methylation analyzed within these studies account for their various findings.
The promoter region within the APP gene is GC rich suggesting that it can be modulated through methylation (Pollwein et al., 1992). APP promoter displays differential methylation within the human brain (Rogaev et al., 1994). Hypomethylation of the APP promoter was reported to correlate with APP overexpression in AD (West et al., 1995). DNA methylation controls BACE and PS1 expression and consequently Aβ levels (Fuso et al., 2005). PS1 expression and methylation is altered in LOAD (Wang et al., 2008a). However, the changes on PS1 gene methylation in AD brains were not significant (Wang et al., 2008a). Another study did not detect significant changes in PS1, APP and tau genes methylation in the cortex and hippocampus of AD patients compared to controls (Barrachina & Ferrer, 2009). Lower paternal age was significantly associated with the increase in LOAD risk which might involve DNA methylation (Farrer et al., 1991). The challenges in acquiring and handling human brain tissues make it difficult to have a large number of matched controls and AD samples, however the available studies along with the sporadic and non-mendelian inheritance nature of the disease suggest that epigenetics is indeed involved in AD. Further research is needed to examine the epigenetic changes affecting AD biomarkers including APP, tau, BACE, PS1 and PS2 among others, as well as global gene methylation patterns which would help with the early diagnosis of the disease.
1.2.3. Non-coding RNA in AD
Non-coding RNA can influence gene expression via epigenetic mechanisms affecting DNA methylation, histone modifications and chromatin remodeling (Costa, 2008). Various microRNAs are differentially expressed in AD and alter the expression of AD pathological intermediates (Cogswell et al., 2008; Nunez-Iglesias et al., 2010; Provost, 2010). An example is microRNA-101 which negatively regulates APP levels and is reduced within the brain cortex of AD patients (Hebert et al., 2008; Vilardo et al., 2010). Another example is microRNA-107 which is lowered early in AD and regulates BACE1 expression (Wang et al., 2008b). BACE1-AS is a long non-coding RNA antisense transcript of BACE1 that improves BACE1 stability and expression and is upregulated in the hippocampus and cortex of AD patients (Faghihi et al., 2008). Additional changes on non-coding RNAs are reported in AD and have been reviewed recently (Schonrock & Gotz, 2012). However, due to the current limitations and the absence of methods that can target or modify non-coding RNAs for therapeutic purposes, only few are mentioned within this review.
2. Epigenetic therapeutic approaches for AD
2.1. HDAC inhibitors
HDAC inhibitors show promise for cognitive improvement and are being considered for drug development for AD (Abel & Zukin, 2008; Fischer et al., 2007; Guan et al., 2009). Epigenetic changes play a role in cognitive decline and reversing such changes by inhibiting HDAC2 improves memory and cognitive functions (Graff et al., 2012). Treatment of hippocampal neurons with Aβ promotes HDAC2 transcription suggesting that the traditional target of Aβ lowering in AD should be complemented with the reversal of epigenetic changes that were caused by increased Aβ levels (Graff et al., 2012). This might explain why Aβ lowering is not always successful in improving memory and cognitive deficits when subsequent epigenetic changes are not reversed as well (Graff et al., 2012). Crebinostat, an HDAC inhibitor, improves memory in mice (Fass et al., 2013). Administration of any of the three Class I HDAC inhibitors sodium valproate, sodium butyrate and vorinostat, which is an HDAC inhibitor approved by the FDA for cancer, improve memory in the APPswe/PS1dE9 AD mouse model (Kilgore et al., 2010). Hence, HDAC inhibitors could be promising therapeutic agents for AD and other disorders associated with dementia and cognitive impairments.
Valproic acid, which is used as an anticonvulsant in epileptic patients and as a mood stabilizer in bipolar disorder patients (Phiel et al., 2001), is a known HDAC inhibitor and has therefore been proposed for use in cancer and AD (Gottlicher et al., 2001; Kramer et al., 2003; Nalivaeva et al., 2009). In addition, valproate seems to have multi-target effects that can be useful for AD including inhibition of the enzyme responsible for tau phosphorylation glycogen synthase kinase 3 beta (GSK3β) (Loy & Tariot, 2002). Valproic acid lowers Aβ in the PDAPP transgenic mouse model of AD (Su et al., 2004). However, in a 2-year clinical trial, valproate did not improve cognitive function or slow memory decline in moderate AD patients and was associated with adverse effects such as somnolence, tremor, weakness and dyspnea (Fleisher et al., 2011; Tariot et al., 2011).
Another HDAC inhibitor, sodium phenylbutyrate was found to improve memory and lower tau phosphorylation by GSK3β in APPswe transgenic AD mice (Ricobaraza et al., 2009). EVP-0334 is an HDAC inhibitor developed for AD by EnVivo Pharmaceuticals that successfully completed phase I clinical trials and was deemed safe for further testing, however, detailed information on the trial have not been made available yet (Arrowsmith et al., 2012; Caraci et al., 2012; Mack, 2010). A class II HDAC inhibitor referred to as W2 lowers Aβ, tau phosphorylated at Thr181 and improves cognition in hAPP transgenic mice (Sung et al., 2013). The authors also found that W2 and I2, a class I and II HDAC inhibitor, both downregulate genes involved in Aβ production and promote genes responsible for Aβ degradation in vitro (Sung et al., 2013).
2.2. Sirtuins
Class III HDACs or SIRTs are epigenetic targets for cancer and AD (Albani et al., 2010; Huber & Superti-Furga, 2011; Outeiro et al., 2008). The natural product found in red grapes skin and wine resveratrol is a SIRT1 activator that improves cognition in mice (Kim et al., 2007). However, resveratrol cognitive benefits involve other mechanisms besides SIRT1 activation and its epigenetic functions (Huber & Superti-Furga, 2011; Kim et al., 2007). A phase II study is currently recruiting mild to moderate AD patients to study the effects of resveratrol on AD biomarkers including cerebrospinal fluid (CSF) tau and Aβ levels as well as memory and daily performance using tests like Mini-Mental State Examination (MMSE) and Alzheimer’s Disease Assessment Scale-Cognitive (ADAS-Cog) (ClinicalTrials.gov., identifier: NCT01504854). Two SIRT activators developed by GSK were in phase I clinical trials, SRT2104 and SRT2379, recently the results from one of the trials were published and showed that SRT2104 was well tolerated by human subjects and suitable for further clinical trials (Hoffmann et al., 2013; Townsend, 2011). Interestingly, the nonselective SIRT inhibitor nicotinamide lowers phosphorylated tau and improves cognition in mice demonstrating that SIRT modulation involves complex mechanisms (Green et al., 2008; Stunkel & Campbell, 2011). Nicotinamide is in phase II clinical trial for AD (ClinicalTrials.gov., identifier: NCT00580931). Furthermore, administration of the SIRT2 inhibitor AK1 directly into the hippocampus protects against neurodegeneration in tau transgenic mice without altering tau tangles (Spires-Jones et al., 2012).
2.3. HATs
Less attention has been given to HAT enzymes as epigenetic targets for AD. Three HATs are involved in memory formation CREB binding protein (CBP), p300 and p300/CBP associated factor (PCAF) which might represent more specific targets than HDACs (Korzus et al., 2004; Selvi et al., 2010). CBP plays an important role in memory as CBP deficient mice display impaired long-term memory formation (Oike et al., 1999; Wood et al., 2005). In humans, mutations on the CBP gene result in Rubinstein-Taybi syndrome which is characterized by mental retardation (Petrij et al., 1995; Rubinstein & Taybi, 1963). Inducing the expression of CBP within the brains of 3xTg-AD triple transgenic AD mouse model recovers the impaired memory functions in these mice (Caccamo et al., 2010). On the other hand, inhibition of the HAT p300 by using the commercially available p300 inhibitor C646 reduces the levels of acetylated tau and phosphorylated tau at Ser202 in vitro (Min et al., 2010). The natural plant product curcumin possesses p300/CBP HAT inhibitor activity and is in phase II clinical trial to study its cognitive effects and Aβ lowering potential in AD patients (Balasubramanyam et al., 2004; ClinicalTrials.gov., identifier: NCT01383161; Marcu et al., 2006). Previous trials with a smaller number of AD subjects reported no significant changes between curcumin- and placebo- treated groups (Hamaguchi et al., 2010; Ringman et al., 2012). While in transgenic animal models, curcumin decreased oxidative damage and Aβ pathology by affecting anti-inflammatory pathways (Begum et al., 2008; Hamaguchi et al., 2010; Lim et al., 2001; Yang et al., 2005).
2.4. DNA methylation
There are multiple ways for targeting DNA methylation for therapeutic purposes (Klose & Bird, 2006). DNA methylation affects the expression of the AD related intermediates APP, PS1 and Aβ (Fuso et al., 2005). It has been hypothesized that hypomethylation of the promoter regions of such genes like PS1 leads to the overexpression of their products including Aβ (Mulder et al., 2005). Overexpression of DNMT3a2 within the hippocampus of old mice increases overall methylation and improves memory (Oliveira et al., 2012).
The levels of the methyl donor SAM are lower in the CSF and within the brains of AD patients (Bottiglieri et al., 1990; Bottiglieri et al., 1994; Morrison et al., 1996). However, in another study, there was no difference in SAM-CSF levels in AD patients vs. healthy subjects (Mulder et al., 2005). Nevertheless, treatment with SAM reduces BACE1, PS1 and Aβ production in vitro in human neuroblastoma cells (Fuso et al., 2005; Scarpa et al., 2003). Moreover, administration of SAM adjunct to antidepressants in depressed patients enhances their cognitive symptoms and ability to remember as determined by cognitive and physical symptoms questionnaire (CPFQ) (Levkovitz et al., 2012).
Betaine, the methyl donor used conventionally for homocystinuria treatment (Key, 2000), was tested in 8 AD patients for 24 weeks and failed to demonstrate cognitive improvement (Craig, 2004; Knopman & Patterson, 2001). However, the small number of patients and the lack of a placebo-treated control group suggest that further trials are needed to properly evaluate betaine’s efficacy in AD (Knopman & Patterson, 2001), especially that elevated homocysteine has been associated with dementia and AD and betaine lowers homocysteine (Seshadri et al., 2002). In a more recent study in mice, betaine was able to improve memory that was compromised by prior lipopolysaccharide administration (Miwa et al., 2011).
2.5. Non-coding RNA
Several non-coding RNAs are involved in AD pathology and could present specific diagnostic and therapeutic targets for the disease (Costa, 2008; Provost, 2010). These include BACE1-AS, microRNA-34c, microRNA-101, and microRNA-107 (Cogswell et al., 2008; Faghihi et al., 2008; Vilardo et al., 2010; Wang et al., 2008b; Zovoilis et al., 2011). However, concerns about ways to alter such targets, off target effects, and delivery methods still need to be adequately addressed before having epigenetic treatments capable of affecting non-coding RNAs. Targeting non-coding RNA regions on APP by the antibiotic erythromycin, the antidepressant paroxetine and N-acetyl cysteine has been found to reduce Aβ in TgCRND8 transgenic mice (Tucker et al., 2005; Tucker et al., 2006).
2.6. Beyond epigenetics: Epigenetics and transcription
Epigenetics is an important mediator that influences DNA transcription and translation. The aim of AD therapy is to enhance the transcription of genes involved in memory formation and reduce the transcription of pathogenic intermediates in the disease process like tau, APP, and BACE1. Hence, transcription factors constitute valid targets for developing novel treatments for AD. One of the important transcription factors for learning and memory is CREB (Silva et al., 1998). CREB is an essential mediator of memory improvement following HDAC inhibition as CREB has histone acetylation activity through recruitment of the histone acetyltransferase CBP (Vecsey et al., 2007). HDAC inhibitors, such as phenylbutyrate or crebinostat promote the transcription of genes involved in memory functions as seen with crebinostat which upregulates the CREB target gene early growth response 1 (egr1), which is involved in memory formation (Fass et al., 2013; Ricobaraza et al., 2009). A clinical trial studying the effects of the antiplatelet drug cilostazol on cognition in AD patients co-administered with donepezil is currently in progress (ClinicalTrials.gov., identifier: NCT01409564). The rationale behind choosing cilostazol is to promote the phosphorylation of CREB which regulates its activity and consequential expression of genes that are controlled by CREB (Bito et al., 1996; ClinicalTrials.gov., identifier: NCT01409564; Silva et al., 1998). Cilostazol protects against Aβ triggered cognitive impairment in mice and improves memory following cerebral hypoperfusion damage in rats (Hiramatsu et al., 2010; Watanabe et al., 2006).
An important transcription factor involved in AD is specificity protein 1 (Sp1). It binds to GC-rich regions within the promoters of APP, tau and BACE1 and upregulates their expression (Docagne et al., 2004; Hoffman & Chernak, 1995; Pollwein et al., 1992). Sp1 is able to bind to CpG sites in genes promoters that have such specific binding motifs and activate their transcription whether they are methylated or non-methylated (Holler et al., 1988). Furthermore, Sp1 can trigger epigenetic modifications as it regulates the expression of DNMT1 (Kishikawa et al., 2002). Tolfenamic acid promotes Sp1 protein degradation and lowers APP, tau, and BACE1 expression as well as Aβ levels and improves cognition in mice (Abdelrahim et al., 2006; Adwan et al., 2011; Adwan et al., unpublished observation; Subaiea et al., in press). Tolfenamic acid is scheduled to be tested in AD patients in the near future.
3. Discussion and conclusions
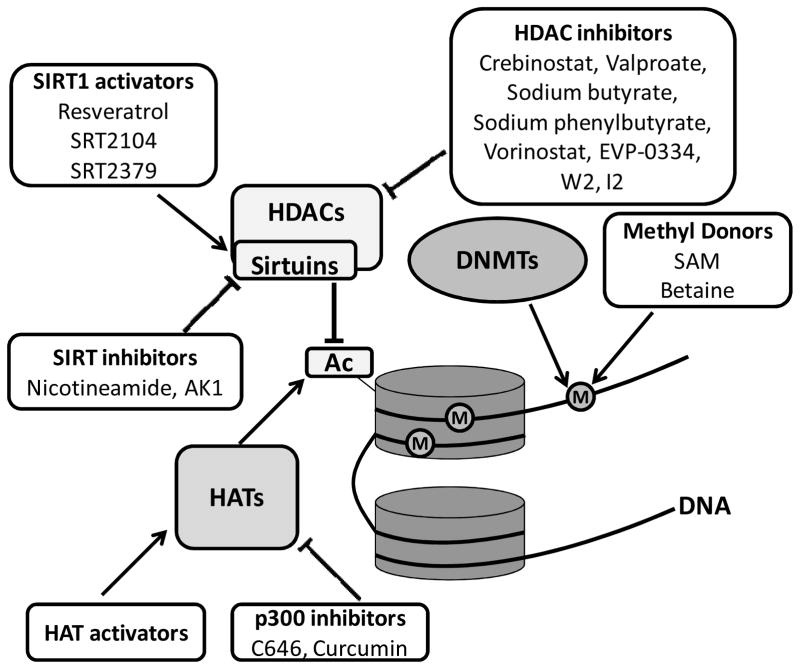
Epigenetic changes that occur early in life can impact our health decades later. Various studies suggest that pathologic changes in AD can be reversed prior to the development of symptoms through epigenetic modifications (Fig. 1). Developmental exposure to lead (Pb) upregulates genes involved in AD late in life through mechanisms that involves DNA methylation and histone acetylation (Bihaqi et al., 2011; Bihaqi & Zawia, 2012; Wu et al., 2008a). Persistent bidirectional changes in DNA methylation in response to earlier Pb exposure are reported with hypermethylation resulting in a latent reduction in gene expression (Alashwal et al., 2012; Dosunmu et al., 2012). Moreover, cognitive impairment accompanies overexpression of Sp1, BACE1, APP and Aβ late in life following early exposure to Pb and consequential epigenetic alterations (Bihaqi et al., in press). Such environmentally-induced changes on AD related intermediates could be reversed via epigenetic mechanisms. Alternatively, active epigenetic changes are involved in memory formation and could be targeted for AD therapy. Modulation of epigenetic intermediates could be a means for upregulation of genes that promote learning and memory, or reversing epigenetic changes that are responsible for the overexpression of genes involved in AD pathology. As neurons have a very limited ability to regenerate, reversing pathological changes through targeting epigenetic intermediates seems to be a promising therapeutic approach.
Fig. 1. Epigenetic targets and therapeutic approaches for AD.
DNA methylation and histone modification mediators involved in AD and potential therapeutic interventions under development reported in literature. Barrels = histones; (M) = methyl group; (Ac) = acetyl group. Abbreviations in the figure are as follows: DNMT, DNA methyltransferase; HAT, histone acetyltransferase; HDAC, histone deacetylase; SIRT, sirtuin.
Interestingly, epigenetic targets in AD are also implicated in the pathophysiology of schizophrenia and depression (Covington et al., 2009; Gavin & Sharma, 2010). Depression is a common comorbidity in demented patients (Alzheimer’s Association, 2012; Holtzer et al., 2005). Psychotic symptoms, especially at later stages of the disease, are also frequent in AD (Alzheimer’s Association, 2012; Ropacki & Jeste, 2005). Besides AD, HDAC inhibitors have been explored for other disorders including schizophrenia and depression. For example, the HDAC inhibitor sodium butyrate shows antidepressant activity in mice (Schroeder et al., 2007). Sodium butyrate also protects against phencyclidine induced psychotic-like behavior in mice (Koseki et al., 2012). It would be interesting to study the effects of HDAC inhibitors as epigenetic modifiers on cognitive as well as depressed and psychotic symptoms in AD patients.
Epigenetic alterations reported in AD are summarized in Table 1. A major challenge for AD management is early diagnosis. Currently, no standard criteria are available for early or accurate detection of AD through reference values of biomarkers from patients CSF and blood samples or imaging results. Epigenetic changes in AD could offer a diagnostic tool for the disease especially that some changes occur long before the molecular pathology of AD develops. If such changes are identified and detected early, reversing them via epigenetic therapeutic approaches would prevent the triggering of alterations in gene expression and transcriptional cascade associated with the neuropathology of AD. Also establishing criteria for epigenetic changes in AD can help administer disease-modifying drugs, once they become available, early in the disease process. The use of epigenetics will likely be even more crucial as the field moves towards early and pre-symptomatic case detection and earlier attempts at intervention (Sperling et al., 2011).
Table 1.
Some epigenetic changes in AD reported in literature.
| Epigenetic Mark | Change | Reference |
|---|---|---|
| HDAC2 | Increase | Graff et al., 2012 |
| HDAC6 | Increase | Ding et al., 2008 |
| SIRT1 | Decrease | Julien et al., 2009 |
| DNMT1 | Decrease | Mastroeni et al., 2010 |
| microRNA-101 | Decrease | Hebert et al., 2008 |
| microRNA-107 | Decrease early in AD | Wang et al., 2008b |
| BACE1-AS | Upregulated | Faghihi et al., 2008 |
| Methylation on APP gene | Hypomethylation | West et al., 1995 |
| Methylation on APP, PS1, tau | No change | Barrachina & Ferrer, 2009 |
The side effects of epigenetic targeting should also be studied. Attention should be made for the consequences of epigenetic modifications that are involved in multiple pathways and which might serve various functions within different cells and organs. Identification of more specific targets and agents could be a way for minimizing toxicity. It is important to realize that drugs with epigenetic effects are already present on the market. Some drugs used for years like the antihypertensive agent hydralazine and the antiepileptic drug valproate are found to interfere with epigenetic pathways which explains their previously unknown mechanisms of action or some of their adverse effects and suggests that their use could be repurposed for other disorders where epigenetic alterations are desired (Csoka & Szyf, 2009).
Our knowledge about epigenetics is still limited, some mechanisms have been studied more thoroughly like histone acetylation and DNA methylation, yet much remains to be revealed, especially when it comes to AD, memory and cognitive functions. Epigenetics is more upstream in AD pathology than the more common or conventional targets such as BACE, γ-secretase, Aβ and tau and thus could be beneficial especially in early stages of the disease to prevent further transcription and accumulation of pathological intermediates. Screening for such modifications and diagnosis of AD at an early stage remain a challenge. Nevertheless, promoting epigenetic mechanisms that trigger memory formation and inhibit pathological events could be a novel and effective therapeutic approach for preventing or at least delaying the development of dementia. Epigenetics offer potential for AD where epigenetic changes are integrated in the disease pathology. While no disease-modifying candidate is available, more research is needed for the refinement of epigenetic targets and identification of specific agents that can improve cognition and prevent or slow AD. Although knowledge is still being gathered about this field of study, there is evidence that epigenetics could provide multi-target therapeutic approaches for AD.
Acknowledgments
Supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences and grant NIH-5RO1ES015867-03 awarded to NHZ. The research core facility was funded by grants from the National Center for Research Resources (5P20RR016457-11) and the National Institute for General Medical Science (8 P20 GM103430-11), components of the National Institutes of Health (NIH).
Abbreviations
- Aβ
amyloid β
- AD
Alzheimer’s disease
- APP
amyloid β precursor protein
- BACE
β-site APP cleaving enzyme
- CBP
CREB binding protein
- CREB
cAMP response element-binding protein
- CSF
cerebrospinal fluid
- DNMT
DNA methyltransferase
- FAD
familial AD
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- LOAD
late onset AD
- MeCP2
methyl CpG binding protein 2
- NFTs
neurofibrillary tangles
- PS
presenilin
- SAM
S-adenosyl methionine
- SIRT
sirtuin
- Sp1
specificity protein 1
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelrahim M, Baker CH, Abbruzzese JL, Safe S. Tolfenamic acid and pancreatic cancer growth, angiogenesis, and Sp protein degradation. J Natl Cancer Inst. 2006;98:855–868. doi: 10.1093/jnci/djj232. [DOI] [PubMed] [Google Scholar]
- Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adwan LI, Basha R, Abdelrahim M, Subaiea GM, Zawia NH. Tolfenamic acid interrupts the de novo synthesis of the beta-amyloid precursor protein and lowers amyloid beta via a transcriptional pathway. Curr Alzheimer Res. 2011;8:385–392. doi: 10.2174/156720511795745285. [DOI] [PubMed] [Google Scholar]
- Alashwal H, Dosunmu R, Zawia NH. Integration of genome-wide expression and methylation data: relevance to aging and Alzheimer’s disease. Neurotoxicology. 2012;33:1450–1453. doi: 10.1016/j.neuro.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani D, Polito L, Forloni G. Sirtuins as novel targets for Alzheimer’s disease and other neurodegenerative disorders: experimental and genetic evidence. J Alzheimers Dis. 2010;19:11–26. doi: 10.3233/JAD-2010-1215. [DOI] [PubMed] [Google Scholar]
- Alonso AD, Grundke-Iqbal I, Barra HS, Iqbal K. Abnormal phosphorylation of tau and the mechanism of Alzheimer neurofibrillary degeneration: sequestration of microtubule-associated proteins 1 and 2 and the disassembly of microtubules by the abnormal tau. Proc Natl Acad Sci U S A. 1997;94:298–303. doi: 10.1073/pnas.94.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer’s Association. 2012 Alzheimer’s disease facts and figures. Alzheimers Dement. 2012;8:131–168. doi: 10.1016/j.jalz.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Alzheimer’s Association. 2013 Alzheimer’s disease facts and figures. Alzheimers Dement. 2013;9:208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Bakulski KM, Dolinoy DC, Sartor MA, Paulson HL, Konen JR, Lieberman AP, et al. Genome-wide DNA methylation differences between late-onset Alzheimer’s disease and cognitively normal controls in human frontal cortex. J Alzheimers Dis. 2012;29:571–588. doi: 10.3233/JAD-2012-111223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- Barneda-Zahonero B, Parra M. Histone deacetylases and cancer. Mol Oncol. 2012;6:579–589. doi: 10.1016/j.molonc.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, Yaffe K. The projected effect of risk factor reduction on Alzheimer’s disease prevalence. Lancet Neurol. 2011;10:819–828. doi: 10.1016/S1474-4422(11)70072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrachina M, Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol. 2009;68:880–891. doi: 10.1097/NEN.0b013e3181af2e46. [DOI] [PubMed] [Google Scholar]
- Begum AN, Jones MR, Lim GP, Morihara T, Kim P, Heath DD, et al. Curcumin structure-function, bioavailability, and efficacy in models of neuroinflammation and Alzheimer’s disease. J Pharmacol Exp Ther. 2008;326:196–208. doi: 10.1124/jpet.108.137455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihaqi SW, Bahmani A, Subaiea GM, Zawia NH. Alzheimers Dement. Infantile exposure to lead (Pb) and late age cognitive decline: relevance to AD. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihaqi SW, Huang H, Wu J, Zawia NH. Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: implications for Alzheimer’s disease. J Alzheimers Dis. 2011;27:819–833. doi: 10.3233/JAD-2011-111013. [DOI] [PubMed] [Google Scholar]
- Bihaqi SW, Zawia NH. Alzheimer’s disease biomarkers and epigenetic intermediates following exposure to Pb in vitro. Curr Alzheimer Res. 2012;9:555–562. doi: 10.2174/156720512800617964. [DOI] [PubMed] [Google Scholar]
- Bird A. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- Bottiglieri T, Godfrey P, Flynn T, Carney MW, Toone BK, Reynolds EH. Cerebrospinal fluid S-adenosylmethionine in depression and dementia: effects of treatment with parenteral and oral S-adenosylmethionine. J Neurol Neurosurg Psychiatry. 1990;53:1096–1098. doi: 10.1136/jnnp.53.12.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottiglieri T, Hyland K, Reynolds EH. The clinical potential of ademetionine (S-adenosylmethionine) in neurological disorders. Drugs. 1994;48:137–152. doi: 10.2165/00003495-199448020-00002. [DOI] [PubMed] [Google Scholar]
- Caccamo A, Maldonado MA, Bokov AF, Majumder S, Oddo S. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107:22687–22692. doi: 10.1073/pnas.1012851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraci F, Leggio GM, Drago F, Salomone S. Epigenetic drugs for Alzheimer’s Disease: hopes and challenges. Br J Clin Pharmacol. 2012;75:1154–1155. doi: 10.1111/j.1365-2125.2012.04443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics. 2012;4:317–324. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DL, Kenis G, Dela Cruz J, Lemmens MA, van Os J, et al. Caloric restriction attenuates age-related changes of DNA methyltransferase 3a in mouse hippocampus. Brain Behav Immun. 2011;25:616–623. doi: 10.1016/j.bbi.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DL, Kenis G, Keitel S, Hof PR, van Os J, et al. Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction. Curr Alzheimer Res. 2012a;9:536–544. doi: 10.2174/156720512800618035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DL, Kenis G, Keitel S, Hof PR, van Os J, et al. Prevention of age-related changes in hippocampal levels of 5-methylcytidine by caloric restriction. Neurobiol Aging. 2012b;33:1672–1681. doi: 10.1016/j.neurobiolaging.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, O’Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron M, Teplow DB, Selkoe DJ. Generation of amyloid beta protein from its precursor is sequence specific. Neuron. 1995;14:661–670. doi: 10.1016/0896-6273(95)90323-2. [DOI] [PubMed] [Google Scholar]
- ClinicalTrials.gov. 18-Month Study of Curcumin: ClinicalTials.gov. identifier NCT01383161. [Google Scholar]
- ClinicalTrials.gov. Cilostazol Augmentation Study in Dementia: ClinicalTrials.gov. identifier NCT01409564. [Google Scholar]
- ClinicalTrials.gov. Resveratrol for Alzheimer’s Disease: ClinicalTrials.gov. identifier NCT01504854. [Google Scholar]
- ClinicalTrials.gov. Safety Study of Nicotinamide to Treat Alzheimer’s Disease: ClinicalTrials.gov. identifier NCT00580931. [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Cohen TJ, Guo JL, Hurtado DE, Kwong LK, Mills IP, Trojanowski JQ, et al. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun. 2011;2:252. doi: 10.1038/ncomms1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Covington HE, 3rd, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig SA. Betaine in human nutrition. Am J Clin Nutr. 2004;80:539–549. doi: 10.1093/ajcn/80.3.539. [DOI] [PubMed] [Google Scholar]
- Csoka AB, Szyf M. Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology. Med Hypotheses. 2009;73:770–780. doi: 10.1016/j.mehy.2008.10.039. [DOI] [PubMed] [Google Scholar]
- Czech C, Tremp G, Pradier L. Presenilins and Alzheimer’s disease: biological functions and pathogenic mechanisms. Prog Neurobiol. 2000;60:363–384. doi: 10.1016/s0301-0082(99)00033-7. [DOI] [PubMed] [Google Scholar]
- Davie JR, Spencer VA. Control of histone modifications. J Cell Biochem. 1999;(Suppl 32–33):141–148. doi: 10.1002/(sici)1097-4644(1999)75:32+<141::aid-jcb17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira RM, Sarkander J, Kazantsev AG, Outeiro TF. SIRT2 as a Therapeutic Target for Age-Related Disorders. Front Pharmacol. 2012;3:82. doi: 10.3389/fphar.2012.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, et al. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- Ding H, Dolan PJ, Johnson GV. Histone deacetylase 6 interacts with the microtubule-associated protein tau. J Neurochem. 2008;106:2119–2130. doi: 10.1111/j.1471-4159.2008.05564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docagne F, Gabriel C, Lebeurrier N, Lesne S, Hommet Y, Plawinski L, et al. Sp1 and Smad transcription factors co-operate to mediate TGF-beta-dependent activation of amyloid-beta precursor protein gene transcription. Biochem J. 2004;383:393–399. doi: 10.1042/BJ20040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dosunmu R, Alashwal H, Zawia NH. Genome-wide expression and methylation profiling in the aged rodent brain due to early-life Pb exposure and its relevance to aging. Mech Ageing Dev. 2012;133:435–443. doi: 10.1016/j.mad.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Connor L, Wolf PA, Growdon JH. Association of decreased paternal age and late-onset Alzheimer’s disease. An example of genetic imprinting? Arch Neurol. 1991;48:599–604. doi: 10.1001/archneur.1991.00530180051017. [DOI] [PubMed] [Google Scholar]
- Fass DM, Reis SA, Ghosh B, Hennig KM, Joseph NF, Zhao WN, et al. Crebinostat: A novel cognitive enhancer that inhibits histone deacetylase activity and modulates chromatin-mediated neuroplasticity. Neuropharmacology. 2013;64:81–96. doi: 10.1016/j.neuropharm.2012.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Epigenetics at the epicenter of modern medicine. JAMA. 2008;299:1345–1350. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Fleisher AS, Truran D, Mai JT, Langbaum JB, Aisen PS, Cummings JL, et al. Chronic divalproex sodium use and brain atrophy in Alzheimer disease. Neurology. 2011;77:1263–1271. doi: 10.1212/WNL.0b013e318230a16c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuso A, Seminara L, Cavallaro RA, D’Anselmi F, Scarpa S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol Cell Neurosci. 2005;28:195–204. doi: 10.1016/j.mcn.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP. Histone modifications, DNA methylation, and schizophrenia. Neurosci Biobehav Rev. 2010;34:882–888. doi: 10.1016/j.neubiorev.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, et al. Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci. 2008;28:11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi T, Ono K, Yamada M. REVIEW: Curcumin and Alzheimer’s disease. CNS Neurosci Ther. 2010;16:285–297. doi: 10.1111/j.1755-5949.2010.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C, Binder EB. Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp Neurol. 2012;233:102–111. doi: 10.1016/j.expneurol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Matzke MA. Exploring and explaining epigenetic effects. Trends Genet. 1997;13:293–295. doi: 10.1016/s0168-9525(97)01219-5. [DOI] [PubMed] [Google Scholar]
- Hiramatsu M, Takiguchi O, Nishiyama A, Mori H. Cilostazol prevents amyloid beta peptide(25–35)-induced memory impairment and oxidative stress in mice. Br J Pharmacol. 2010;161:1899–1912. doi: 10.1111/j.1476-5381.2010.01014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PW, Chernak JM. DNA binding and regulatory effects of transcription factors SP1 and USF at the rat amyloid precursor protein gene promoter. Nucleic Acids Res. 1995;23:2229–2235. doi: 10.1093/nar/23.12.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Wald J, Lavu S, Roberts J, Beaumont C, Haddad J, et al. Pharmacokinetics and tolerability of SRT2104, a first-in-class small molecule activator of SIRT1, after single and repeated oral administration in man. Br J Clin Pharmacol. 2013;75:186–196. doi: 10.1111/j.1365-2125.2012.04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler M, Westin G, Jiricny J, Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- Holtzer R, Scarmeas N, Wegesin DJ, Albert M, Brandt J, Dubois B, et al. Depressive symptoms in Alzheimer’s disease: natural course and temporal relation to function and cognitive status. J Am Geriatr Soc. 2005;53:2083–2089. doi: 10.1111/j.1532-5415.2005.00535.x. [DOI] [PubMed] [Google Scholar]
- Houston I, Peter CJ, Mitchell A, Straubhaar J, Rogaev E, Akbarian S. Epigenetics in the human brain. Neuropsychopharmacology. 2013;38:183–197. doi: 10.1038/npp.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol Cell Biol. 1999;19:8211–8218. doi: 10.1128/mcb.19.12.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber K, Superti-Furga G. After the grape rush: sirtuins as epigenetic drug targets in neurodegenerative disorders. Bioorg Med Chem. 2011;19:3616–3624. doi: 10.1016/j.bmc.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Inkster B, Strijbis EM, Vounou M, Kappos L, Radue EW, Matthews PM, et al. Histone deacetylase gene variants predict brain volume changes in multiple sclerosis. Neurobiol Aging. 2013;34:238–247. doi: 10.1016/j.neurobiolaging.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Liu F, Gong CX, Alonso Adel C, Grundke-Iqbal I. Mechanisms of tau-induced neurodegeneration. Acta Neuropathol. 2009;118:53–69. doi: 10.1007/s00401-009-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;(33 Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Julien C, Tremblay C, Emond V, Lebbadi M, Salem N, Jr, Bennett DA, et al. Sirtuin 1 reduction parallels the accumulation of tau in Alzheimer disease. J Neuropathol Exp Neurol. 2009;68:48–58. doi: 10.1097/NEN.0b013e3181922348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis TC, Ververis K. Potential of chromatin modifying compounds for the treatment of Alzheimer’s disease. Pathobiol Aging Age Relat Dis. 2012:2. doi: 10.3402/pba.v2i0.14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key NS. Hyperhomocyst(e)inemia. Current treatment options in cardiovascular medicine. 2000;2:65–72. doi: 10.1007/s11936-000-0029-7. [DOI] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Thompson LA, Wenger SD, O’Bryant CL. Romidepsin: a histone deacetylase inhibitor for refractory cutaneous T-cell lymphoma. Ann Pharmacother. 2012;46:1340–1348. doi: 10.1345/aph.1R036. [DOI] [PubMed] [Google Scholar]
- Kishikawa S, Murata T, Kimura H, Shiota K, Yokoyama KK. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur J Biochem. 2002;269:2961–2970. doi: 10.1046/j.1432-1033.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Knopman D, Patterson M. An open-label, 24-week pilot study of the methyl donor betaine in Alzheimer disease patients. Alzheimer Dis Assoc Disord. 2001;15:162–165. doi: 10.1097/00002093-200107000-00008. [DOI] [PubMed] [Google Scholar]
- Korzus E. Manipulating the brain with epigenetics. Nat Neurosci. 2010;13:405–406. doi: 10.1038/nn0410-405. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseki T, Mouri A, Mamiya T, Aoyama Y, Toriumi K, Suzuki S, et al. Exposure to enriched environments during adolescence prevents abnormal behaviours associated with histone deacetylation in phencyclidine-treated mice. Int J Neuropsychopharmacol. 2012;15:1489–1501. doi: 10.1017/S1461145711001672. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kramer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth S, Thon N, Kreth FW. Epigenetics in human gliomas. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.04.008. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Zawia NH. The LEARn model: an epigenetic explanation for idiopathic neurobiological diseases. Mol Psychiatry. 2009;14:992–1003. doi: 10.1038/mp.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Zawia NH, Greig NH, Sambamurti K, Maloney B. Early-life events may trigger biochemical pathways for Alzheimer’s disease: the “LEARn” model. Biogerontology. 2008;9:375–379. doi: 10.1007/s10522-008-9162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Levkovitz Y, Alpert JE, Brintz CE, Mischoulon D, Papakostas GI. Effects of S-adenosylmethionine augmentation of serotonin-reuptake inhibitor antidepressants on cognitive symptoms of major depressive disorder. J Affect Disord. 2012;136:1174–1178. doi: 10.1016/j.jad.2011.04.059. [DOI] [PubMed] [Google Scholar]
- Lim GP, Chu T, Yang F, Beech W, Frautschy SA, Cole GM. The curry spice curcumin reduces oxidative damage and amyloid pathology in an Alzheimer transgenic mouse. J Neurosci. 2001;21:8370–8377. doi: 10.1523/JNEUROSCI.21-21-08370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li Y, Tollefsbol TO. Gene-environment interactions and epigenetic basis of human diseases. Curr Issues Mol Biol. 2008;10:25–36. [PMC free article] [PubMed] [Google Scholar]
- Liu L, van Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiol Aging. 2009;30:549–560. doi: 10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy R, Tariot PN. Neuroprotective properties of valproate: potential benefit for AD and tauopathies. J Mol Neurosci. 2002;19:303–307. doi: 10.1385/jmn:19:3:301. [DOI] [PubMed] [Google Scholar]
- Mack GS. To selectivity and beyond. Nat Biotechnol. 2010;28:1259–1266. doi: 10.1038/nbt.1724. [DOI] [PubMed] [Google Scholar]
- Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, et al. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169–174. doi: 10.2174/157340606776056133. [DOI] [PubMed] [Google Scholar]
- Maric NP, Svrakic DM. Why schizophrenia genetics needs epigenetics: a review. Psychiatr Danub. 2012;24:2–18. [PubMed] [Google Scholar]
- Martin KC, Sun YE. To learn better, keep the HAT on. Neuron. 2004;42:879–881. doi: 10.1016/j.neuron.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiol Aging. 2010;31:2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic mechanisms in Alzheimer’s disease. Neurobiol Aging. 2011;32:1161–1180. doi: 10.1016/j.neurobiolaging.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One. 2009;4:e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Cheng B, Davis D, Bryant K, Lieberburg I, Rydel RE. beta-Amyloid peptides destabilize calcium homeostasis and render human cortical neurons vulnerable to excitotoxicity. J Neurosci. 1992;12:376–389. doi: 10.1523/JNEUROSCI.12-02-00376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Nestler EJ. The epigenetic landscape of addiction. Ann N Y Acad Sci. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud KR, Gutierrez-Mecinas M, Trollope AF, Collins A, Saunderson EA, Reul JM. Epigenetic mechanisms in stress and adaptation. Brain Behav Immun. 2011;25:1305–1315. doi: 10.1016/j.bbi.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Mill J. Toward an integrated genetic and epigenetic approach to Alzheimer’s disease. Neurobiol Aging. 2011;32:1188–1191. doi: 10.1016/j.neurobiolaging.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M, Tsuboi M, Noguchi Y, Enokishima A, Nabeshima T, Hiramatsu M. Effects of betaine on lipopolysaccharide-induced memory impairment in mice and the involvement of GABA transporter 2. J Neuroinflammation. 2011;8:153. doi: 10.1186/1742-2094-8-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molfese DL. Advancing neuroscience through epigenetics: molecular mechanisms of learning and memory. Dev Neuropsychol. 2011;36:810–827. doi: 10.1080/87565641.2011.606395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Curr Opin Genet Dev. 2006;16:276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Morrison LD, Smith DD, Kish SJ. Brain S-adenosylmethionine levels are severely decreased in Alzheimer’s disease. J Neurochem. 1996;67:1328–1331. doi: 10.1046/j.1471-4159.1996.67031328.x. [DOI] [PubMed] [Google Scholar]
- Mulder C, Schoonenboom NS, Jansen EE, Verhoeven NM, van Kamp GJ, Jakobs C, et al. The transmethylation cycle in the brain of Alzheimer patients. Neurosci Lett. 2005;386:69–71. doi: 10.1016/j.neulet.2005.03.073. [DOI] [PubMed] [Google Scholar]
- Nalivaeva NN, Belyaev ND, Turner AJ. Sodium valproate: an old drug with new roles. Trends Pharmacol Sci. 2009;30:509–514. doi: 10.1016/j.tips.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Naruse S, Thinakaran G, Luo JJ, Kusiak JW, Tomita T, Iwatsubo T, et al. Effects of PS1 deficiency on membrane protein trafficking in neurons. Neuron. 1998;21:1213–1221. doi: 10.1016/s0896-6273(00)80637-6. [DOI] [PubMed] [Google Scholar]
- Nebbioso A, Carafa V, Benedetti R, Altucci L. Trials with ‘epigenetic’ drugs: an update. Mol Oncol. 2012;6:657–682. doi: 10.1016/j.molonc.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New M, Olzscha H, La Thangue NB. HDAC inhibitor-based therapies: can we interpret the code? Mol Oncol. 2012;6:637–656. doi: 10.1016/j.molonc.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS One. 2010;5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oike Y, Hata A, Mamiya T, Kaname T, Noda Y, Suzuki M, et al. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum Mol Genet. 1999;8:387–396. doi: 10.1093/hmg/8.3.387. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Bading H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci. 2012;15:1111–1113. doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- Orr BA, Haffner MC, Nelson WG, Yegnasubramanian S, Eberhart CG. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS One. 2012;7:e41036. doi: 10.1371/journal.pone.0041036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro TF, Marques O, Kazantsev A. Therapeutic role of sirtuins in neurodegenerative disease. Biochim Biophys Acta. 2008;1782:363–369. doi: 10.1016/j.bbadis.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- Pollwein P, Masters CL, Beyreuther K. The expression of the amyloid precursor protein (APP) is regulated by two GC-elements in the promoter. Nucleic Acids Res. 1992;20:63–68. doi: 10.1093/nar/20.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P. MicroRNAs as a molecular basis for mental retardation, Alzheimer’s and prion diseases. Brain Res. 2010;1338:58–66. doi: 10.1016/j.brainres.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Epigenetic mechanisms underlying human epileptic disorders and the process of epileptogenesis. Neurobiol Dis. 2010;39:53–60. doi: 10.1016/j.nbd.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan A, Shah ZA. Sirtuins in neurodegenerative diseases: a biological-chemical perspective. Neurodegener Dis. 2012;9:1–10. doi: 10.1159/000329724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- Ringman JM, Frautschy SA, Teng E, Begum AN, Bardens J, Beigi M, et al. Oral curcumin for Alzheimer’s disease: tolerability and efficacy in a 24-week randomized, double blind, placebo-controlled study. Alzheimers Res Ther. 2012;4:43. doi: 10.1186/alzrt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaev EI, Lukiw WJ, Lavrushina O, Rogaeva EA, St George-Hyslop PH. The upstream promoter of the beta-amyloid precursor protein gene (APP) shows differential patterns of methylation in human brain. Genomics. 1994;22:340–347. doi: 10.1006/geno.1994.1393. [DOI] [PubMed] [Google Scholar]
- Ropacki SA, Jeste DV. Epidemiology of and risk factors for psychosis of Alzheimer’s disease: a review of 55 studies published from 1990 to 2003. Am J Psychiatry. 2005;162:2022–2030. doi: 10.1176/appi.ajp.162.11.2022. [DOI] [PubMed] [Google Scholar]
- Rubinstein JH, Taybi H. Broad thumbs and toes and facial abnormalities. A possible mental retardation syndrome. Am J Dis Child. 1963;105:588–608. doi: 10.1001/archpedi.1963.02080040590010. [DOI] [PubMed] [Google Scholar]
- Scarpa S, Fuso A, D’Anselmi F, Cavallaro RA. Presenilin 1 gene silencing by S-adenosylmethionine: a treatment for Alzheimer disease? FEBS Lett. 2003;541:145–148. doi: 10.1016/s0014-5793(03)00277-1. [DOI] [PubMed] [Google Scholar]
- Schonrock N, Gotz J. Decoding the non-coding RNAs in Alzheimer’s disease. Cell Mol Life Sci. 2012;69:3543–3559. doi: 10.1007/s00018-012-1125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder FA, Lin CL, Crusio WE, Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol Psychiatry. 2007;62:55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Physiological production of the beta-amyloid protein and the mechanism of Alzheimer’s disease. Trends Neurosci. 1993;16:403–409. doi: 10.1016/0166-2236(93)90008-a. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Preventing Alzheimer’s disease. Science. 2012;337:1488–1492. doi: 10.1126/science.1228541. [DOI] [PubMed] [Google Scholar]
- Selvi BR, Cassel JC, Kundu TK, Boutillier AL. Tuning acetylation levels with HAT activators: therapeutic strategy in neurodegenerative diseases. Biochim Biophys Acta. 2010;1799:840–853. doi: 10.1016/j.bbagrm.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Kabat J, Novak M, Wu Q, Grundke-Iqbal I, Iqbal K. Phosphorylation of tau at both Thr 231 and Ser 262 is required for maximal inhibition of its binding to microtubules. Arch Biochem Biophys. 1998;357:299–309. doi: 10.1006/abbi.1998.0813. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Beiser A, Selhub J, Jacques PF, Rosenberg IH, D’Agostino RB, et al. Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. N Engl J Med. 2002;346:476–483. doi: 10.1056/NEJMoa011613. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Zoghbi HY. Rett syndrome and MeCP2: linking epigenetics and neuronal function. Am J Hum Genet. 2002;71:1259–1272. doi: 10.1086/345360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Shimojo M, Sahara N, Murayama M, Ichinose H, Takashima A. Decreased Abeta secretion by cells expressing familial Alzheimer’s disease-linked mutant presenilin 1. Neurosci Res. 2007;57:446–453. doi: 10.1016/j.neures.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Shoji M, Golde TE, Ghiso J, Cheung TT, Estus S, Shaffer LM, et al. Production of the Alzheimer amyloid beta protein by normal proteolytic processing. Science. 1992;258:126–129. doi: 10.1126/science.1439760. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spires-Jones TL, Fox LM, Rozkalne A, Pitstick R, Carlson GA, Kazantsev AG. Inhibition of Sirtuin 2 with Sulfobenzoic Acid Derivative AK1 is Non-Toxic and Potentially Neuroprotective in a Mouse Model of Frontotemporal Dementia. Front Pharmacol. 2012;3:42. doi: 10.3389/fphar.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkel W, Campbell RM. Sirtuin 1 (SIRT1): the misunderstood HDAC. J Biomol Screen. 2011;16:1153–1169. doi: 10.1177/1087057111422103. [DOI] [PubMed] [Google Scholar]
- Su Y, Ryder J, Li B, Wu X, Fox N, Solenberg P, et al. Lithium, a common drug for bipolar disorder treatment, regulates amyloid-beta precursor protein processing. Biochemistry. 2004;43:6899–6908. doi: 10.1021/bi035627j. [DOI] [PubMed] [Google Scholar]
- Subaiea GM, Adwan LI, Ahmed AH, Stevens KE, Zawia NH. Short-term treatment with tolfenamic acid improves cognitive functions in AD mice. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2013.04.002. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Day JJ. Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011;3:157–181. doi: 10.2217/epi.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YM, Lee T, Yoon H, Dibattista AM, Song JM, Sohn Y, et al. Mercaptoacetamide-based class II HDAC inhibitor lowers Abeta levels and improves learning and memory in a mouse model of Alzheimer’s disease. Exp Neurol. 2013;239:192–201. doi: 10.1016/j.expneurol.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE. The genetics of Alzheimer disease. Cold Spring Harb Perspect Med. 2012:2. doi: 10.1101/cshperspect.a006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE, Kovacs DM, Kim TW, Moir RD, Guenette SY, Wasco W. The gene defects responsible for familial Alzheimer’s disease. Neurobiol Dis. 1996;3:159–168. doi: 10.1006/nbdi.1996.0016. [DOI] [PubMed] [Google Scholar]
- Taqi MM, Bazov I, Watanabe H, Sheedy D, Harper C, Alkass K, et al. Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict Biol. 2011;16:499–509. doi: 10.1111/j.1369-1600.2011.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariot PN, Schneider LS, Cummings J, Thomas RG, Raman R, Jakimovich LJ, et al. Chronic divalproex sodium to attenuate agitation and clinical progression of Alzheimer disease. Arch Gen Psychiatry. 2011;68:853–861. doi: 10.1001/archgenpsychiatry.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Genda Y, Ukitsu M. Reduction with age in methylcytosine in the promoter region -224 approximately -101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res. 1999;70:288–292. doi: 10.1016/s0169-328x(99)00163-1. [DOI] [PubMed] [Google Scholar]
- Townsend M. When will Alzheimer’s disease be cured? A pharmaceutical perspective. J Alzheimers Dis. 2011;24(Suppl 2):43–52. doi: 10.3233/JAD-2011-110020. [DOI] [PubMed] [Google Scholar]
- Tricco AC, Vandervaart S, Soobiah C, Lillie E, Perrier L, Chen MH, et al. Efficacy of cognitive enhancers for Alzheimer’s disease: protocol for a systematic review and network meta-analysis. Syst Rev. 2012;1:31. doi: 10.1186/2046-4053-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S, Ahl M, Bush A, Westaway D, Huang X, Rogers JT. Pilot study of the reducing effect on amyloidosis in vivo by three FDA pre-approved drugs via the Alzheimer’s APP 5′ untranslated region. Curr Alzheimer Res. 2005;2:249–254. doi: 10.2174/1567205053585855. [DOI] [PubMed] [Google Scholar]
- Tucker S, Ahl M, Cho HH, Bandyopadhyay S, Cuny GD, Bush AI, et al. RNA therapeutics directed to the non coding regions of APP mRNA, in vivo anti-amyloid efficacy of paroxetine, erythromycin, and N-acetyl cysteine. Curr Alzheimer Res. 2006;3:221–227. doi: 10.2174/156720506777632835. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E, Barbato C, Ciotti M, Cogoni C, Ruberti F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J Biol Chem. 2010;285:18344–18351. doi: 10.1074/jbc.M110.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fivecoat H, Ho L, Pan Y, Ling E, Pasinetti GM. The role of Sirt1: at the crossroad between promotion of longevity and protection against Alzheimer’s disease neuropathology. Biochim Biophys Acta. 2010;1804:1690–1694. doi: 10.1016/j.bbapap.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008a;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, et al. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008b;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Zhang N, Liu M, Tanaka R, Mizuno Y, Urabe T. Cilostazol protects against brain white matter damage and cognitive impairment in a rat model of chronic cerebral hypoperfusion. Stroke. 2006;37:1539–1545. doi: 10.1161/01.STR.0000221783.08037.a9. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, et al. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci U S A. 1975;72:1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J Mol Neurosci. 1995;6:141–146. doi: 10.1007/BF02736773. [DOI] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, et al. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Basha MR, Brock B, Cox DP, Cardozo-Pelaez F, McPherson CA, et al. Alzheimer’s disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J Neurosci. 2008a;28:3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Basha MR, Zawia NH. The environment, epigenetics and amyloidogenesis. J Mol Neurosci. 2008b;34:1–7. doi: 10.1007/s12031-007-0009-4. [DOI] [PubMed] [Google Scholar]
- Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- Yang F, Lim GP, Begum AN, Ubeda OJ, Simmons MR, Ambegaokar SS, et al. Curcumin inhibits formation of amyloid beta oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J Biol Chem. 2005;280:5892–5901. doi: 10.1074/jbc.M404751200. [DOI] [PubMed] [Google Scholar]
- Yen RW, Vertino PM, Nelkin BD, Yu JJ, el-Deiry W, Cumaraswamy A, et al. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992;20:2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawia NH, Basha MR. Environmental risk factors and the developmental basis for Alzheimer’s disease. Rev Neurosci. 2005;16:325–337. doi: 10.1515/revneuro.2005.16.4.325. [DOI] [PubMed] [Google Scholar]
- Zawia NH, Lahiri DK, Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic Biol Med. 2009;46:1241–1249. doi: 10.1016/j.freeradbiomed.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, et al. microRNA-34c is a novel target to treat dementias. EMBO J. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]