Abstract
Quantification of plasma HIV-1 RNA below the limit of FDA-approved assays by a single copy quantitative PCR assays (SCA) has provided significant insights into HIV-1 persistence despite potent antiretroviral therapy as well as a means to assess the impact of therapeutic strategies, such as treatment intensification, on residual viremia. In this review, we discuss insights gained from plasma HIV-1 RNA SCA and highlight the need for additional assays to characterize better the cellular and tissue reservoirs of HIV-1. Accurate, reproducible, and sensitive assays to quantify HIV-1 reservoirs, before and after therapeutic interventions, are essential tools in the quest for a cure of HIV-1 infection.
Keywords: Single copy assay, Residual viremia, HIV reservoirs, HIV quantification, Ultrasensitive PCR, HIV treatment intensification
Introduction
Current antiretroviral therapy (ART) inhibits HIV-1 replication and markedly lowers plasma viremia (HIV-1 RNA), often below the limit of detection (LOD) of FDA-approved assays (typically 50 copies/mL of plasma). Despite this profound antiviral effect, low level residual viremia persists in most patients on current antiretroviral therapy [1, 2••]. To gain further insight into residual viremia, a number of PCR-based assays have been developed with enhanced sensitivity for HIV-1 RNA. Initially, these assays achieved a LOD of <10 copies/mL through “home brew” methods [1] or through modifications to the commercially available Amplicor HIV-1 Monitor test to accommodate larger plasma volumes [3–6]. Subsequently, the sensitivity of quantitative PCR (qPCR) assays was further improved such that detection of a single copy of HIV-1 RNA in plasma was possible [7••]. Application of this single copy plasma HIV-1 RNA assay (HIV-1 RNA SCA) has provided critical new insights into HIV-1 persistence.
In this article, we review the methods behind HIV-1 RNA SCA and the insights made possible through its uses. Expected advances in single copy assays will then be highlighted along with their anticipated uses to further characterize HIV-1 persistence.
Single Copy HIV-1 RNA Detection
The single copy assay was designed to detect as few as one copy of HIV-1 RNA in 7.5 mL of plasma. Plasma is first isolated from whole blood by two sequential rounds of centrifugation (400 g×10 min and 1350 g×15 min), which are critical steps for removing cells and substances that interfere with PCR. Plasma is then spiked with an internal control virus derived from the avian Rous sarcoma virus (RSV) SR-A strain (RCAS) to assess virion recovery. Virions are pelleted by ultracentrifugation at 170,000 g for 30 min, digested with proteinase K, and homogenized with guanidinium isothiocyanate supplemented with glycogen. HIV-1 RNA is precipitated with isopropanol, washed with ethanol, and resuspended in Tris-HCl supplemented with dithiothreitol and an RNase inhibitor. HIV-1 RNA and RCAS RNA are converted to cDNA and quantified by real-time PCR utilizing external standard curves generated from serial dilutions of RNA transcripts synthesized in vitro. Positive and negative control plasma standards, containing 5 copies/mL and 0 copies/mL of HIV-1 RNA, respectively, are included in each assay to assess run to run variation and to exclude false-positive results due to contamination. The RCAS internal standard is used to evaluate the efficiency of virion recovery. As such, the development of the HIV-1 RNA SCA allowed quantification of plasma virus below that which was achieved previously, providing important insights into viremia decay and low-level residual viremia in patients on suppressive antiretroviral therapy.
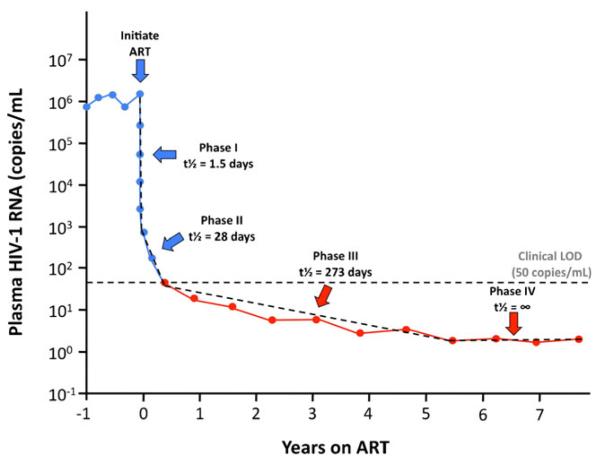
Viremia Decay Kinetics and Residual Viremia (Fig. 1)
Fig. 1.
Decay dynamics of plasma HIV-1 RNA during ART. Upon initiation of ART, viremia decays in multiple overlapping phases, which reflects the turnover of cells infected prior to ART with different half-lives. The first phase of decay has a half-life of approximately 1.5 days and represents the turnover of free virus and productively infected T cells [8, 9, 10••]. The second phase of decay, with a half-life of approximately 28 days, represents the attrition of cells more resistant to HIV cytopathicity, such as partially activated T cells and cells of the monocyte-macrophage lineage [11, 12••]. The third phase of decay, which has a half-life of approximately 273 days, levels off to a stable set point that represents a fourth phase showing no evidence of further decay [2••]. Viremia persists at this stable set point for at least 7 years following the initiation of ART and reflects the remarkable stability of the long-lived cellular reservoirs that maintain residual viremia [16••]. Blue = above clinical limit of detection (LOD). Red = below clinical LOD (ie, detectable by SCA). Dotted lines = theoretical decay slopes
Following the advent of potent antiretroviral therapy, plasma HIV-1 RNA was initially shown to undergo biphasic decay kinetics. The first phase of decay reflects the clearance of free virus (t½=6 h) and short-lived productively infected cells (t½=1–2 days) [8, 9, 10••]. The second phase of decay (t½=1–4 weeks) is thought to reflect the loss of infected cells that are more resistant to HIV-1 cytopathicity, such as partially activated T cells or macrophages [11, 12••]. Based on the trajectory of the second phase, some believed decay would continue until HIV-1 was eradicated. These hopes were shattered by the discovery of long-lived, inducible reservoirs of HIV-1 in latently infected resting CD4+ memory cells with an estimated half-life of 44 months [13, 14, 15••].
Studies using HIV-1 RNA SCA revealed a third phase of decay (t½=39 weeks) that continues below the LOD of commercial assays [2••, 16••], followed by a fourth phase in which viremia does not decay further (t½=infinity) for at least 7 years of suppressive ART [16••]. Figure 1 provides an overview of the four phases of viremia decay after initiation of antiretroviral therapy, the third and fourth of which were by HIV-1 RNA SCA.
Characteristics and Predictors of Residual Viremia on ART
In addition to decay dynamics, HIV-1 RNA SCA has revealed other notable properties of residual viremia. The set point to which residual viremia decays correlates with several parameters including 1) the level of pre-ART viremia [2••, 17], 2) the length of time between infection and treatment [5, 17, 18], 3) the duration of treatment [19, 20], and possibly 4) age [19], but not with CD4+ T-cell count or ART regimen [2••]. Immune activation does not appear to correlate with the residual viremia set point [20], but this issue has yet to be resolved fully. Together parameters that correlate with the residual viremia set point likely reflect the infection frequency of long-lived cells; and subsequently, the rate of virus release from these reservoirs during suppressive ART. This hypothesis is consistent with a recent study that showed the frequency of infected CD4+ T cells, as measured by HIV-1 proviral DNA, correlated with the level of residual viremia [21•].
SCA Utility in Optimizing ART and Assessing Simplified Regimens
Residual viremia is thought to arise from cells that were infected before ART and that cannot be eliminated by ART alone. Such long-lived cells may contribute to residual viremia by 1) the occasional activation of latently infected cells to produce virus and/or 2) the continuous or intermittent production of virus by cells resistant to HIV-1 cytopathicity. Whether HIV-1 continues to undergo complete replicative cycles with infection of new cells during suppressive ART has been the subject of much debate. In this regard, sensitive HIV-1 RNA assays, including HIV-1 RNA SCA, have been used to compare the efficacy of different treatment regimens and to assess the impact of treatment intensification with an additional, potent antiretroviral agent on residual viremia.
The efficacy of different ART regimens in suppressing viremia has been studied extensively. An initial report suggested that tenofovir (TDF) was superior to stavudine (d4T) when administered in combination with efavirenz (EFV) and lamivudine (3TC) [22]. In the TFV arm, the mean post-treatment HIV-1 RNA was lower (3.8 vs 4.1 copies/mL) and the fraction of patients with undetectable viremia was higher (47% vs 29%) than in the d4T arm. In a more recent retrospective study, nevirapine (NVP) was claimed to be superior to EFV in suppressing residual viremia below 1 copy/mL (81% vs 56%) when combined with emtricitabine (FTC) and TDF [23]. However, randomized groups were not compared in this analysis. A large randomized clinical trial of stavudine, lamivudine, and either nelfinavir or lopinavir/ritonavir found residual viremia was not different between treatment arms [2••]. The latter finding is consistent with maximal suppression of residual viremia independent of suppressive regimen.
Aside from achieving optimal virus suppression, it is often desirable to change or simplify treatment regimens to reduce pill burden, treatment cost, drug toxicities, and drug-drug interactions. Two recent studies investigated regimen simplification of standard ART (typically two nucleotide reverse transcriptase inhibitors [NRTI] and one ritonavir-boosted protease inhibitor [PI] to a single ritonavir-boosted PI alone). When continuous standard ART was compared to regimen simplification with ritonavir-boosted lopinavir (LPV/r), 81% (17 of 21) of patients’ viremia remained suppressed with no change in residual viremia among those with suppressed viremia on the simplified regimen [24]. Similarly, when a ritonavir-boosted atazanavir regimen (ATV/r) was initiated and NRTI were discontinued, 88% (30 of 34) of patients’ viremia remained suppressed with no changes in the level of residual viremia among those with continued suppression of viremia [25, 26]. In the latter study, viral rebound was predicted by an increase in residual viremia 4–12 weeks before plasma HIV-1 RNA was detectable by commercial assays, showing the potential utility of SCA in revealing early evidence of virologic failure. Overall, these studies indicate that residual viremia remains unchanged in most patients after regimen simplification to a ritonavir-boosted PI alone.
SCA Utility in Assessing Impact of Treatment Intensification (Table 1)
Table 1.
Summary of antiviral intensification trials
| Study | Drug(s) added | n | Duration (weeks) |
Residual viremia |
CD4 count | T-cell activation | Other major findings |
|---|---|---|---|---|---|---|---|
| Hatano [29] | RALa | 30 | 24 | No changeb | No change | No change | — |
| Yukl [31••] | RAL | 7 | 12 | No change | Increased in the ileum | Decreased in CD4 and CD8 cells |
Decrease in unspliced HIV RNA in CD4 T cells in ileum |
| Gandhi [20] | RAL | 25 | 12 | No change | Slight increase | No change | — |
| Buzon [27•] | RAL | 45 | 24 | No change | No change | No changec | Transient increase in 2-LTR circles in PBMCs |
| McMahon [30] | RAL | 10 | 4 | No change | No change | — d | — |
| Hilldorfere [33] | MVCf | 34 | 24 | No change | — | — | No change in 2-LTR circles |
| Hunte [34] | MVC | 23 | 24 | No change | No change | Increased, notably in GALT |
CD8 cells seemed to relocate from GALT to blood |
| Wilkine [35] | MVC | 34 | 24 | — | No change | Decreased in CD4 and CD8 cells |
Observed improvement in markers of apoptosis |
| Gutiérreze [32] | MVC | 9 | 48 | Increased | No change | Increased in CD4 cells only |
Reduction in latent reservoir and increase in 2-LTR circles |
| Dinoso [28] | EFVg, LPV/rh, or ATV/ri |
9 | 4–7 | No change | — | — | — |
| Hammer [37] | ABCj | 116 | 272k | No change | No change | — | — |
| Havlir [36] | ABC | 8 | 24l | Decreased | No change | Decreased in CD4 and CD8 cells |
Reduced CD8 HIV Gag and p24 antigen responses |
Raltegravir
No clinically significant change among aggregate data or difference between intensified and placebo groups (when available)
A “normalization” of immune activation was observed in the 29% of patients with detectable 2-LTR circles
Parameter was not evaluated
Data has been presented at conferences, but has not yet been published in peer-reviewed journals
Maraviroc
Efavirenz
Lopinavir/ritonavir
Atazanavir/ritonavir
Abacavir
The median duration of treatment was 272 weeks (range, 3–283 weeks)
Post-intensification data was represented as the geometric mean of 6 samples drawn at 4-week intervals beginning 4 weeks after adding ABC
If ongoing cycles of viral replication persist during ART and contribute to residual viremia, adding a new, potent drug to an existing suppressive regimen should further lower residual viremia. Several studies report that intensification of therapy with a ritonavir-boosted PI, efavirenz, or the integrase inhibitor raltegravir (RAL) does not reduce viremia [20, 27•, 28–30, 31••]. Interestingly, one RAL intensification study showed 2-LTR circles increased transiently in circulating mononuclear cells [27•], and another showed unspliced HIV-1 RNA in CD4+ T cells decreased in the ileum in 5 of 7 patients studied [31••]. These two studies support ongoing HIV-1 replication because 1) RAL prevents integration of viral cDNA, thereby promoting formation of viral episomes, including 2-LTR circles, and 2) inhibition of integration would results in reduced unspliced HIV-1 RNA levels only if replicative cycles are ongoing. Additional studies are warranted to assess whether the observed effects of RAL intensification on 2-LTR circles and HIV-1 RNA in ileal tissue are reproducible. Recently, several intensification studies have been conducted using the entry inhibitor maraviroc (MVC). Although one study found an increase in residual viremia during intensification [32], later studies with larger sample sizes were unable to reproduce this finding [33–35]. Overall, intensification studies suggest that if ongoing replication occurs during suppressive ART, it is not reflected by plasma viremia. This conclusion contradicts an earlier study that showed a reduction in viremia with abacavir intensification of a two-drug regimen (EFV and indinavir) [36], however, this result may be a consequence of incomplete viral suppression by the two-drug regimen. Indeed, a later studied showed that abacavir intensification of a three-drug regimen (zidovudine/lamivudine/indinavir) had no effect on viremia and no clinical benefit [37]. Table 1 provides a summary of intensification studies and their results.
Other Applications of HIV-1 RNA SCA
In addition to quantification of residual viremia in patients on ART, HIV-1 RNA SCA has been used to evaluate viremia in elite controllers. Elite controllers are rare individuals capable of suppressive HIV-1 in the absence of ART. Most elite controllers (81%) have residual viremia detectable by SCA [38], and no statistically significant difference was found between the frequency of undetectable residual viremia (<1 copy/mL) in elite controllers compared with patients on suppressive ART [39]. In contrast to patients on ART, HIV-1 replication continues in elite controllers as evidenced by evolution of plasma virus over time [40••]. Remarkably, virus evolution continues in elite controllers without progressive escape from the immune response. The control mechanisms by which virus escape is thwarted are the focus of intense investigation.
Beyond HIV-1 RNA SCA: Future Technologies and Their Applications
Despite the insights provided by plasma HIV-1 RNA SCA, a more complete picture of residual HIV-1 infection is needed since viremia represents only one aspect of HIV-1 persistence. Indeed, the source of residual plasma viremia has not been well defined. To fully characterize the location of HIV-1 reservoirs and their relative contribution to plasma viremia, it will be necessary to go well beyond quantifying plasma HIV-1 RNA. Table 2 provides a list of assays that should be applied to identify and characterize persistent HIV-1 reservoirs.
Table 2.
Molecular assays to quantify relevant HIV DNA and RNA forms
| HIV form | Detection method | Pathogenetic relevance |
|---|---|---|
| Unintegrated, linear DNA | Cannot be specifically detecteda | Transient viral DNA intermediate |
| Integrated, linear DNA | qRT-PCR after gel separation of HMW (≥20 kB) DNAb |
Stably integrated provirus; potential long-lived reservoir |
| 1-LTR and 2-LTR circles (episomes) |
Primer/probe set that detects circle junctions | Dead-end products from failed integration; possible marker of recent infection |
| Promoter proximal RNA | qRT-PCR | Indicative of stalled elongation of HIV transcripts |
| 2-kb RNA transcripts (cell-associated) |
qRT-PCR across 2-kb splice junctions | Early transcription products coding for accessory genes |
| 4-kb RNA transcripts (cell-associated) |
qRT-PCR of vpu and gag/polc | Late transcription products coding for accessory genes and envelope |
| 9-kb RNA transcripts (cell-associated) |
qRT-PCR of gag/pol | Late transcription product encoding all genes |
| Virion-associated RNA | qRT-PCR | Viremia |
PCR-based assays cannot specifically quantify unintegrated HIV linear DNA. Rather, it is derived by first quantifying total HIV DNA, then subtracting integrated linear and 2-LTR DNA (1-LTR DNA is considered negligible)
High molecular weight
Vpu is encoded by both 4-kb and 9-kb transcripts, whereas gag and pol are encoded by only 9-kb transcripts. Therefore, 4-kb transcripts are quantified by subtracting the number of transcripts encoding gag and pol from the number of transcripts encoding vpu
HIV-1 DNA Assays
To define the origins of residual viremia, it will first be necessary to develop and apply assays to enumerate and characterize HIV-infected cells. Although HIV-1 DNA quantification in PBMCs and other cell sources has been reported [13, 14, 15••], quantification of the different forms of HIV-1 DNA within cells remains a contemporary research goal. The different forms of HIV-1 DNA include integrated HIV-1 DNA, linear unintegrated HIV-1 DNA, and 1 or 2-LTR circles. HIV-1 DNA forms that can be quantified include total HIV-1 DNA, 2-LTR circles, and integrated HIV-1 DNA, although methods for the latter DNA form are still being optimized [41, 42]. Currently, no technique exists to selectively quantify 1-LTR circles; however, they likely constitute a negligible portion of HIV-1 DNA. Linear, unintegrated HIV-1 DNA is not amenable to specific quantification because of near complete overlap with integrated DNA. Consequently, the difference between total HIV-1 DNA and the sum of integrated HIV-1 DNA and 2-LTR DNA can provide a reasonable estimate of unintegrated linear DNA (assuming a negligible quantity of 1-LTR DNA).
Quantification the different forms of HIV-1 DNA will provide insight into tissue variation in integrated versus episomal DNA and their relation to intracellular HIV-1 RNA levels as well as persistent viremia. Furthermore, quantification HIV-1 DNA in immune cell subsets sorted by flow cytometry will help characterize HIV-infected cellular reservoirs [43••].
Cell-associated HIV-1 RNA Assays
The next assay needed to characterize HIV-1 reservoirs is a cell-associated HIV-1 RNA assay with single copy sensitivity. The objective of this assay would be to quantify the expression of different splicing variants (both 2 kb and 4 kb transcripts) as well as aborted transcripts and full length genomes (9 kb). This can be achieved by utilizing primer and probe sets specific for distinct HIV-1 RNA forms, providing important information about transcriptional activity in infected cells. It is expected that the majority of integrated HIV-1 DNA will be defective, “dead” DNA that does not lead to the production of full-length transcripts. In addition to detecting distinct RNA forms, a cell-associated HIV-1 RNA assay with single copy sensitivity would also be useful in investigating the amount of RNA that is produced by infected cells during interventions designed to activate latent virus. Intracellular HIV-1 RNA assays have been developed and implemented [31••], but were designed to detect HIV-1 RNA in a population of cells. Methods to detect HIV-1 RNA expression in single infected cells are thus a priority.
Sequencing of HIV-1 Genomes
In addition to qPCR-based techniques, single genome sequencing (SGS) will also be of utility to understand better the genetics of the HIV-1 populations that persist despite antiretroviral therapy. SGS has shown that sequences present in less than 10% of single genomes are not readily detectable by population sequencing methods [44]. Even though SGS was originally developed to sequence low-frequency genomes from plasma, the method can be readily applied to genetically characterize HIV-1 DNA. Other groups have used large volume blood draws to compare the sequences of plasma HIV-1 RNA over time on suppressive ART to determine whether there are changes in the genetics of persistent virus populations [45].
Similarly, new sequencing technologies are being applied to characterize the genetics of persistent HIV-1 populations. Pyrosequencing (Life Sciences 454 platform) has emerged as the initial leader in high-throughput technology [46]; however, the 454 platform uses the same primer set to amplify all PCR product for sequencing, leading to the possibility of contamination and false-positive sequences. This limitation can be overcome with the advent of individualized”dog tagged” primers that can be used to uniquely mark amplicons arising from specific primers [47]. These unique “dog tag” identifiers can then be used to compile consensus sequences, effectively screening out the background of contaminating sequences and sequencing errors. Thus, high-throughput sequencing is likely to be a useful technique to characterize persistent HIV-1 populations in different tissues and cells types.
Sensitive sequencing methods were used by Bailey and colleagues to demonstrate that sequences found in the low-level residual viremia of patients on suppressive ART are infrequently found in circulating resting CD4+ T cells, and that identical sequences (predominant plasma clones) can emerge after long-term viral suppression on ART [48••]. However, Anderson and colleagues recently showed that replicating virus isolated from circulating, resting CD4+ T cells matched cell-free plasma HIV-1 RNA sequences (i.e. virions) in two patients on suppressive ART [49••]. The source of residual viremia thus remains a critical question, and its identification is an essential step toward its elimination. Sensitive sequencing methods applied to plasma and tissues have the potential to identify such sources.
Surrogate for Infectious Virus Recovery?
The ability to quantify different HIV-1 DNA and RNA forms raises the key question about which measure, if any, is indicative of the potential to produce infectious virus. A cell culture–based assay for infectious virus recovery from CD4+ T cells, developed by the Siliciano group [50], has provided critical insights into HIV-1 persistence. Although informative, the infectious virus recovery assay is limited in that throughput is low and that the assay is time consuming and resource intensive. A relatively high-throughput surrogate for infectious virus recovery would allow for larger-scale studies of the impact of therapeutic interventions.
Assessments of HIV-1 Reservoirs in Non-Blood Compartments
The blood compartment contains approximately 2% of total body lymphocytes. Therefore, application of single copy assays to tissue samples is essential for identifying HIV-1 reservoirs that are not reflected in blood. An example of differences between blood and tissue is evident in immune activation; the GI tract contains the majority of CD4+ T cells, and chronic immune activation is more evident in cells from GI biopsies than those in blood. Yukl and colleagues have demonstrated the feasibility of quantifying HIV-1 DNA and RNA from various locations in the GI tract and also showed that levels of T-cell activation markers in the gut are (paradoxically) negatively correlated with HIV-1 DNA levels in the gut [31••]. This indicates that different mechanisms may contribute to HIV-1 persistence in peripheral blood compared to the gut and should thus be evaluated further.
Additionally, bone marrow, cerebral spinal fluid (CSF), and both male and female reproductive tracts represent other anatomical compartments that have been shown to harbor HIV-1 infected cells. Regarding the bone marrow, multipotent hematopoietic progenitor cells (CD34+ cells) capable of clonal expansion and differentiation into hematopoietic lineages have long been know to be susceptible to HIV-1 infection [51]. These progenitor cells are susceptible in vitro [52–54] and a compelling case has been made for infection of these cells in vivo [55••]. As such, the quantification of HIV-1 DNA and RNA, as well as sequence information from cells isolated from bone marrow, is likely to provide key clues about the role of progenitor cells in the clonal expansion of HIV-1 as infected cells proliferate and differentiate into different cell lineages, as suggested by Carter and colleagues [55••].
Regarding the central nervous system, HIV-1 has been of great interest since the recognition of AIDS-related dementia. While there are major barriers to direct sampling of tissue from human brain, sampling of CSF for free virus is feasible. Additionally, markers of inflammation in the CNS, such as neopterin, could be correlated with low-level CNS HIV-1 RNA, allowing conclusions about the extent of HIV-1 infection and immune-related inflammation in the CNS [56]. In this regard, studies utilizing RT-SHIV in non-human primate (NHP) systems may be informative for the study of reservoirs in tissues that are intrinsically difficult to sample from human subjects [57].
HIV-1 Infection and Expression in Immune Cell Subsets
Quantification of HIV-1 nucleic acids in immune cell subsets isolated from peripheral blood is expected to be more tractable than quantification in other anatomical compartments. A typical bone marrow aspirate yields on the order of 105 CD34+ cells, which include hematopoietic stem cells as well as early progenitor cell populations [55••]. Alternatively, CD34+ hematopoietic progenitor cells can also be isolated from peripheral blood, where they constitute approximately 0.02% of PBMCs [55••]. CD4+ T-cell yield from gut-associated lymphoid tissue (GALT) is expected to be similarly limited. Infection of these populations in the bone marrow and GALT is sufficiently extensive to detect HIV-1 DNA [31••, 55••]; however, it is anticipated that viral RNA expression will be rare and may not be detected in all patients. Therefore, larger quantities of cells, such as those obtained during autopsy, gut resections, or from NHP studies may be required to detect rare RNA expression, particularly when examining discrete immune cell subsets.
The distinct subsets of immune cells that constitute HIV-1 reservoirs and the methods by which these reservoirs are maintained is of great interest. Seminal work by Chomont and colleagues has demonstrated that within the CD4+ T-cell compartment, transitional and central memory T cells are more frequently infected by HIV-1 (i.e. HIV-1 DNA positive) than effector memory or naïve T cells [43••]. Sequencing and phylogenetic comparisons were used to infer that the transitional memory T-cell reservoir is replenished by antigen-driven proliferation from the central memory T-cell reservoir and the central memory T-cell reservoir is maintained through IL-7–mediated homeostatic proliferation [43••]. Expression of the surface receptor programmed death-1 (PD-1) was also shown to be associated with integrated HIV-1 DNA. Blockage of the PD-1 and PDL1 interaction with monoclonal antibodies is currently being investigated as a means by which to induce HIV-1 expression in an effort to reduce size of the HIV-1 reservoir [58].
Single Cell Technologies
A single cell sequencing method was recently reported by Josefsson and colleagues [59••] in which cells were first sorted by flow cytometry and then diluted to the point where each PCR reaction contained far fewer than one infected cell. HIV-1 DNA was sequenced, and the genomes were then compared to HIV-1 DNA from other individual infected cells as well as RNA sequences from plasma as determined by SGS [59••]. This single cell sequencing analysis led to the conclusion that the large majority of infected CD4+ T cell contain only one integrated HIV-1 DNA molecule, and that there is concordances between the sequences from plasma virus and HIV-infected CD4+ T cells. Similar methods to assess other HIV-1 DNA and RNA forms on a single-cell basis will be useful in quantifying and characterizing HIV-1 reservoirs before and after therapeutic interventions.
Conclusions
In order to reduce the number HIV-infected cells, it will first be necessary to understand better the biology of HIV-1 reservoirs in suppressed patients. This will require the application of highly specific and sensitive assays that are accurate and reproducible. Single copy assays and single infected cell assays will be crucial in describing the size and location of HIV-1 reservoirs, the extent of HIV-1 RNA expression, and the sources of persistent residual viremia. Such assays, in addition to a molecular surrogate for infectious virus recovery, will also be essential tools in evaluating the impact of interventions on HIV-1 reservoirs. Indeed, single copy assays have been used effectively to determine that the intensification of ART does not generally result in a decrease in residual viremia. In the search for a cure for HIV-1 infection, the ability to accurately assess the impact of interventions designed to eradicate HIV-1 will be critical, and small but significant changes may offer insights into the appropriate path toward a cure.
Acknowledgment
Supported by award T32 AI065380 from the National Institute of Allergy and Infectious Disease, NIAID, Division of AIDS (University of Pittsburgh CTU Grant 1U01 AI069494-01 and supplement), a Virology Support Laboratory subcontract (204VC009) of the ACTG Central Group Grant (1U01AI068636-01), and by the NCI (SAIC contract 25XS119).
Footnotes
Disclosure No potential conflicts of interest relevant to this article were reported.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L, Jr, Ingerman MJ, et al. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282(17):1627–32. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 2••.Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 2007;3(4):e46. doi: 10.1371/journal.ppat.0030046. This paper demonstrated that patients on suppressive ART had residual viremia (median, 3.1 copies/mL) that fell to a stable set point and did not decrease significantly between weeks 60 and 110 of suppressive ART.
- 3.Havlir DV, Bassett R, Levitan D, Gilbert P, Tebas P, Collier AC, et al. Prevalence and predictive value of intermittent viremia with combination hiv therapy. JAMA. 2001;286(2):171–9. doi: 10.1001/jama.286.2.171. [DOI] [PubMed] [Google Scholar]
- 4.Schockmel GA, Yerly S, Perrin L. Detection of low HIV-1 RNA levels in plasma. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14(2):179–83. doi: 10.1097/00042560-199702010-00013. [DOI] [PubMed] [Google Scholar]
- 5.Yerly S, Kaiser L, Perneger TV, Cone RW, Opravil M, Chave JP, et al. Time of initiation of antiretroviral therapy: impact on HIV-1 viraemia. The Swiss HIV Cohort Study. AIDS. 2000;14(3):243–9. doi: 10.1097/00002030-200002180-00006. [DOI] [PubMed] [Google Scholar]
- 6.Yerly S, Perneger TV, Vora S, Hirschel B, Perrin L. Decay of cell-associated HIV-1 DNA correlates with residual replication in patients treated during acute HIV-1 infection. AIDS. 2000;14(18):2805–12. doi: 10.1097/00002030-200012220-00001. [DOI] [PubMed] [Google Scholar]
- 7••.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41(10):4531–6. doi: 10.1128/JCM.41.10.4531-4536.2003. This paper described the single copy assay, the first technique capable of detecting a single copy of plasma HIV-1 RNA.
- 8.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373(6510):123–6. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 9.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell lifespan, and viral generation time. Science. 1996;271(5255):1582–6. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 10••.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373(6510):117–22. doi: 10.1038/373117a0. References 8–10 were the first reports describing the decay of plasma HIV-1 RNA after initiating ART. They provided the framework for future decay analyses, and characterize the first and most profound phase of decay.
- 11.Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233(4760):215–9. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 12••.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387(6629):188–91. doi: 10.1038/387188a0. This study described the second phase of plasma HIV-1 RNA decay during ART, providing evidence of long-lived infected cells in vivo that persist despite suppression of viral replication.
- 13.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–8. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 14.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 15••.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–5. doi: 10.1126/science.278.5341.1291. References 13–15 were seminal publications that provide evidence that resting CD4+ T cells harbored latent, inducible provirus, preventing eradication of HIV-1 by ART alone.
- 16••.Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2008;105(10):3879–84. doi: 10.1073/pnas.0800050105. This study revealed a third phase of HIV-1 RNA decay and a fourth phase of no further decay that persists for at least 7 years following initiation of ART, expanding upon the findings in reference 2.
- 17.Pascual-Pareja JF, Martinez-Prats L, Luczkowiak J, Fiorante S, Rubio R, Pulido F, et al. Detection of HIV-1 at between 20 and 49 copies per milliliter by the Cobas TaqMan HIV-1 v2.0 assay is associated with higher pretherapy viral load and less time on antiretroviral therapy. J Clin Microbiol. 2010;48(5):1911–2. doi: 10.1128/JCM.02388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chun TW, Justement JS, Murray D, Hallahan CW, Maenza J, Collier AC, et al. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS. 2010;24(18):2803–8. doi: 10.1097/QAD.0b013e328340a239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandhi RT, BR, Aga E, Albrecht M, Demeter L, Bastow B, Siliciano R, Siliciano J, Eron J, ATCG A5173 No evidence for decay in the latent reservoir in HIV-infected patients receiving intensive enfuvirtide-containing ART. 17th Conference on Retroviruses and Opportunistic Infections; San Francisco. 2010. [Google Scholar]
- 20.Gandhi RT, Zheng L, Bosch RJ, Chan ES, Margolis DM, Read S, et al. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med. 2010 doi: 10.1371/journal.pmed.1000321. doi:10.1371/journal.pmed/1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Chun TW, Murray D, Justement JS, Hallahan CW, Moir S, Kovacs C, et al. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J Infect Dis. 2011;204(1):135–8. doi: 10.1093/infdis/jir208. This paper showed a direct correlation between the size of CD4+ T-cell reservoir (ie, HIV-1 DNA) and residual viremia; however, no correlation was found between reservoir size and immune activation.
- 22.Havlir DV, Koelsch KK, Strain MC, Margot N, Lu B, Ignacio CC, et al. Predictors of residual viremia in HIV-infected patients successfully treated with efavirenz and lamivudine plus either tenofovir or stavudine. J Infect Dis. 2005;191(7):1164–8. doi: 10.1086/428588. [DOI] [PubMed] [Google Scholar]
- 23.Haim-Boukobza S, Morand-Joubert L, Flandre P, Valin N, Fourati S, Sayon S, et al. Higher efficacy of nevirapine than efavirenz to achieve HIV-1 plasma viral load below 1 copy/ml. AIDS. 2011;25(3):341–4. doi: 10.1097/QAD.0b013e3283427de3. [DOI] [PubMed] [Google Scholar]
- 24.McKinnon JE, Arribas JR, Pulido F, Delgado R, Mellors JW. The level of persistent HIV viremia does not increase after successful simplification of maintenance therapy to lopinavir/ritonavir alone. AIDS. 2006;20(18):2331–5. doi: 10.1097/QAD.0b013e32801189f6. [DOI] [PubMed] [Google Scholar]
- 25.Swindells S, DiRienzo AG, Wilkin T, Fletcher CV, Margolis DM, Thal GD, et al. Regimen simplification to atazanavir-ritonavir alone as maintenance antiretroviral therapy after sustained virologic suppression. JAMA. 2006;296(7):806–14. doi: 10.1001/jama.296.7.806. [DOI] [PubMed] [Google Scholar]
- 26.Wilkin TJ, McKinnon JE, DiRienzo AG, Mollan K, Fletcher CV, Margolis DM, et al. Regimen simplification to atazanavir-ritonavir alone as maintenance antiretroviral therapy: final 48-week clinical and virologic outcomes. J Infect Dis. 2009;199(6):866–71. doi: 10.1086/597119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Buzon MJ, Massanella M, Llibre JM, Esteve A, Dahl V, Puertas MC, et al. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med. 2010;16(4):460–5. doi: 10.1038/nm.2111. This study showed a transient increase in 2-LTR circles in a fraction of subjects when suppressive ART was intensified with the integrase inhibitor raltegravir, suggesting low-level replication continues despite therapy.
- 28.Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange SJ, Cranmer L, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 2009;106(23):9403–8. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatano H, Hayes TL, Dahl V, Sinclair E, Lee TH, Hoh R, et al. A randomized, controlled trial of raltegravir intensification in antiretroviral-treated, HIV-infected patients with a suboptimal CD4+ T cell response. J Infect Dis. 2011;203(7):960–8. doi: 10.1093/infdis/jiq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMahon D, Jones J, Wiegand A, Gange SJ, Kearney M, Palmer S, et al. Short-course raltegravir intensification does not reduce persistent low-level viremia in patients with HIV-1 suppression during receipt of combination antiretroviral therapy. Clin Infect Dis. 2010;50(6):912–9. doi: 10.1086/650749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Yukl SA, Shergill AK, McQuaid K, Gianella S, Lampiris H, Hare CB, et al. Effect of raltegravir-containing intensification on HIV burden and T-cell activation in multiple gut sites of HIV-positive adults on suppressive antiretroviral therapy. AIDS. 2010;24(16):2451–60. doi: 10.1097/QAD.0b013e32833ef7bb. This study revealed unspliced HIV-1 RNA decrease in gut CD4+ cells during raltegravir intensification, indicating suppressive ART may not completely block replication in tissue compartments.
- 32.Gutierrez C, DL Hernandez, Novoa B, Vallejo A, Page C, Lorente R, Madrid N, Palmer S, Munoz-Fernandez MA, Moreno S. Effect of the intensification with a CCR5-antagonist on the decay of the HIV-1 latent reservoir and residual viremia. 17th Conference on Retroviruses & Opportunistic Infections; San Francisco. 2010. [Google Scholar]
- 33.Hilldorfer B, LC, McKinnon J, Coombs B, Tenorio A, Fox L, Gandhi R, Ribauldo H, Currier J, Gulick R, Wilkins TJ, Mellors JW. Effects of Maraviroc (MVC) on residual low-level viremia in patients on suppressive Antiretroviral Therapy (ART): Results from ACTG 5256. International AIDS Association; Rome: 2011. [Google Scholar]
- 34.Hunt PW, SN, Hayes T, Dahl V, Somsouk M, Funderburg N, Landay AL, Adeyemi O, Shafer R, Clagett B, Rodriguez B, Martin JN, Shacklett B, Palmer S, Lederman MM, Deeks SG. The immunomodulatory effects of Maraviroc intensification among ART-suppressed patients with incomplete CD4 recovery. 18th Conference for Retroviruses and Opportunistic Infections; San Francisco. 2011. [Google Scholar]
- 35.Wilkin TJ, LC, Landay A, Ribaudo H, McKinnon J, Gandhi R, Mellors J, Currier J, Gulick R. Maraviroc (MVC) intensification for suboptimal CD4+ response despite sustained virologic suppression: ACTG 5256. Conference on Retroviruses and Opportunistic Infections; San Francisco. 2010. [Google Scholar]
- 36.Havlir DV, Strain MC, Clerici M, Ignacio C, Trabattoni D, Ferrante P, et al. Productive infection maintains a dynamic steady state of residual viremia in human immunodeficiency virus type 1-infected persons treated with suppressive antiretroviral therapy for five years. J Virol. 2003;77(20):11212–9. doi: 10.1128/JVI.77.20.11212-11219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammer SM, Ribaudo H, Bassett R, Mellors JW, Demeter LM, Coombs RW, et al. A randomized, placebo-controlled trial of abacavir intensification in HIV-1-infected adults with virologic suppression on a protease inhibitor-containing regimen. HIV Clin Trials. 2010;11(6):312–24. doi: 10.1310/hct1106-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereyra F, Palmer S, Miura T, Block BL, Wiegand A, Rothchild AC, et al. Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J Infect Dis. 2009;200(6):984–90. doi: 10.1086/605446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinoso JB, Kim SY, Siliciano RF, Blankson JN. A comparison of viral loads between HIV-1-infected elite suppressors and individuals who receive suppressive highly active antiretroviral therapy. Clin Infect Dis. 2008;47(1):102–4. doi: 10.1086/588791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Mens H, Kearney M, Wiegand A, Shao W, Schonning K, Gerstoft J, et al. HIV-1 continues to replicate and evolve in patients with natural control of HIV infection. J Virol. 2010;84(24):12971–81. doi: 10.1128/JVI.00387-10. Low-level, residual viremia from elite controls was sequenced in this study and was found to evolve, in contrast to residual viremia observed during suppressive ART that does not show evolution.
- 41.Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Gunthard HF, et al. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis. 2008;197(3):411–9. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 42.Liszewski MK, Yu JJ, O’Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods. 2009;47(4):254–60. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. This study demonstrated the presence of two distinct HIV-1 DNA reservoirs in the memory T-cell compartment. The reservoir in central memory T cells is maintained by antigen-induced proliferation, whereas the reservoir in transitional memory T cells is maintained by homeostatic proliferation.
- 44.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43(1):406–13. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kearney M, SJ, Yu S, Shao W, O’Shea A, Rehm C, Poethke C, Mellors J, Coffin J, Maldarelli F. The genetic diversity of HIV-1 in plasma persists despite suppression with ART. Conference on Retroviruses and Opportunistic Infections; San Francisco. 2010. [Google Scholar]
- 46.Rothberg JM, Leamon JH. The development and impact of 454 sequencing. Nat Biotechnol. 2008;26(10):1117–24. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- 47.Jabara CJC, Anderson J, Swanstrom R. Accurate sampling and deep sequencing HIV-1 protease using primer ID. Conference on Retroviruses and Opportunistic Infections; Boston. 2011. [Google Scholar]
- 48••.Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80(13):6441–57. doi: 10.1128/JVI.00591-06. This report provided extensive genotypic analyses of HIV-1 provirus in circulating CD4+ T cells from patients on suppressive ART. It revealed that the sequences in CD4+ T cells were distinct from those in plasma, providing evidence that residual viremia arises, at least in part, from sources other than circulating CD4+ T cells.
- 49••.Anderson JA, Archin NM, Ince W, Parker D, Wiegand A, Coffin JM, et al. Clonal sequences recovered from plasma from patients with residual HIV-1 viremia and on intensified antiretroviral therapy are identical to replicating viral RNAs recovered from circulating resting CD4+ T cells. J Virol. 2011;85(10):5220–3. doi: 10.1128/JVI.00284-11. This study of two patients on suppressive ART found that HIV-1 sequences isolated from virus outgrowth assays of resting CD4+ T cells matched those of the predominant plasma clone. This is consistent with the hypothesis that residual viremia arises in part from reactivation of latent provirus in resting CD4+ T cells.
- 50.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 51.Folks TM, Kessler SW, Orenstein JM, Justement JS, Jaffe ES, Fauci AS. Infection and replication of HIV-1 in purified progenitor cells of normal human bone marrow. Science. 1988;242(4880):919–22. doi: 10.1126/science.2460922. [DOI] [PubMed] [Google Scholar]
- 52.Kitano K, Abboud CN, Ryan DH, Quan SG, Baldwin GC, Golde DW. Macrophage-active colony-stimulating factors enhance human immunodeficiency virus type 1 infection in bone marrow stem cells. Blood. 1991;77(8):1699–705. [PubMed] [Google Scholar]
- 53.Ruiz ME, Cicala C, Arthos J, Kinter A, Catanzaro AT, Adelsberger J, et al. Peripheral blood-derived CD34+ progenitor cells: CXC chemokine receptor 4 and CC chemokine receptor 5 expression and infection by HIV. J Immunol. 1998;161(8):4169–76. [PubMed] [Google Scholar]
- 54.Steinberg HN, Crumpacker CS, Chatis PA. In vitro suppression of normal human bone marrow progenitor cells by human immunodeficiency virus. J Virol. 1991;65(4):1765–9. doi: 10.1128/jvi.65.4.1765-1769.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.Carter CC, Onafuwa-Nuga A, McNamara LA, Riddell JT, Bixby D, Savona MR, et al. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat Med. 2010;16(4):446–51. doi: 10.1038/nm.2109. This report utilized flow cytometry to provide evidence for infection of early progenitor cells in the bone marrow of some patients on suppressive ART, implicating such cells as long-lived reservoirs of HIV-1.
- 56.Hagberg L, Cinque P, Gisslen M, Brew BJ, Spudich S, Bestetti A, et al. Cerebrospinal fluid neopterin: an informative biomarker of central nervous system immune activation in HIV-1 infection. AIDS Res Ther. 2010;7:15. doi: 10.1186/1742-6405-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ambrose Z, Palmer S, Boltz VF, Kearney M, Larsen K, Polacino P, et al. Suppression of viremia and evolution of human immunodeficiency virus type 1 drug resistance in a macaque model for antiretroviral therapy. J Virol. 2007;81(22):12145–55. doi: 10.1128/JVI.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DaFonseca S. In: Purging the HIV-1 reservoir through the disruption of the PD-1 pathway. Them TaCHRaStC., editor. International AIDS Society; Rome: 2011. [Google Scholar]
- 59••.Josefsson L, King MS, Makitalo B, Brannstrom J, Shao W, Maldarelli F, et al. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc Natl Acad Sci U S A. 2011;108(27):11199–204. doi: 10.1073/pnas.1107729108. This study utilized single cell analyses to demonstrate that most HIV-1-infected cells contain a single provirus. Similar single cell analyses of HIV-1 expression are likely to be important for future studies of HIV-1 reservoirs.