Abstract
Autism spectrum disorders (ASDs) are highly heritable, and six genome-wide association studies (GWASs) of ASDs have been published to date. In this study, we have integrated the findings from these GWASs with other genetic data to identify enriched genetic networks that are associated with ASDs. We conducted bioinformatics and systematic literature analyses of 200 top-ranked ASD candidate genes from five published GWASs. The sixth GWAS was used for replication and validation of our findings. Further corroborating evidence was obtained through rare genetic variant studies, that is, exome sequencing and copy number variation (CNV) studies, and/or other genetic evidence, including candidate gene association, microRNA and gene expression, gene function and genetic animal studies. We found three signaling networks regulating steroidogenesis, neurite outgrowth and (glutamatergic) synaptic function to be enriched in the data. Most genes from the five GWASs were also implicated—independent of gene size—in ASDs by at least one other line of genomic evidence. Importantly, A-kinase anchor proteins (AKAPs) functionally integrate signaling cascades within and between these networks. The three identified protein networks provide an important contribution to increasing our understanding of the molecular basis of ASDs. In addition, our results point towards the AKAPs as promising targets for developing novel ASD treatments.
Keywords: A-kinase anchor proteins, autism spectrum disorders, genetics, genome-wide association studies
Introduction
Autism spectrum disorders (ASDs) are a group of pervasive neurodevelopmental disorders that are characterized by qualitative impairments in reciprocal social interaction and communication as well as restricted, repetitive and stereotyped patterns of behavior, interests and activities. The ASDs include autism, Asperger's syndrome and pervasive developmental disorder, not otherwise specified (Diagnostic and Statistical Manual of Mental Disorders-fourth edition-text revision (DSM-IV-TR)). Individuals with autism show impairments in all three core symptom domains and an onset of symptoms before 3 years of age, whereas Asperger's syndrome is characterized by social and behavioral impairments in the presence of a normal language development before age 3 years. Pervasive developmental disorder, not otherwise specified, is diagnosed in people who do not meet autism criteria for all three symptom domains and/or show a later age of onset (DSM-IV-TR). The prevalence of ASDs is estimated to be 0.5–1%,1, 2, 3 with ∼4 times more males than females being affected by these disorders.4, 5 Family and twin studies show that ASDs are highly heritable, and ∼90% of the phenotypic variance that is observed in these disorders can be explained by genetic factors.5, 6, 7 Despite this high heritability, the identification and replication of genetic susceptibility factors for ASDs has proven challenging, as the genetic architecture of these disorders is highly heterogeneous (see recent reviews8, 9, 10). Rare genetic variants that predispose individuals to ASDs are currently thought to account for 10–20% of all ASD cases.8, 10 These variants include rare mutations in single genes that can lead to monogenic disorders (such as fragile X syndrome and tuberous sclerosis) associated with ASD symptoms, or genes identified through exome sequencing studies and copy number variations (CNVs), with the latter constituting deletions or duplications of a large variety of genes and potentially contributing to oligogenic forms of ASDs.8, 9, 10 Common genetic variants for ASDs—for example, single-nucleotide polymorphism (SNPs)—have also been identified through candidate gene and genome-wide association studies (GWASs), and each variant is assumed to contribute only a small increase in disease risk.7, 8, 9 To date, six GWASs for ASDs have been published.11, 12, 13, 14, 15, 16 In addition to many protein-coding genes that are implicated in ASD pathophysiology, increasing evidence indicates that microRNAs may also be involved in ASD aetiology.17, 18, 19, 20 In recent years, three main biological ‘themes' have emerged from genetic ASD findings, namely steroid synthesis,21, 22 growth and neurite outgrowth of developing neurons,9, 15, 21, 22, 23 and synaptic function.8, 9, 23, 24, 25 In this article, we integrate the most important findings from ASD GWASs into protein signaling networks that are linked to these biological themes, and provide corroborating evidence for these networks from exome sequencing/mutation, CNV, microRNA and other genetic data. Importantly, we find that signaling cascades within and between the three networks are functionally connected by A-kinase anchor proteins (AKAPs), which seem to be promising targets for the development of novel psychopharmacological treatment for ASDs.
Materials and methods
Identification of candidate genes and finding corroborating evidence
The main characteristics of the six published GWASs of ASDs published to date are presented in Table 1. We used SNP data from five of these studies to compile a list of top-ranked genes associated with ASDs (the cutoff for association was P<1 × 10−4).11, 12, 13, 14, 15 The sixth GWAS16 was used for validation and replication processes (see below), as it was conducted on a discovery sample that was independent of the discovery samples that were used in the five other GWASs. The used SNPs included those that were located within exonic, intronic or untranslated regions of genes or found within 100 kilobases (kb) of downstream or upstream, potentially regulatory26, 27, 28, 29 sequences flanking a gene. Of note, for the selection of SNPs from two of the GWASs, we used the complete and detailed association results for the combined discovery samples, rather than the lists of SNPs as they were published, which included replication samples.11, 13 In addition, although our chosen statistical cutoff for association was essentially an arbitrary threshold, it was used in four of the six published ASD GWASs to designate ‘suggestive' evidence of association,11, 12, 13, 16 and application of this cutoff provided us with a manageable number of genes for our subsequent analyses. Applying the above described SNP selection criteria yielded a total of 200 genes from the five GWASs11, 12, 13, 14, 15(Supplementary Table 1).
Table 1. Overview of the clinical samples from six published GWASs of ASDs11, 12, 13, 14, 15, 16.
| GWAS | Clinical sample | Genotyping platform | Phenotype | Diagnosis |
|---|---|---|---|---|
| Wang et al.11 | Combined discovery sample (AE): AGRE (943 families) a + ACC cohort (1453 cases and 7070 controls) from CHOP and several other US clinical sites | Illumina | ASD | ADOS+ADI-R |
| Ma et al.12 | CAP discovery sample (AE): four US clinical sites (438 families) | Illumina | Autism | ADI-R+DSM-IV |
| Weiss et al.13 | Combined discovery sample (AE), consisting of 1031 multiplex families with 1553 affected offspring from: AGRE (801 families) a + NIMH (341 families) | Affymetrix | ASD | ADI-R |
| Salyakina et al.14 | Combined sample: discovery sample from two US clinical sites (124 families) + AGRE validation sample (110 families) | Illumina | ASP | ADI-R+DSM-IV |
| Hussman et al.15 | Combined sample: sample from two US clinical sites (597 families) + AGRE sample (696 families) | Illumina | Autism | ADI-R+DSM-IV |
| Anney et al.16 | AGP Consortium discovery sample (AE): Autism (809 families) + ASD (1369 families) | Illumina | Autism+ASD | ADOS+ADI-R |
Abbreviations: ACC, Autism Case Control; ADOS, Autism Diagnostic Observation Schedule; ADI-R, Autism Diagnostic Interview-Revised; AE, all ethnicities; AGP, Autism Genome Project; AGRE, Autism Genetic Resource Exchange; ASD, autism spectrum disorder; ASP, Asperger's syndrome; CAP, Collaborative Autism Project; CHOP, Children's Hospital of Philadelphia; DSM-IV, Diagnostic and Statistical Manual of Mental Disorders-fourth edition; GWAS, genome-wide association study; NIMH, National Institute for Mental Health.
Subsequently, we searched the literature for rare genetic variants—mutations and/or CNVs—that have been found in people with ASDs to find genes that were also identified through the five GWASs (Supplementary Table 1). We searched for mutations that were identified in one or more recently published exome sequencing studies30, 31, 32, 33, 34 and/or candidate gene mutation studies in people with ASDs. In addition, we searched for overlapping CNVs that were identified in people with ASDs through a genome-wide array-based hybridization approach and/or that were ‘recurrent', in that they were observed in case studies of at least two unrelated people with ASDs. Furthermore, we searched for GWAS candidate genes that are targeted by ASD-implicated microRNAs. MicroRNAs are very small RNAs that negatively regulate the expression of multiple genes.17, 18, 19, 20 We compiled a list of microRNAs that have been repeatedly implicated in the aetiology of ASDs (Supplementary Table 2), after which we used Targetscan to identify which of the 200 ASD candidate genes are targeted and hence downregulated by ASD-implicated microRNAs. Targetscan is a web-based tool (http://www.targetscan.org/) that predicts possible gene targets of microRNAs and ranks the predicted efficacy of microRNA targeting using a calculated context score.
Finally, we searched the literature for ‘other' genetic evidence implicating the GWAS candidate genes in ASDs, including data from candidate gene association studies, gene expression studies, gene/protein function studies and genetic animal studies. In addition, we compiled a list of 33 additional ASD candidate genes that were not directly observed in the top-ranked GWAS findings but are implicated in ASDs through at least two independent lines of (strong) genetic evidence (see Supplementary Table 3).
Bioinformatics analysis
To detect enriched gene categories in the 200 top-ranked candidate genes from the five GWASs, we performed a bioinformatics analysis using the Ingenuity Pathway Analysis software package (http://www.ingenuity.com) (see Supplementary Table 4). Based on the Ingenuity Knowledge Base—which draws on information from the published literature as well as many other sources, including gene expression and gene annotation databases—genes are assigned to different categories and subcategories of functionally related genes. The Ingenuity Pathway Analysis program calculates single P-values for the enrichment of each gene category and subcategory using the right-tailed Fisher's exact test. For each category and subcategory, a multiple testing corrected P-value, calculated using the Benjamini–Hochberg correction, is also provided.
Literature analysis and identification of protein networks
Subsequently, we systematically searched the literature for the (proposed) function of all proteins derived from the ASD candidate genes using two databases: the Uniprot Protein Knowledgebase (UniProtKB) (http://www.uniprot.org/uniprot) and PubMed (http://www.ncbi.nlm.nih.gov/sites/entrez).
For each ASD candidate from the five GWASs, we first looked at the available information in UniProtKB, which in most cases already provided a general indication of the (putative) function of the gene or encoded protein in question. Based on the Ingenuity analysis, UniProtKB and the three main biological ‘themes' linking the genetic ASD findings (see Introduction), we subsequently searched PubMed using the search terms ‘Leydig', ‘steroid', ‘steroid synthesis', ‘steroidogenesis', ‘testosterone', ‘estradiol', ‘estrogen', ‘brain', ‘neuron', ‘neuronal growth', ‘neurite', ‘neurite outgrowth', ‘synapse', ‘synaptic', ‘glutamate' and ‘glutamate receptor' in combination with the name of each candidate gene (or encoded protein). Guided by the literature we found, we also searched PubMed for functional interactions between the candidate genes/encoded proteins.
Replication and validation
Using the above described SNP selection criteria, we compiled a list of top-ranked genes from GWAS 6,16 and we subsequently searched for overlapping genes between this study and the five GWASs that were considered in the aforementioned analyses.11, 12, 13, 14, 15 We also systematically searched the literature using UniProtKb and PubMed (see above) for the (proposed) function of the proteins encoded by all top-ranked genes from the sixth GWAS, and based on this information, we then tried to (putatively) place each protein in one or more of the three identified networks.
Results
Identification of candidate genes and finding corroborating evidence
As stated, six GWASs of ASDs have been published to date (Table 1). Applying the SNP selection criteria described in the Materials and methods section to data from the five GWASs11, 12, 13, 14, 15 yielded a total of 200 ASD candidate genes (Supplementary Table 1). Corroborating evidence for a role of these genes in ASD aetiology (see Materials and methods for criteria), was found through rare variant studies. We found that 11 of the 200 candidate genes from the GWASs (5.5%) were reported to contain mutations in people with ASDs (Supplementary Table 1), including findings from 5 recently published ASD exome sequencing studies.30, 31, 32, 33, 34 In addition, 93 of the 200 GWAS candidate genes (46.5%) have been observed in CNVs in people with ASDs (Supplementary Table 1). Furthermore, we found that 100 (50%) of the GWAS-identified candidate genes were targets of ASD-implicated microRNAs (see Materials and methods, Supplementary Table 1 and Supplementary Table 2). Lastly, 28 of the 200 GWAS candidate genes (14%) are additionally implicated in ASD aetiology through other genetic evidence, including data from candidate gene association studies, gene expression studies, gene/protein function studies and genetic animal studies (Supplementary Table 1). Through literature study, we also added 33 strong ASD candidate genes, including CNTNAP2,35, 36 MET37, 38 and NRXN1,39, 40 implicated in the disorder through at least two independent lines of strong genetic evidence other than GWASs (Supplementary Table 3).
Bioinformatics analysis
For the 200 top-ranked ASD GWAS genes, the Ingenuity pathway software revealed a significant enrichment (multiple testing corrected P=1.64 × 10−4) of the gene category ‘neurological disease', with 81 of the 200 genes falling into this category. Moreover, 8 of the 9 other significant categories in the top 10 of nervous system-related gene categories could be directly linked to neurite outgrowth or synaptic function (Supplementary Table 4).
Literature analysis and identification of protein networks
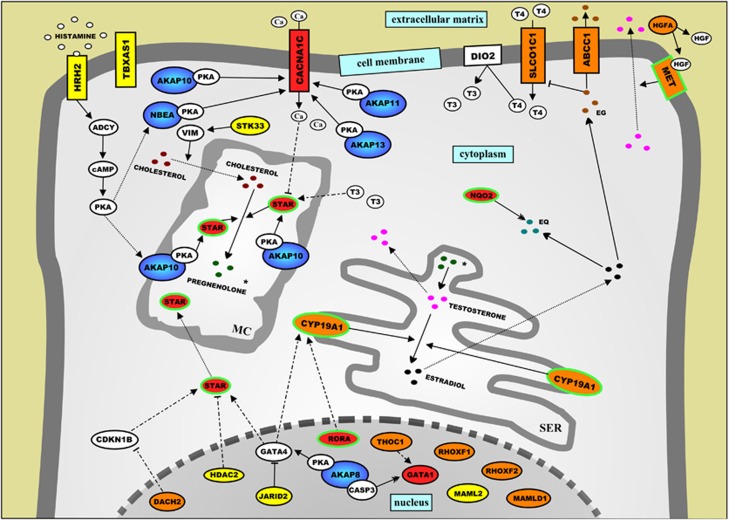
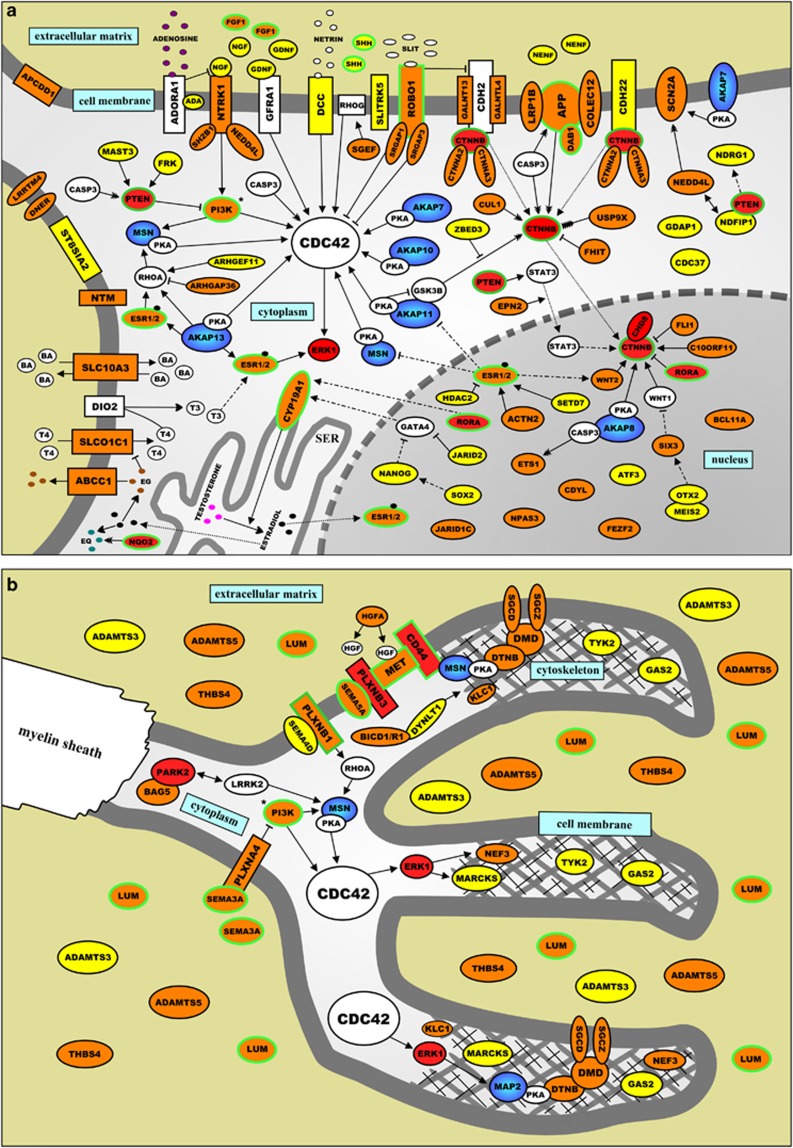
On the basis of the findings from the Ingenuity analysis and our systematic literature analysis (see Materials and methods), we found that the proteins encoded by 150 of the 233 above mentioned ASD candidate genes (64%) fit into three distinct signaling networks that correspond very well to the three aforementioned biological themes for ASDs. The first network is involved in regulating the production and metabolism of the steroid hormones testosterone and estradiol in testicular Leydig cells (Figure 1), whereas the second network relates to directed neurite outgrowth, and integrates molecular signaling cascades in the extracellular matrix, cell membrane, cytoplasm and nucleus of developing neurons (Figure 2a) with signaling cascades that modulate the cytoskeleton in and the extracellular matrix that surrounds the growth cone of these neurons (Figure 2b).
Figure 1.
Schematic representation of a protein network that is implicated in autism spectrum disorders (ASDs) through regulating the production and metabolism of the steroid hormones testosterone and estradiol in testicular Leydig cells. The proteins encoded by genes implicated in ASDs through common genetic variants—single-nucleotide polymorphisms (SNPs) from five published genome-wide association studies (GWASs) (Supplementary Table 1) and/or ASD candidate gene association studies—are indicated in yellow. The proteins encoded by genes implicated in ASD aetiology through rare genetic variants—one or more mutations and/or copy number variations (CNVs) affecting the gene—are indicated in red, whereas the proteins encoded by genes implicated in ASDs through both common and rare genetic variants are indicated in orange. In addition, all A-kinase anchor proteins (AKAPs) are dark blue and the proteins encoded by genes that have been implicated in ASD aetiology through ‘other' genetic evidence—including gene expression studies, gene/protein function studies and genetic animal studies—have a green border. In the Supplementary Information, the network is described in detail, and the current knowledge about the function of the network proteins is presented.
Figure 2.
(a) Schematic representation of a protein network that is implicated in autism spectrum disorders (ASDs) and leads to neurite outgrowth through molecular signaling cascades located in the extracellular matrix/compartment, cell membrane, cytoplasm and nucleus of developing neurons. The proteins encoded by genes implicated in ASDs through common genetic variants—single-nucleotide polymorphisms (SNPs) from five published genome-wide association studies (GWASs) (Supplementary Table 1) and/or ASD candidate gene association studies—are indicated in yellow. The proteins encoded by genes implicated in ASD aetiology through rare genetic variants—one or more mutations and/or copy number variations (CNVs) affecting the gene—are indicated in red, whereas the proteins encoded by genes implicated in ASDs through both common and rare genetic variants are indicated in orange. In addition, all A-kinase anchor proteins (AKAPs) are dark blue and the proteins encoded by genes that have been implicated in ASD aetiology through ‘other' genetic evidence—including gene expression studies, gene/protein function studies and genetic animal studies—have a green border. In the Supplementary Information, the network is described in detail, and the current knowledge about the function of the network proteins is presented. (b) Schematic representation of a protein network that is implicated in ASDs and leads to neurite outgrowth through molecular signaling cascades located in the growth cone and surrounding extracellular matrix/compartment of developing neurons. The proteins encoded by genes implicated in ASDs through common genetic variants—SNPs from five published GWASs (Supplementary Table 1) and/or ASD candidate gene association studies—are indicated in yellow. The proteins encoded by genes implicated in ASD aetiology through rare genetic variants—one or more mutations and/or copy number variations (CNVs) affecting the gene—are indicated in red, whereas the proteins encoded by genes implicated in ASDs through both common and rare genetic variants are indicated in orange. In addition, all AKAPs are dark blue and the proteins encoded by genes that have been implicated in ASD aetiology through ‘other' genetic evidence—including gene expression studies, gene/protein function studies and genetic animal studies—have a green border. In the Supplementary Information, the network is described in detail, and the current knowledge about the function of the network proteins is presented.
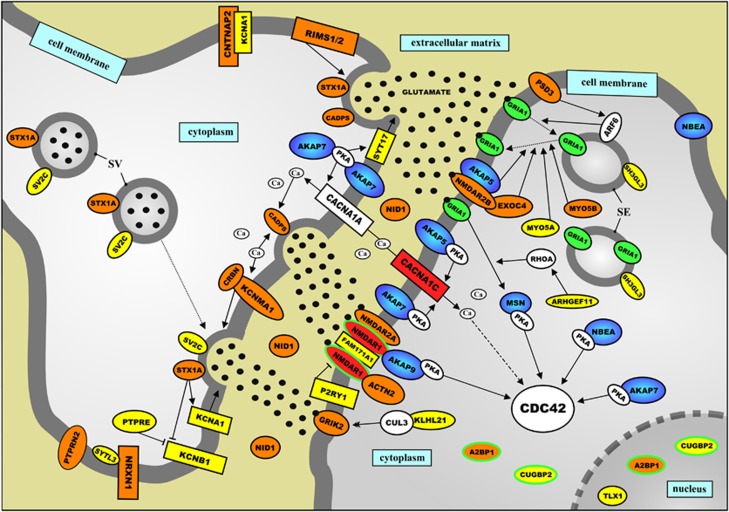
The third network regulates and modulates (glutamatergic) neurotransmission across the synapse (Figure 3). A detailed description of the evidence linking all the genes and encoded proteins in the networks shown in Figures 1, 2, 3 is provided in the Supplementary Information. Importantly, signaling cascades within all three networks are modulated and regulated by the proteins encoded by 10 ASD-implicated AKAP genes (Supplementary Table 5).
Figure 3.
Schematic representation of a protein network that is located in the neuronal synapse and implicated in autism spectrum disorders (ASDs) by its involvement in modulating glutamatergic neurotransmission. The proteins encoded by genes implicated in ASDs through common genetic variants—single-nucleotide polymorphisms (SNPs) from five published genome-wide association studies (GWASs) (Supplementary Table 1) and/or ASD candidate gene association studies—are indicated in yellow. The proteins encoded by genes implicated in ASD aetiology through rare genetic variants—one or more mutations and/or copy number variations (CNVs) affecting the gene—are indicated in red, whereas the proteins encoded by genes implicated in ASDs through both common and rare genetic variants are indicated in orange. In addition, all A-kinase anchor proteins (AKAPs) are dark blue and the proteins encoded by genes that have been implicated in ASD aetiology through ‘other' genetic evidence—including gene expression studies, gene/protein function studies and genetic animal studies—are green/have a green border. In the Supplementary Information, the network is described in detail, and the current knowledge about the function of the network proteins is presented.
Replication and validation
As indicated in the Materials and methods section, the sixth ASD GWAS by Anney et al.16 was used for validation and replication purposes, as it was entirely independent of the five other published ASD GWASs.11, 12, 13, 14, 15 A total of 15 genes from GWASs 1–5 (7.5%) were directly replicated—they were implicated through SNPs that meet the selection criteria of this study in 2 of the 6 published GWASs—and 13 of these genes were found in GWAS 6 (Supplementary Table 6). In addition, the proteins encoded by 111 of the 333 top-ranked (protein coding) genes from GWAS 6 (33%) could be placed in one or more of the three identified networks (Supplementary Table 7), which provides a strong validation of the involvement of these networks in ASD aetiology. In this respect, it is also very intriguing that an intronic SNP in NQO2, a gene that had already been implicated in ASD aetiology (Supplementary Table 3) and that encodes a protein with an important function in the identified steroidogenesis and neurite outgrowth networks (see Figures 1 and 2a, and Supplementary Information), yields genome-wide significant association (P=2 × 10−15) with ASD in GWAS 6 (Supplementary Table 7).
Potential statistical bias
An important potential bias in the analysis of the top findings from GWASs is the fact that large genes, which are often brain expressed, may be more likely found as associated with a phenotype in a GWAS as a result of chance, as more SNPs are present in these large genes.41 Indeed, the genes found among the top findings of ASD GWASs 1–5 were considerably larger than the average gene size of the human genome, with the 200 (unique) ASD candidate genes having an average size of 221 kb (Supplementary Table 8) versus an average gene size in the human genome of 27 kb.42
In order to test whether the GWAS genes included in this study were likely to constitute false positive findings, we looked for confirmation from other types of genetic studies and we performed a comparative analysis of the top ranked GWAS findings from unrelated disorders. First, a total of 111 of the 200 candidate genes from GWASs 1–5 were also implicated in ASDs through rare genetic variants—that is, mutations and/or CNVs affecting the gene in people with ASDs—and/or other genetic evidence, which includes SNPs/common genetic variants from classic candidate gene association studies, gene expression and function studies and genetic animal studies (Supplementary Table 1).
Most importantly, 42 of the 47 (unique) large genes (size >250 kb) from GWASs 1–5 (89%) and all of the 26 very large genes (size >500 kb) were implicated in ASD aetiology through at least one additional line of genetic evidence (Supplementary Tables 1 and 8). Second, arguing that if large genes are more likely found to be associated with a phenotype of a GWAS because of chance, this would be expected to be the case for GWASs of nonpsychiatric disorders as well. Therefore, we compared the results of our Ingenuity analyses of the 20 top ASD candidate genes from GWASs 1–5 with those for the top 20 candidate genes from four published GWASs for diabetes mellitus type I and Crohn's disease (Supplementary Tables 9 and 10), two polygenic disorders that are assumed not to primarily originate in the brain. This comparison showed that the neurological disease category was significantly enriched in the ASD GWAS findings (with 9 of the 20 top genes falling into this category), only contained a single gene in diabetes type I and was not enriched at all in Crohn's disease (Supplementary Table 10).
Discussion
In this study, we used both bioinformatics and extensive manual literature mining to integrate the top-ranked findings of six published GWASs of ASDs into three biological processes. In addition, we have comprehensively addressed the subject of possible gene size-based bias.
Importantly, we found that many of the genes encoding proteins in the three networks have been implicated in ASD aetiology through both common genetic variants (that is, SNPs from GWASs and classic candidate gene association studies) and rare genetic variants (that is, one or more mutations and/or CNVs affecting the gene in people with ASDs). All these proteins are indicated in orange in Figures 1, 2, 3, and this finding suggests that the often made distinction between rare and common genetic ASD variants (see above) is somewhat artificial: to a considerable extent, multifactorial and oligogenic forms of ASDs—caused by common and rare genetic variants, respectively—seem to share aetiologic pathways.
On the matter of potential gene size-based bias, and based on the corroborating evidence from other sources as well as the analysis of unrelated nonpsychiatric disorders, we would like to submit that the (vast) majority of the ASD candidate genes from the GWASs were not identified because of gene size bias and therefore represent true associations. The corroborating evidence for many of the large(r) GWAS genes comes from mutation, CNV, microRNA and other genetic data that are not subjected to potential gene size bias, as well as from the replication and validation through an entirely independent additional GWAS (GWAS 6). We have come to our conclusion about the absence of gene size-based bias despite the fact that our chosen statistical cutoff for association (P<1 × 10−4) is essentially an arbitrary threshold. It is below the commonly used threshold for genome-wide significant association (P<5 × 10−8),11, 12, 13, 14, 15, 16 but was chosen because it is often used to designate ‘suggestive' association (for example, in four published ASD GWASs11, 12, 13, 16) and because it provided us with a manageable number of genes for subsequent analyses.
Using a threshold below the genome-wide significance level (in conjunction with validation through other genetic data) is also supported by polygenic analyses showing that many signals from GWASs, although not reaching genome-wide significance individually, contribute to a given trait or disease.43, 44 That being said, we explicitly do not wish to exclude the possibility that a number of GWAS-identified ASD candidate genes are spurious findings.
The three ASD networks we describe fit very well with the three biological themes that have already emerged from diverse types of ASD studies (see above), which adds further weight to our findings. The identified networks have important implications for advancing our understanding of the molecular basis of ASDs. First, the testicular steroidogenesis network provides additional proof for both the excess of males in ASDs (see above) and the ‘extreme male brain' theory of ASDs. This theory integrates several lines of evidence and states that ASD behaviors are associated with increased fetal testosterone levels and are therefore more common in males, as well as in females with disorders characterized by an overproduction of testosterone.45, 46 Furthermore, our findings relating to the involvement of disturbed neurite outgrowth and synaptic function in the genetic aetiology of ASDs fit very well with literature reports describing aberrant structural and functional brain connectivity in people with ASDs.47 Hence, ASDs can be seen as disorders of ‘neuronal communication' that, at the level of individual neurons, could be caused by disturbances or alterations in the efficiency, direction and/or timing of neurite outgrowth (during brain development) and/or synaptic function (throughout adult life). This is further corroborated by a study reporting enrichment of several neurite outgrowth-related and ASD-implicated genes in a set of genes that were correlated with structural connectivity in the adult rat brain through gene expression patterns during brain development.48
Despite being important contributing factors, impairments of neurite outgrowth and synaptic function are not specific to the aetiology of ASDs, and our research group has previously reported that (impaired) neurite outgrowth is also important in the genetic aetiology of dyslexia49 and attention deficit hyperactivity disorder,50 two neurodevelopmental disorders that show high comorbidity and are genetically correlated with each other51 and with ASDs.52, 53, 54 Impaired synaptic function is also implicated in the aetiology of schizophrenia,55 a neurodevelopmental disorder that manifests in late adolescence or early adulthood and shares genetic factors with ASDs.56 Nevertheless, we believe that the clinical specificity of these neurodevelopmental disorders could be at least partially explained by the different functional consequences of disturbed or abnormal neurite outgrowth and/or synaptic function in the most affected brain region(s) for each disorder. The cerebellum and, more specifically, cerebellar Purkinje cells have been consistently found to be among the most affected brain regions in ASDs, with the disorders often linked to an actual loss of these cells.57, 58
As several of the ASD-implicated microRNAs show dysregulated expression in post-mortem cerebellar cortex of people with ASDs,17 and many of the proteins encoded by identified ASD candidate genes—including the ASD-implicated AKAP5, AKAP9 and AKAP11 (see below)—are highly expressed and/or have a specific function in cerebellar Purkinje cells (see Supplementary Tables 1,3,4 and 6), cerebellar dysfunction of the identified genetic networks may be very important in the aetiology of ASDs.
As already indicated above, the proteins encoded by 10 AKAP genes (AKAP5, AKAP7, AKAP8, AKAP9, AKAP10, AKAP11, AKAP13, MAP2, MSN and NBEA) constitute a common functional theme within the identified networks. The AKAPs constitute a growing protein family of currently more than 50 members that share an ability to bind protein kinase A (PKA) to its substrates, that is, the proteins that are phosphorylated and hence regulated in their activity by PKA. AKAPs are found in many subcellular locations and expressed in many tissues, including testis and brain. Apart from anchoring PKA, AKAPs can also directly bind and/or regulate many other proteins, including kinases other than PKA and phosphatases.59 As shown and described in Figures 1, 2, 3 and the Supplementary Information, ASD-implicated AKAPs are involved in signaling cascades regulating Leydig cell-based steroidogenesis, neurite outgrowth of developing neurons and glutamatergic synaptic transmission. Within the neurite outgrowth network, AKAPs also regulate the two main signaling ‘hubs', by regulating both the activity of CDC42 and (the transcription factor function of) CTNNB. Furthermore, in a study using weighted gene coexpression analysis in human neuronal progenitor cells, it was reported that AKAP7 and AKAP11 are coexpressed in a ‘coexpression module' containing genes—including previously implicated ASD candidate genes—that are likely to be involved in the onset of differentiation as well as the cessation of proliferation of these cells.60 Lin et al.61 reported that MAP2 and NBEA, two AKAP genes that have been strongly implicated in ASD aetiology (Supplementary Table 4), as well as a large number of (candidate) genes from the identified networks, including the two AKAP-regulated network ‘hubs' CDC42 and CTNNB, undergo significant changes in their expression during the transition from pluripotent stem cells to early differentiating neurons. All these data provide further evidence for the functional involvement of the identified protein networks as well as specific ASD-implicated AKAPs in early brain development.
In addition, there is ample genetic evidence for the involvement of the 10 AKAPs in the aetiology of ASDs (Supplementary Table 5). First, six AKAP genes (AKAP7, AKAP10, AKAP11, MAP2, MSN and NBEA) are implicated to have a role in these disorders through SNP associations that have been detected in the published GWASs11, 16 and/or a recent candidate gene association study.62 In this respect, rs5918959, a SNP located 66 kb downstream of the moesin (MSN) gene, showed a genome-wide significant P-value of 1.22 × 10−10 for association with ASDs in the combined discovery samples from the GWAS by Wang et al.11 Furthermore, eight AKAP genes (AKAP5, AKAP8, AKAP9, AKAP10, AKAP13, MAP2, MSN and NBEA) have been found in CNVs in people with ASDs63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73 and NBEA was found to be disrupted by a chromosomal translocation in a male with autism.73 AKAP9 is also present in a locus linked to autism.74
Moreover, five AKAP genes are targets of ASD-implicated microRNAs (AKAP5, AKAP8, AKAP11, AKAP13 and NBEA) (Supplementary Tables 2 and 5) or expressed differentially in lymphoblastoid cells of people with ASDs (AKAP7).20 Expression studies also implicate MSN and MAP2 with increased75 and decreased76 expression, respectively, found in the post-mortem brain of autistic people. In addition, exposure to low levels of polychlorinated biphenyls in rats leads to increased cerebellar expression of Akap11 (ref. 77) and autism-like behavior.78 Recently, it was also reported that Nbea+/− mice—that is, mice that are haploinsufficient for the Nbea gene—exhibit several ASD-like features, including changes in social behaviors as well as spatial learning and memory.79
An eleventh AKAP gene is implicated in ASDs, in that rs10038113, a SNP yielding genetic association with ASDs in the GWASs by Wang et al.11(P=7.40 × 10−8) and Ma et al.12(P=3.40 × 10−6), maps only 7 kb upstream of the moesin pseudogene 1 (MSNP1) gene (http://www.ensembl.org). Although Wang et al.11 speculate that CDH10 and/or CDH9—two cadherin genes that are located 1257 kb upstream and 978 kb downstream of rs10038113, respectively—may be implicated in this association, MSNP1 is the only gene in a linkage disequilibrium block of ∼100 kb in size that contains rs10038113 as well as several SNPs yielding genome-wide significant associations with ASDs for the combined discovery and replication samples in the GWAS they published.11 Interestingly, rs10038113 and rs1896731, a SNP present in the same linkage disequilibrium block and mapping 11 kb upstream of MSNP1, were recently also found to be nominally associated with autistic traits,80 whereas rs4307059, a SNP from the linkage disequilibrium block that is located 56 kb downstream of MSNP1, associates with ASD-like social communication difficulties in the general population.81 Moreover and most importantly, MSNP1 shows a very high degree of sequence homology to the MSN gene82 and was recently demonstrated to regulate the expression of MSN in the human cerebral cortex through expression of its antisense strand.83
Additional functional evidence points to a role for the AKAP genes (and the encoded proteins) in the aetiology of ASDs. AKAP10,84 AKAP13,85 and MSN86 are involved in innate immunity, known to be disturbed in at least a subset of people with ASDs,87, 88 through regulation of the activity of Toll-like receptors. AKAP5 and AKAP8 are both involved in the biosynthesis of the circadian hormone melatonin,89 which is markedly disturbed in people with ASDs.90 Thus, it is interesting that both anti-inflammatory treatments and melatonin have been proposed as possible new treatments of ASDs.91 Neuronal AKAPs also play an important role in the serotonin-induced release of neurotransmitters.92 AKAP5 is involved in the recycling of postsynaptic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type glutamate receptors (Figure 3) and the regulation of long-term potentiation and depression through glutamate-bound N-Methyl-𝒟-aspartate (NMDA)-type receptors.93, 94 AKAP7 regulates signaling cascades downstream of D1-like dopamine receptors.95, 96 Hence, it is interesting and intriguing that risperidone, one of only two drugs approved by the Food and Drug Administration (FDA) for the treatment of ASDs (http://www.fda.gov/), functions both as a modulator of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid- and NMDA-type glutamate receptor expression97 and regulates the serotonin-induced release of glutamate and dopamine.98, 99
Risperidone is also involved in upregulating the brain expression of microtubule-associated protein 2 (MAP2)100—an AKAP that is implicated in neurite outgrowth (Figure 2b)—as well as CTNNB and GSK3B,101 the two proteins that are regulated by AKAP8 and AKAP11, respectively (Figure 2a). RHOA, a protein that is positively regulated by AKAP13 and has an important role in the neurite outgrowth and synaptic networks (Figures 2a, Figures 2b and Figures 3), also modulates the effects of risperidone treatment for autistic symptoms.102
In summary, 8 of the 10 ASD-implicated AKAPs that we identified in our analyses—AKAP5, AKAP7, AKAP8, AKAP10, AKAP11, AKAP13, MAP2 and MSN—regulate two ASD-implicated biological processes (innate immunity and melatonin synthesis) for which new treatments have been proposed and/or are involved in directly or indirectly modulating the downstream effects of risperidone treatment for ASDs. The effects of existing ASD drugs on these and other AKAPs should be investigated in future studies. In this respect, several AKAP-dependent protein–protein interactions and the AKAPs themselves are also increasingly considered and investigated as potential drug targets,59 and several molecules that specifically inhibit the binding of PKA to its AKAP-binding site have been patented (http://www.wipo.int/patentscope/search/en/detail.jsf?docId=EP14889042&recNum=2&office=&queryString=EN_ALLTXT%3A%28AKAP%29&prevFilter=&sortOption=Relevance&maxRec=255). One of these molecules is Ht31, a peptide that is derived from AKAP13. Interestingly, administration of Ht31 to rats effectively blocks the ‘masculinization' of a part of their brain,103 which suggests an additional and more direct role of AKAPs in mediating the masculinizing effects of testosterone—or rather, estradiol, which is converted from testosterone in the brain104, 105(see Figure 2a)—on brain development. Furthermore, transgenic mice that conditionally express Ht31 in their hippocampus show spatial learning and memory impairments106 that resemble cognitive deficits in ASDs.107, 108 For these reasons, we think that the ASD-implicated AKAPs and/or protein–protein interactions dependent on these AKAPs are excellent targets for developing novel ASD drugs.
Conclusions
In this study, we used bioinformatics and literature analyses to investigate and integrate the top-ranked findings of five published GWASs of ASDs. We were able to place the proteins encoded by 117 of the 200 ASD candidate genes from these GWASs and 33 additional strong ASD candidates into three signaling networks regulating steroidogenesis in Leydig cells, neurite outgrowth and glutamatergic synaptic function. Importantly, the sixth published GWAS of ASDs strongly replicated and validated the identified networks. Several members of the AKAP family functionally integrate signaling cascades within and between the three identified networks. Thus, these AKAPs—that are themselves strongly genetically and functionally linked to ASDs—seem ideally suited to be studied further as possible ‘druggable' targets for the treatment of ASDs.
There is also a strong overlap between rare and common ASD-implicated genetic variants, which implies that multifactorial and oligogenic forms of ASDs partially share aetiologic pathways. Furthermore, we have provided compelling evidence against gene size-based bias within the ASD GWAS data.
Acknowledgments
The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under grant agreement n°278948. We thank the families who made all the genetic studies of ASDs possible and the many investigators whose work drives the ASD genetics field forward. In particular, we are grateful to K Wang, H Hakonarson, LA Weiss and their collaborators for providing us with the detailed results from their GWASs.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children: confirmation of high prevalence. Am J Psychiatry. 2005;162:1133–1141. doi: 10.1176/appi.ajp.162.6.1133. [DOI] [PubMed] [Google Scholar]
- Baird G, Simonoff E, Pickles A, Chandler S, Loucas T, Meldrum D, et al. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP) Lancet. 2006;368:210–215. doi: 10.1016/S0140-6736(06)69041-7. [DOI] [PubMed] [Google Scholar]
- Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr Res. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, Pauls D. Autism. Lancet. 2003;362:1133–1141. doi: 10.1016/S0140-6736(03)14471-6. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, Lord C, Bailey A, Schultz RT, Klin A. Autism and pervasive developmental disorders. J Child Psychol Psychiatry. 2004;45:135–170. doi: 10.1046/j.0021-9630.2003.00317.x. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag CM, Staal W, Klauck SM, Duketis E, Waltes R. Genetics of autistic disorders: review and clinical implications. Eur Child Adolesc Psychiatry. 2010;19:169–178. doi: 10.1007/s00787-009-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Salyakina D, Jaworski JM, Konidari I, Whitehead PL, Andersen AN, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14. Ann Hum Genet. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Brune CW, West K, O'Connor A, Hilton G, et al. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyakina D, Ma DQ, Jaworski JM, Konidari I, Whitehead PL, Henson R, et al. Variants in several genomic regions associated with asperger disorder. Autism Res. 2010;3:303–310. doi: 10.1002/aur.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussman JP, Chung RH, Griswold AJ, Jaworski JM, Salyakina D, Ma D, et al. A noise-reduction GWAS analysis implicates altered regulation of neurite outgrowth and guidance in autism. Mol Autism. 2011;2:1. doi: 10.1186/2040-2392-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- Talebizadeh Z, Butler MG, Theodoro MF. Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Res. 2008;1:240–250. doi: 10.1002/aur.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarachana T, Zhou R, Chen G, Manji HK, Hu VW. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med. 2010;2:23. doi: 10.1186/gm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahramani Seno MM, Hu P, Gwadry FG, Pinto D, Marshall CR, Casallo G, et al. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 2011;1380:85–97. doi: 10.1016/j.brainres.2010.09.046. [DOI] [PubMed] [Google Scholar]
- Chakrabarti B, Dudbridge F, Kent L, Wheelwright S, Hill-Cawthorne G, Allison C, et al. Genes related to sex steroids, neural growth, and social-emotional behavior are associated with autistic traits, empathy, and Asperger syndrome. Autism Res. 2009;2:157–177. doi: 10.1002/aur.80. [DOI] [PubMed] [Google Scholar]
- Hu VW, Nguyen A, Kim KS, Steinberg ME, Sarachana T, Scully MA, et al. Gene expression profiling of lymphoblasts from autistic and nonaffected sib pairs: altered pathways in neuronal development and steroid biosynthesis. PLoS One. 2009;4:e5775. doi: 10.1371/journal.pone.0005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A, Gauthier J, Hamdan FF, Lafrenière RG, Yang Y, Henrion E, et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2011;16:867–880. doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuscó I, Medrano A, Gener B, Vilardell M, Gallastegui F, Villa O, et al. Autism-specific copy number variants further implicate the phosphatidylinositol signaling pathway and the glutamatergic synapse in the etiology of the disorder. Hum Mol Genet. 2009;18:1795–1804. doi: 10.1093/hmg/ddp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherman A, Wang R, Avramopoulos D. Orientation, distance, regulation and function of neighbouring genes. Hum Genomics. 2009;3:143–156. doi: 10.1186/1479-7364-3-2-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GC, Doke TF, Ashburner J, Wood NW, Frackowiak RS. Normal variation in fronto-occipital circuitry and cerebellar structure with an autism-associated polymorphism of CNTNAP2. Neuroimage. 2010;53:1030–1042. doi: 10.1016/j.neuroimage.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2:56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, Warren D, Sutcliffe JS, Lee EB, Levitt P. Association of MET with social and communication phenotypes in individuals with autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:438–446. doi: 10.1002/ajmg.b.30998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, D'Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, et al. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann Neurol. 2007;62:243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, et al. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T, Read AP.Human Molecular Genetics Garland Publishing: New York, USA; 2004. p253 [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Decandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auyeung B, Taylor K, Hackett G, Baron-Cohen S. Foetal testosterone and autistic traits in 18 to 24-month-old children. Mol Autism. 2010;1:11. doi: 10.1186/2040-2392-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Guest PC, Rahmoune H, Wang L, Levin Y, Ingudomnukul E, et al. Sex-specific serum biomarker patterns in adults with Asperger's syndrome. Mol Psychiatry. 2011;16:1213–1220. doi: 10.1038/mp.2010.102. [DOI] [PubMed] [Google Scholar]
- Schipul SE, Keller TA, Just MA. Inter-regional brain communication and its disturbance in autism. Front Syst Neurosci. 2011;5:10. doi: 10.3389/fnsys.2011.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L, Pavlidis P. Relationships between gene expression and brain wiring in the adult rodent brain. PLoS Comput Biol. 2011;7:e1001049. doi: 10.1371/journal.pcbi.1001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelmans G, Buitelaar JK, Pauls DL, Franke B. A theoretical molecular network for dyslexia: integrating available genetic findings. Mol Psychiatry. 2011;16:365–382. doi: 10.1038/mp.2010.105. [DOI] [PubMed] [Google Scholar]
- Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated genome-wide association study findings: identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am J Psychiatry. 2011;168:365–377. doi: 10.1176/appi.ajp.2010.10070948. [DOI] [PubMed] [Google Scholar]
- Chadwick O, Taylor E, Taylor A, Heptinstall E, Danckaerts M. Hyperactivity and reading disability: a longitudinal study of the nature of the association. J Child Psychol Psychiatry. 1999;40:1039–1050. [PubMed] [Google Scholar]
- Jones CR, Happé F, Golden H, Marsden AJ, Tregay J, Simonoff E, et al. Reading and arithmetic in adolescents with autism spectrum disorders: peaks and dips in attainment. Neuropsychology. 2009;23:718–728. doi: 10.1037/a0016360. [DOI] [PubMed] [Google Scholar]
- Miniscalco C, Dahlgren SA. Basic reading skills in Swedish children with late developing language and with or without autism spectrum disorder or ADHD. Res Dev Disabil. 2010;31:1054–1061. doi: 10.1016/j.ridd.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Rommelse NN, Franke B, Geurts HM, Hartman CA, Buitelaar JK. Shared heritability of attention-deficit/hyperactivity disorder and autism spectrum disorder. Eur Child Adolesc Psychiatry. 2010;19:281–295. doi: 10.1007/s00787-010-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, Vanleeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingason A, Kirov G, Giegling I, Hansen T, Isles AR, Jakobsen KD, et al. Maternally derived microduplications at 15q11-q13: implication of imprinted genes in psychotic illness. Am J Psychiatry. 2011;168:408–417. doi: 10.1176/appi.ajp.2010.09111660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ER, Kemper TL, Rosene DL, Bauman ML, Blatt GJ. Density of cerebellar basket and stellate cells in autism: evidence for a late developmental loss of Purkinje cells. J Neurosci Res. 2009;87:2245–2254. doi: 10.1002/jnr.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LA, Goldowitz D, Mittleman G. Repetitive behavior and increased activity in mice with Purkinje cell loss: a model for understanding the role of cerebellar pathology in autism. Eur J Neurosci. 2010;31:544–555. doi: 10.1111/j.1460-9568.2009.07073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skroblin P, Grossmann S, Schafer G, Rosenthal W, Klussmann E. Mechanisms of protein kinase a anchoring. Int Rev Cell Mol Biol. 2010;283:235–330. doi: 10.1016/S1937-6448(10)83005-9. [DOI] [PubMed] [Google Scholar]
- Konopka G, Wexler E, Rosen E, Mukamel Z, Osborn GE, Chen L, et al. Modeling the functional genomics of autism using human neurons. Mol Psychiatry. 2012;17:202–214. doi: 10.1038/mp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R, et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet. 2012;131:565–579. doi: 10.1007/s00439-011-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung RH, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder associated pathways. Hum Mol Genet. 2012;21:3513–3523. doi: 10.1093/hmg/dds164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Merico D, Thiruvahindrapuram B, Wei J, Lionel AC, Sato D, et al. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda) 2012;2:1665–1685. doi: 10.1534/g3.112.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Ballif BC, Torchia BS, Sahoo T, Ravnan JB, Schultz R, et al. Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet Med. 2010;12:694–702. doi: 10.1097/GIM.0b013e3181f0c5f3. [DOI] [PubMed] [Google Scholar]
- Bremer A, Giacobini M, Eriksson M, Gustavsson P, Nordin V, Fernell E, et al. Copy number variation characteristics in subpopulations of patients with autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet. 2011;156:115–124. doi: 10.1002/ajmg.b.31142. [DOI] [PubMed] [Google Scholar]
- Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, Friedman EM, et al. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am J Hum Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya N, Colak D, Albakheet A, Al-Owain M, Abu-Dheim N, Al-Younes B, et al. A novel X-linked disorder with developmental delay and autistic features. Ann Neurol. 2012;71:498–508. doi: 10.1002/ana.22673. [DOI] [PubMed] [Google Scholar]
- Celestino-Soper PB, Shaw CA, Sanders SJ, Li J, Murtha MT, Ercan-Sencicek AG, et al. Use of array CGH to detect exonic copy number variants throughout the genome in autism families detects a novel deletion in TMLHE. Hum Mol Genet. 2011;20:4360–4370. doi: 10.1093/hmg/ddr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castermans D, Volders K, Crepel A, Backx L, De Vos R, Freson K, et al. SCAMP5, NBEA and AMISYN: three candidate genes for autism involved in secretion of large dense-core vesicles. Hum Mol Genet. 2010;19:1368–1378. doi: 10.1093/hmg/ddq013. [DOI] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, et al. An autosomal genomic screen for autism. Collaborative linkage study of autism. Am J Med Genet. 1999;88:609–615. doi: 10.1002/(sici)1096-8628(19991215)88:6<609::aid-ajmg7>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaetova-Ladinska EB, Arnold H, Jaros E, Perry R, Perry E. Depletion of MAP2 expression and laminar cytoarchitectonic changes in dorsolateral prefrontal cortex in adult autistic individuals. Neuropathol Appl Neurobiol. 2004;30:615–623. doi: 10.1111/j.1365-2990.2004.00574.x. [DOI] [PubMed] [Google Scholar]
- Royland JE, Kodavanti PR. Gene expression profiles following exposure to a developmental neurotoxicant, Aroclor 1254: pathway analysis for possible mode(s) of action. Toxicol Appl Pharmacol. 2008;231:179–196. doi: 10.1016/j.taap.2008.04.023. [DOI] [PubMed] [Google Scholar]
- Jolous-Jamshidi B, Cromwell HC, McFarland AM, Meserve LA. Perinatal exposure to polychlorinated biphenyls alters social behaviors in rats. Toxicol Lett. 2010;199:136–143. doi: 10.1016/j.toxlet.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuytens K, Gantois I, Stijnen P, Iscru E, Laeremans A, Serneels L, et al. Haploinsufficiency of the autism candidate gene Neurobeachin induces autism-like behaviors and affects cellular and molecular processes of synaptic plasticity in mice. Neurobiol Dis. 2012;51:144–151. doi: 10.1016/j.nbd.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Hu VW, Addington A, Hyman A. Novel autism subtype-dependent genetic variants are revealed by quantitative trait and subphenotype association analyses of published GWAS data. PLoS One. 2011;6:e19067. doi: 10.1371/journal.pone.0019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pourcain B, Wang K, Glessner JT, Golding J, Steer C, Ring SM, et al. Association between a high-risk autism locus on 5p14 and social communication spectrum phenotypes in the general population. Am J Psychiatry. 2010;167:1364–1372. doi: 10.1176/appi.ajp.2010.09121789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilgenbus KK, Hsieh CL, Lankes WT, Milatovich A, Francke U, Furthmayr H. Structure and localization on the X chromosome of the gene coding for the human filopodial protein moesin (MSN) Genomics. 1994;19:326–333. doi: 10.1006/geno.1994.1065. [DOI] [PubMed] [Google Scholar]
- Kerin T, Ramanathan A, Rivas K, Grepo N, Coetzee GA, Campbell DB. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci Transl Med. 2012;4:128ra40. doi: 10.1126/scitranslmed.3003479. [DOI] [PubMed] [Google Scholar]
- Kim SH, Serezani CH, Okunishi K, Zaslona Z, Aronoff DM, Peters-Golden M. Distinct protein kinase a anchoring proteins direct prostaglandin E2 modulation of toll-like receptor signaling in alveolar macrophages. J Biol Chem. 2011;286:8875–8883. doi: 10.1074/jbc.M110.187815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibolet O, Giallourakis C, Rosenberg I, Mueller T, Xavier RJ, Podolsky DK. AKAP13, a RhoAGTPase-specific guanine exchange factor, is a novel regulator of TLR2 signaling. J Biol Chem. 2007;282:35308–35317. doi: 10.1074/jbc.M704426200. [DOI] [PubMed] [Google Scholar]
- Zawawi KH, Kantarci A, Schulze-Späte U, Fujita T, Batista EL, Jr, Amar S, et al. Moesin-induced signaling in response to lipopolysaccharide in macrophages. J Periodontal Res. 2010;45:589–601. doi: 10.1111/j.1600-0765.2010.01271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyonouchi H, Geng L, Cushing-Ruby A, Quraishi H. Impact of innate immunity in a subset of children with autism spectrum disorders: a case control study. J Neuroinflammation. 2008;5:52. doi: 10.1186/1742-2094-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Samra D, Agrawal S. Adaptive and innate immune responses in autism: rationale for therapeutic use of intravenous immunoglobulin. J Clin Immunol. 2010;30:S90–S96. doi: 10.1007/s10875-010-9402-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Korf HW. Distribution of regulatory subunits of protein kinase A and A kinase anchor proteins (AKAP 95, 150) in rat pinealocytes. Cell Tissue Res. 2002;310:331–338. doi: 10.1007/s00441-002-0633-9. [DOI] [PubMed] [Google Scholar]
- Melke J, Goubran Botros H, Chaste P, Betancur C, Nygren G, Anckarsäter H, et al. Abnormal melatonin synthesis in autism spectrum disorders. Mol Psychiatry. 2008;13:90–98. doi: 10.1038/sj.mp.4002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol DA. Novel and emerging treatments for autism spectrum disorders: a systematic review. Ann Clin Psychiatry. 2009;21:213–236. [PubMed] [Google Scholar]
- Liu J, Hu JY, Schacher S, Schwartz JH. The two regulatory subunits of aplysia cAMP-dependent protein kinase mediate distinct functions in producing synaptic plasticity. J Neurosci. 2004;24:2465–2474. doi: 10.1523/JNEUROSCI.4331-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Génin A, French P, Doyère V, Davis S, Errington ML, Maroun M, et al. LTP but not seizure is associated with up-regulation of AKAP-150. Eur J Neurosci. 2003;17:331–340. doi: 10.1046/j.1460-9568.2003.02462.x. [DOI] [PubMed] [Google Scholar]
- Smith KE, Gibson ES, Dell'Acqua ML. cAMP-dependent protein kinase postsynaptic localization regulated by NMDA receptor activation through translocation of an A-kinase anchoring protein scaffold protein. J Neurosci. 2006;26:2391–2402. doi: 10.1523/JNEUROSCI.3092-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell AR, Tibbs VC, Westenbroek RE, Scheuer T, Catterall WA. Dopaminergic modulation of voltage-gated Na+ current in rat hippocampal neurons requires anchoring of cAMP-dependent protein kinase. J Neurosci. 1999;19:RC21. doi: 10.1523/JNEUROSCI.19-17-j0003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell AR, Tibbs VC, Yu FH, Murphy BJ, Sharp EM, Qu Y, et al. Molecular mechanism of convergent regulation of brain Na(+) channels by protein kinase C and protein kinase A anchored to AKAP-15. Mol Cell Neurosci. 2002;21:63–80. doi: 10.1006/mcne.2002.1162. [DOI] [PubMed] [Google Scholar]
- Tarazi FI, Baldessarini RJ, Kula NS, Zhang K. Long-term effects of olanzapine, risperidone, and quetiapine on ionotropic glutamate receptor types: implications for antipsychotic drug treatment. J Pharmacol Exp Ther. 2003;306:1145–1151. doi: 10.1124/jpet.103.052597. [DOI] [PubMed] [Google Scholar]
- Pehek EA, Nocjar C, Roth BL, Byrd TA, Mabrouk OS. Evidence for the preferential involvement of 5-HT2A serotonin receptors in stress- and drug-induced dopamine release in the rat medial prefrontal cortex. Neuropsychopharmacology. 2006;31:265–277. doi: 10.1038/sj.npp.1300819. [DOI] [PubMed] [Google Scholar]
- Kuroki T, Nagao N, Nakahara T. Neuropharmacology of second-generation antipsychotic drugs: a validity of the serotonin-dopamine hypothesis. Prog Brain Res. 2008;172:199–212. doi: 10.1016/S0079-6123(08)00910-2. [DOI] [PubMed] [Google Scholar]
- Law AJ, Hutchinson LJ, Burnet PW, Harrison PJ. Antipsychotics increase microtubule-associated protein 2 mRNA but not spinophilin mRNA in rat hippocampus and cortex. J Neurosci Res. 2004;76:376–382. doi: 10.1002/jnr.20092. [DOI] [PubMed] [Google Scholar]
- Alimohamad H, Rajakumar N, Seah YH, Rushlow W. Antipsychotics alter the protein expression levels of beta-catenin and GSK-3 in the rat medial prefrontal cortex and striatum. Biol Psychiatry. 2005;57:533–542. doi: 10.1016/j.biopsych.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Quincozes-Santos A, Abib RT, Leite MC, Bobermin D, Bambini-Junior V, Gonçalves CA, et al. Effect of the atypical neuroleptic risperidone on morphology and S100B secretion in C6 astroglial lineage cells. Mol Cell Biochem. 2008;314:59–63. doi: 10.1007/s11010-008-9765-x. [DOI] [PubMed] [Google Scholar]
- Lenz KM, Wright CL, Martin RC, McCarthy MM. Prostaglandin E regulates AMPA receptor phosphorylation and promotes membrane insertion in preoptic area neurons and glia during sexual differentiation. PLoS One. 2011;6:e18500. doi: 10.1371/journal.pone.0018500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean SL, McCarthy MM. Steroids, sex and the cerebellar cortex: implications for human disease. Cerebellum. 2008;7:38–47. doi: 10.1007/s12311-008-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel-Wagner B. Neurosteroid biosynthesis in the human brain and its clinical implications. Ann NY Acad Sci. 2003;1007:64–78. doi: 10.1196/annals.1286.007. [DOI] [PubMed] [Google Scholar]
- Nie T, McDonough CB, Huang T, Nguyen PV, Abel T. Genetic disruption of protein kinase A anchoring reveals a role for compartmentalized kinase signaling in theta-burst long-term potentiation and spatial memory. J Neurosci. 2007;27:10278–10288. doi: 10.1523/JNEUROSCI.1602-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheh MA, Millonig JH, Roselli LM, Ming X, Jacobsen E, Kamdar S, et al. En2 knockout mice display neurobehavioral and neurochemical alterations relevant to autism spectrum disorder. Brain Res. 2006;1116:166–176. doi: 10.1016/j.brainres.2006.07.086. [DOI] [PubMed] [Google Scholar]
- Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.