Abstract
Protein palmitoylation is a critical post-translational modification important for membrane compartmentalization, trafficking, and regulation of many key signaling proteins. Recent non-radioactive chemo-proteomic labeling methods have enabled a new focus on this emerging regulatory modification. Palmitoylated proteins can now be profiled in complex biological systems by mass spectrometry for direct annotation and quantification. Based on these analyses, palmitoylation is clearly widespread and broadly influences the function of many cellular pathways. The recent introduction of selective chemical labeling approaches has opened new opportunities to revisit long-held questions about the enzymatic regulation of this widespread post-translational modification. In this review we will discuss the impact of new chemical labeling approaches and future challenges for the dynamic global analysis of protein palmitoylation.
Keywords: Palmitoylation, thioesterase, proteomics, post-translational modifications, mass spectrometry
Introduction
Protein palmitoylation was first discovered over 30 years ago in classic experiments using tritiated palmitate for metabolic labeling of virus particles and virus infected cells [1–2]. Fatty acid modification of viral proteins was shown to be post-translational [3], covalent [1–2], and hydroxylamine-sensitive [4]. Further studies demonstrated this modification occurred on cellular proteins and involved an acyl-thioester linkage to specific cysteines [4–5]. It was quickly evident that palmitoylation is a universal post-translational modification important for the regulation of trafficking, localization, and activity of many membrane proteins. Additionally, given the labile properties of the thioester linkage, palmitoylation is potentially reversible and may be regulated in a manner analogous to other widely studied post-translational modifications, such as acetylation, phosphorylation, or ubiquination. Until the last few years, the lack of robust enrichment methods prevented a more thorough understanding of palmitoylation and its regulation.
For decades, the only method to detect palmitoylation relied on metabolic incorporation of radioactive palmitate derivates, followed by immunoprecipitation, and long exposures from days to weeks. Using this approach, H-ras de-palmitoylation was shown to be accelerated >15 times upon activation [6]. Similarly, G-alpha-s palmitoylation turnover is accelerated 50-fold following stimulation [7]. These key findings demonstrate specific, signal-dependent de-palmitoylation of key signaling proteins involved in cell growth and hormonal signaling, but left many longstanding questions. First, is the de-palmitoylation of key signaling proteins enzymatic? Which palmitoylated proteins are regulated by de-palmitoylating enzymes? What are the enzymes that catalyze palmitoylation and de-palmitoylation?
Chemo-proteomic enrichment of palmitoylated proteins
The introduction of new labeling approaches has revolutionized the enrichment, annotation, and quantification of protein palmitoylation. The first systematic annotation of palmitoylated proteins was accomplished using a novel enrichment technique based on selective thioester hydrolysis by hydroxylamine, termed acyl biotin exchange (ABE) [8–10]. Free cysteines are first alkylated, and then half the sample is treated with neutral hydroxylamine to selectively cleave thioesters. The newly exposed cysteines are then disulfide-bonded to a biotin analog, affinity purified, and digested into peptides, leaving the labeled peptides on the affinity beads. The eluant is then analyzed by Multidimensional Protein Identification Technology (MudPIT) using in-line liquid chromatography mass spectrometry (LC-MS) [11]. The ratio of hydroxylamine-dependent enrichment is used to annotate specific palmitoylated proteins. This method has been widely used to annotate hundreds of palmitoylated proteins from diverse tissues, cells, and organisms [9, 12–16]. Non-specific capture of endogenous stable thioesters, such as acyl carrier proteins and ubiquitin ligases, and incomplete cysteine alkylation reduce the fidelity of the method. More recent modifications of this method introduce a thiol affinity resin to bypass the biotin-streptavidin enrichment [17], reducing the enrichment steps and eliminating non-specific enrichment of endogenous biotinylated carboxylases. After elution of tryptic peptides, labeled peptides are released by disulfide reduction for LC-MS annotation. This approach allows specific sites of palmitoylation to be inferred. Overall, ABE provides a static profile of palmitoylation without metabolic labeling.
The second enrichment approach uses metabolic labeling with the commercial alkynyl fatty acid analogue 17-octadecynoic acid (17-ODYA) [18]. The cellular palmitoylation machinery readily incorporates 17-ODYA into endogenous sites of palmitoylation within the native cellular environment. After sufficient labeling, the cells are lysed and coupled to azide-linked reporter tags by Cu(I) catalyzed click chemistry. Palmitoylated proteins are easily detected after rhodamine-azide conjugation for fluorescent gel-based analysis, or selectively enriched after biotin-azide conjugation. This approach has several unique advantages, including temporal control of probe incorporation for pulse chase analysis, and a significant reduction in non-specific labeling. Most importantly, the bioorthogonal approach does not require thiol reduction and alkylation, and small samples can be easily analyzed without multiple precipitation steps. This method was first reported using azido-fatty acids [19–20], but click chemistry with alkyne-linked reporters generates significantly higher background labeling [21]. Systematic analysis of the acyl chain length has shown optimal incorporation of 17-ODYA, while shorter chain lengths are better substrates for N-myristoyltransferases [22]. After prolonged incubation, β-oxidation of 17-ODYA generates shorter acyl variants for incorporation into N-myristoylation sites [18]. Probe elongation and β-oxidation can be prevented by incubation with acid (HDYOA) [23], which resists fatty acid remodeling. Several studies have indirectly compared the overlap of 17-ODYA and ABE methods, and general conclusion is they both identify a similar profile of palmitoylated proteins, and are best used in combination for orthogonal validation [12].
Quantitative Annotation of Palmitoylated Proteins
The earliest palmitoylation proteomics studies were qualitatively measured by spectral counting, a semi-quantitative method correlated with protein abundance of a limited dynamic range [24]. Spectral counts are assigned as the sum of the number of triggered MS2 peptide fragmentations for a specific protein. Ions are analyzed by data dependent acquisition, where the most abundant precursors at a given retention time are selected for ion trap isolation, fragmentation, and low resolution m/z assignment. Typically, an exclusion algorithm is included to prevent sequential fragmentation of the same precursor, and to appropriately time a second fragmentation at the apex of the elution profile. Larger, wider peaks are fragmented more times than smaller, narrower peaks. Spectral count numbers are highly dependent on the length of the protein and the complexity of the sample, and require multiple replicates for statistical significance. For annotating sites of palmitoylation, separate samples are analyzed with or without addition of the enrichment probe, generally with multiple replicates for further validation. The probe-dependent enrichment ratio is then used to assign confidence to the resulting dataset. Overall, spectral counting is acceptable for comparing large changes from abundant proteins, but cannot measure subtle changes.
These initial studies led to the first large scale analysis describing the extent of protein palmitoylation in the proteome, but are not sufficiently sensitive to quantitatively compare dynamic palmitoylation events between different cell types or within the same population of cells following perturbation. To achieve this goal, we sought to measure small changes in palmitoylation that may result from temporally or spatially restricted forms of regulation. SILAC (Stable Isotope Labeling with Amino acids in Cell culture) is a preferred method for proteome-wide quantification [25], since complete heavy isotope labeling is achieved metabolically, with no errors associated with in vitro modification. Cells are cultured for several passages in media supplemented with stable isotopes of lysine and/or arginine. Lysates from perturbed cells grown in light media are mixed with lysates from control cells grown in heavy media for high resolution LC-MS analysis. After click chemistry and enrichment, the samples are digested with trypsin for shotgun proteomics analysis [26]. Importantly, trypsin cleaves at each arginine and lysine, leaving a C-terminal isotopic tag with a matching chromatographic retention time. The resulting isotopic SILAC precursor ions are precisely quantified based on the ion current integration across the chromatographic elution peak.
SILAC analysis solves many of the deficiencies of semi-quantitative spectral counting. First, each sample has its own internal reference with matching sample preparation, elution time, and instrument analysis. Additional algorithms for peak shape and predicted isotopic distribution provide an additional filter to exclude ions with significant interference. Second, the number of replicates is reduced, since the perturbed sample is run at the same time as the internal reference sample. Third, the absolute abundance of each peptide is quantified by the precursor peak area, and matched to the corresponding SILAC isotopic pair. Fourth, each peptide registers an individual SILAC ratio, which allows calculation of a corresponding standard deviation for each ratio assignment to a specific protein. Finally, reciprocal replicates with inversed isotopic labeling provide a final validation of the selective enrichment, eliminating small variances in expression levels and non-specific enrichment. Altogether, this approach quadruples the number of high confidence palmitoylated proteins annotated in mouse T-cells by reproducibly and accurately quantifying fractional changes in protein enrichment [26] (Figure 1).
Figure 1.
SILAC quantitative proteomic annotation of palmitoylated proteins. Light and heavy cells are separately labeled with or without 17-ODYA, and lysates are mixed equally for click chemistry enrichment, protease digestion, and quantitative LC-MS analysis [26].
Proteomic Profiling of Dynamic Palmitoylation
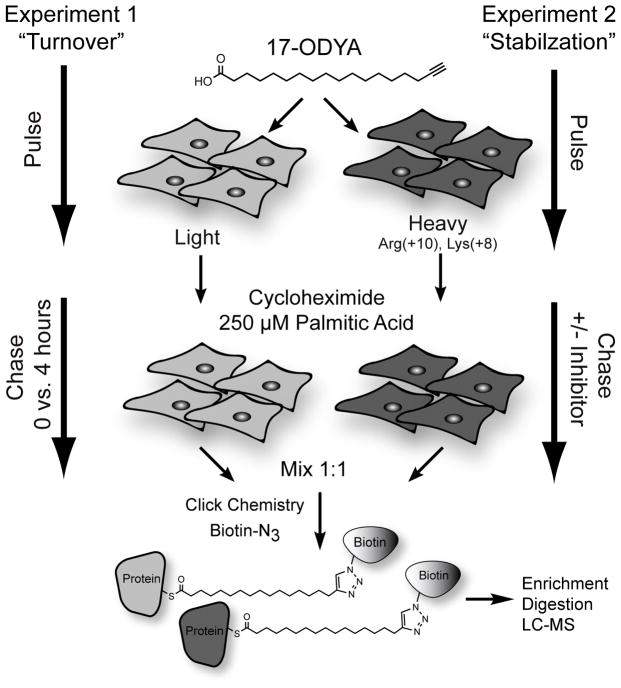
Ras GTPases and G-proteins are rapidly de-palmitoylated after stimulation, yet the broader dynamics of palmitoylation have remained largely uncharacterized. Several candidate thioesterases have been identified able to de-palmitoylate protein substrates in vitro [27–28], or over-expressed substrates in single cells [29–30], yet these experiments do not validate the endogenous role of thioesterases in the dynamic turnover of palmitoylation on native proteins. In order to establish the role of thioesterases in dynamic palmitoylation turnover, SILAC proteomics methods and 17-ODYA pulse-chase labeling [31] were used together to in two separate experiments to quantitatively profile dynamic palmitoylation in mouse T-cell hybridoma cells [26] (Figure 2). In the first experiment, cells were pulsed with 17-ODYA, and exchanged for media with 10-fold excess palmitic acid. Cells were then harvested immediately or after 4 hours to profile individual turnover rates. In the second experiment, cells were pulse labeled with 17-ODYA, and then incubated with or without a non-selective lipase inhibitor for 4 hours with excess palmitic acid. This experiment identified palmitoylated proteins that are stabilized by the generic lipase inhibitor hexadecylfluorophosphonate (HDFP), which targets approximately 20 serine hydrolases in mouse T-cells, but does not inhibit peptidases, proteases, or small molecule hydrolases [26]. An unenriched SILAC proteomics experiments was also preformed to identify any protein abundance changes occurring during the course of the pulse-chase experiment. All together, more than 300 palmitoylated proteins were assayed for accelerated palmitoylation turnover and palmitoylation stabilization by lipase inhibition.
Figure 2.
Experimental approach to profile palmitoylation dynamics. Two separate pulse-chase experiments were designed to identify the palmitoylated proteins with rapid turnover kinetics (Experiment 1), and those stabilized after lipase inhibition (Experiment 2) [26]. Proteomics analysis showed nearly complete overlap in both experiments, signifying the role serine hydrolase family thioesterases in the regulation of dynamics palmitoylation.
These chemo-proteomics experiments revealed several interesting findings. First, the majority of palmitoylated proteins show relatively stable palmitoylation over the 4 hour chase. Of course, this observed stability is relative, since there must be a basal level of palmitoylation turnover in the short pulse labeling time in order to achieve sufficient 17-ODYA labeling. Second, a small subset of palmitoylated proteins demonstrated accelerated palmitoylation turnover. This group includes Ras family small GTPases, G-proteins, MAGUKs family scaffolding proteins that have all been previously demonstrated to undergo rapid, activity-dependent palmitoylation turnover using traditional biochemical methods [6–7, 32]. This study also identified several other proteins with rapid palmitoylation dynamics, including nucleoporin (Nup210), the metastasis related protein metadherin (Mtdh), the ubiquitin-like modifier Ubl3, and the unannotated protein FAM49b. Third, palmitoylated proteins that are stabilized by lipase inhibition are the same proteins with accelerated palmitoylation turnover. This data validates the role of thioesterases in the dynamic regulation of protein palmitoylation on native proteins, and suggests that HDFP inhibits all of the enzymes capable of regulating dynamic palmitoylation. The candidate thioesterases LYPLA1, LYPLA2, and PPT1 are strongly inhibited by HDFP, although the contribution of other unannotated lipases cannot be excluded. Finally, this experimental approach is ideal for profiling activity-dependent de-palmitoylation in other biological contexts.
Palmitoyl Acyl Transferase substrate annotation
Protein palmitoylation is enzymatic and catalyzed by 23 individual protein acyl transferases (PATs), commonly referred to as the DHHC enzyme family due the conserved Asp-His-His-Cys catalytic motif [33]. The size of the DHHC enzyme family may explain why no consensus palmitoylation sites have emerged, since each enzyme presumably palmitoylated different substrates. Not surprisingly, PATs localize to different sub-cellular compartments [34] and have additional domains involved in protein-protein interactions and substrate recognition. Several DHHC PATs have been genetically linked to human diseases, including schizophrenia (DHHC8) [35], mental retardation (DHHC9, DHHC15) [36–37], Huntington’s disease (DHHC17) [38], and cancer (DHHC2, DHHC7, DHHC11, DHHC14, and DHHC20) [39–43]. Understanding the molecular role of palmitoylation in human disease will require a detailed analysis of the regulation and substrates of each DHHC enzyme.
In order to identify which specific PAT(s) palmitoylate a given protein, the each DHHC is separately co-expressed with the candidate substrate, and assayed for enhanced substrate palmitoylation. This approach led to the initial identification of four candidate PATs (DHHC2, DHHC3, DHHC7, and DHHC15) able to palmitoylate PSD-95 [33]. While this approach is relatively straightforward, heterologous expression studies may bias towards DHHC enzymes with high expression or function autonomously. DHHC9 activity, for instance, requires association with GCP16 (GOLGA7) and is inactive when over-expressed alone [44–45]. Furthermore, many PATs are highly post-translationally modified, suggesting additional levels of regulation. Genetic approaches coupled with proteomic profiling promises a more direct route to annotate DHHC substrates.
The palmitoyl transferase DHHC5 is significantly expressed in the brain, and localizes to the post-synaptic density [46] and dendritic endosomes [47], suggesting it may be responsible for palmitoylating synaptic proteins. Mice homozygous for a hypomorphic allele of the ZDHHC5 gene are impaired in context-dependent learning and memory, yet the molecular mechanism of this deficiency remains unclear [46]. DHHC5 directly interacts with PSD-95 at the post-synaptic density, but has no effect on its palmitoylation [46]. DHHC5 clearly plays an important role at synapses, but more advance approaches were needed to understand its functional role at the synapse.
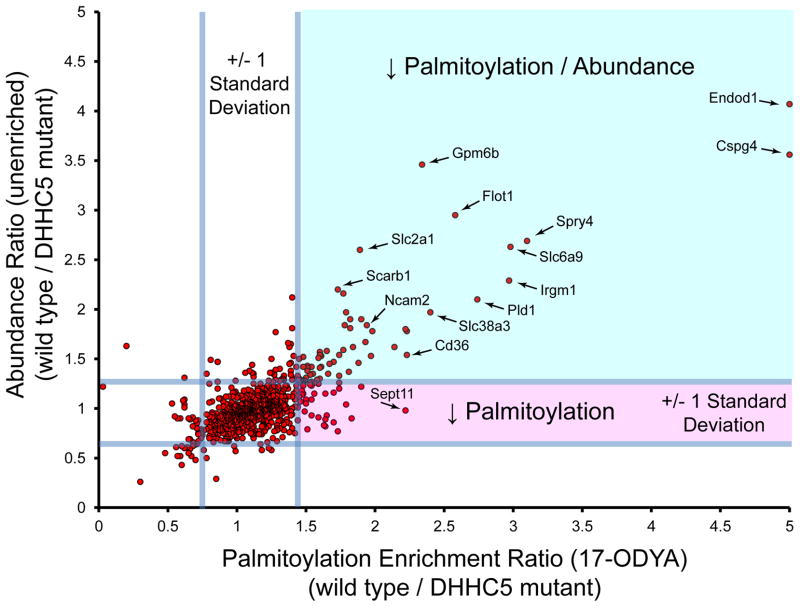
In order to identify potential substrates of DHHC5, neuronal stem cells were isolated from wild type and homozygous DHHC5 mutant mice for SILAC isotopic labeling [48]. Heavy and light wild type cells were labeled with or without 17-ODYA (Experiment 1), and analyzed with or without hydroxylamine hydrolysis (Experiment 2). SILAC pairs were mixed, enriched, and analyzed by mass spectrometry. Altogether, more than 300 proteins were identified in both experimental groups, as well as several hundred additional proteins only validated in one experimental group. Next, SILAC labeled wild type and mutant cells were analyzed, revealing about 3 dozen candidates with > 2-fold reduction in palmitoylation in DHHC5 mutant cells. This list includes the AMPA receptor subunit Gria4, as well as other neuronal channels, receptors, and signaling proteins. To correct for potential changes in protein abundance, unenriched SILAC proteomics experiments were performed in parallel. Surprisingly, essentially all of these palmitoylated proteins demonstrated a corresponding reduction in abundance, suggesting protein palmitoylation may be a critical factor in maintaining protein stability (Figure 3). This finding is not surprising, but highlights an inherent biological consequence that will likely emerge in future broad profiling studies. The caveolae organizing proteins flotillins 1 and 2 (Flot1 and Flot2) were both identified as potential DHHC5 targets. Interestingly, N-terminally myristoylated Flot2 resists degradation in the absence of DHHC5. Overall, this study annotates hundreds of novel palmitoylated proteins in neuronal stem cells, and demonstrates the unfortunate role of palmitoylation in the stabilization of candidate substrates.
Figure 3.
Profiling DHHC5 substrates by quantitative proteomics. SILAC labeled wild type and DHHC5 mutant neuronal stem cells were profiled directly by unenriched proteomics or after 17-ODYA enrichment [48]. Median SILAC ratios of validated palmitoylated proteins observed in both experiments are shown with reference lines signifying 2 standard deviations centered at the mean. The unenriched proteomics experiment is more complex, and hence many palmitoylated proteins were not detected and therefore not shown. Ratios <5 are displayed, which excludes several proteins, including Gjc2, Mydam, Gstk1, and Glb1. Proteins displayed in the upper right quadrant (green shading) display reduced abundance, which corresponds to reduced 17-ODYA enrichment. Essentially no proteins show a singular reduction in palmitoylation (lower right quadrant, red shading) without a corresponding drop in abundance. These results highlight difficulties in direct annotation of DHHC substrates by proteomic approaches.
Conclusions
New chemical enrichment methods have greatly accelerated the annotation and analysis of protein palmitoylation. These approaches are critical for moving away from over-expressed substrates and moving towards experiments in native cells. SILAC and other isotopic mass spectrometry methods facilitate accurate and sensitive profiling across all palmitoylated proteins in biological samples, and will undoubtedly lead to a greater annotation of DHHC substrates and sites of activity-dependent dynamic palmitoylation. A critical future step is to optimize chromatography methods for direct analysis of unmodified palmitoylated peptides, which is fundamentally the best validation for annotating sites of palmitoylation. Altogether, new profiling methods open new opportunities to profile the dynamics of this poorly characterized post-translational modification.
Acknowledgments
Funding is provided by the National Institutes of Health (R00CA151460) and the University of Michigan.
Abbreviations
- ABE
acyl-biotin exchange
- SILAC
Stable Isotope Labeling with Amino acids in Cell culture
- 17-ODYA
17-Octadecynoic acid
- LC-MS
Liquid chromatography mass spectrometry
- HDFP
Hexadecylfluorophosphonate
References
- 1.Schmidt MF, Bracha M, Schlesinger MJ. Evidence for covalent attachment of fatty acids to Sindbis virus glycoproteins. Proc Natl Acad Sci U S A. 1979;76:1687–1691. doi: 10.1073/pnas.76.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt MFG, Schlesinger MJ. Fatty acid binding to vesicular stomatitis virus glycoprotein: a new type of post-translational modification of the viral glycoprotein. Cell. 1979;17:813–819. doi: 10.1016/0092-8674(79)90321-0. [DOI] [PubMed] [Google Scholar]
- 3.Schlesinger M, Magee A, Schmidt M. Fatty acid acylation of proteins in cultured cells. J Biol Chem. 1980;255:10021–10024. [PubMed] [Google Scholar]
- 4.Magee AI, Koyama AH, Malfer C, Wen D, Schlesinger MJ. Release of fatty acids from virus glycoproteins by hydroxylamine. Biochimica et Biophysica Acta (BBA) -General Subjects. 1984;798:156–166. doi: 10.1016/0304-4165(84)90298-8. [DOI] [PubMed] [Google Scholar]
- 5.Rose JK, Adams GA, Gallione CJ. The presence of cysteine in the cytoplasmic domain of the vesicular stomatitis virus glycoprotein is required for palmitate addition. Proc Natl Acad Sci U S A. 1984;81:2050–2054. doi: 10.1073/pnas.81.7.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker TL, Zheng H, Walker J, Coloff JL, Buss JE. Distinct Rates of Palmitate Turnover on Membrane-bound Cellular and Oncogenic H-Ras. J Biol Chem. 2003;278:19292–19300. doi: 10.1074/jbc.M206956200. [DOI] [PubMed] [Google Scholar]
- 7.Wedegaertner PB, Bourne HR. Activation and depalmitoylation of Gs[alpha] Cell. 1994;77:1063–1070. doi: 10.1016/0092-8674(94)90445-6. [DOI] [PubMed] [Google Scholar]
- 8.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 9.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, 3rd, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat Protocols. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 11.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotech. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 12.Jones Matthew L, Collins Mark O, Goulding D, Choudhary Jyoti S, Rayner Julian C. Analysis of Protein Palmitoylation Reveals a Pervasive Role in Plasmodium Development and Pathogenesis. Cell Host & Microbe. 2012;12:246–258. doi: 10.1016/j.chom.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, Green WN, Yates JR, III, Davis NG, El-Husseini A. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome Scale Characterization of Human S-Acylated Proteins in Lipid Raft-enriched and Non-raft Membranes. Molecular & Cellular Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivaldi C, Martin BR, Kieffer-Jaquinod S, Chapel A, Levade T, Garin J, Journet A. Proteomic Analysis of S-Acylated Proteins in Human B Cells Reveals Palmitoylation of the Immune Regulators CD20 and CD23. PLoS One. 2012;7:e37187. doi: 10.1371/journal.pone.0037187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marin EP, Derakhshan B, Lam TT, Davalos A, Sessa WC. Endothelial Cell Palmitoylproteomic Identifies Novel Lipid-Modified Targets and Potential Substrates for Protein Acyl Transferases/Novelty and Significance. Circulation Research. 2012;110:1336–1344. doi: 10.1161/CIRCRESAHA.112.269514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostiuk MA, Corvi MM, Keller BO, Plummer G, Prescher JA, Hangauer MJ, Bertozzi CR, Rajaiah G, Falck JR, Berthiaume LG. Identification of palmitoylated mitochondrial proteins using a bio-orthogonal azido-palmitate analogue. FASEB J. 2008;22:721–732. doi: 10.1096/fj.07-9199com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. Chemical probes for the rapid detection of Fatty-acylated proteins in Mammalian cells. J Am Chem Soc. 2007;129:2744–2745. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 21.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 23.Yount JS, Charron G, Hang HC. Bioorthogonal proteomics of 15-hexadecynyloxyacetic acid chemical reporter reveals preferential targeting of fatty acid modified proteins and biosynthetic enzymes. Bioorganic & Medicinal Chemistry. 2012;20:650–654. doi: 10.1016/j.bmc.2011.03.062. [DOI] [PubMed] [Google Scholar]
- 24.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duncan JA, Gilman AG. Characterization of Saccharomyces cerevisiae Acyl-protein Thioesterase 1, the Enzyme Responsible for G Protein alpha Subunit Deacylation in Vivo. J Biol Chem. 2002;277:31740–31752. doi: 10.1074/jbc.M202505200. [DOI] [PubMed] [Google Scholar]
- 28.Camp L, Hofmann S. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J Biol Chem. 1993;268:22566–22574. [PubMed] [Google Scholar]
- 29.Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, Rusch M, Kramer JW, Rauh D, Coates GW, Brunsveld L, Bastiaens PI, Waldmann H. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449–456. doi: 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- 30.Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, Waldmann H, Bastiaens PI. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141:458–471. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Zhang MM, Tsou LK, Charron G, Raghavan AS, Hang HC. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proceedings of the National Academy of Sciences. 2010;107:8627–8632. doi: 10.1073/pnas.0912306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- 33.Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2006;1761:474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Mukai J, Liu H, Burt RA, Swor DE, Lai WS, Karayiorgou M, Gogos JA. Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nat Genet. 2004;36:725–731. doi: 10.1038/ng1375. [DOI] [PubMed] [Google Scholar]
- 36.Raymond FL, Tarpey PS, Edkins S, Tofts C, O’Meara S, Teague J, Butler A, Stevens C, Barthorpe S, Buck G, Cole J, Dicks E, Gray K, Halliday K, Hills K, Hinton J, Jones D, Menzies A, Perry J, Raine K, Shepherd R, Small A, Varian J, Widaa S, Mallya U, Moon J, Luo Y, Shaw M, Boyle J, Kerr B, Turner G, Quarrell O, Cole T, Easton DF, Wooster R, Bobrow M, Schwartz CE, Gecz J, Stratton MR, Futreal PA. Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid habitus. Am J Hum Genet. 2007;80:982–987. doi: 10.1086/513609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansouri MR, Marklund L, Gustavsson P, Davey E, Carlsson B, Larsson C, White I, Gustavson K-H, Dahl N. Loss of ZDHHC15 expression in a woman with a balanced translocation t(X;15)(q13.3;cen) and severe mental retardation. Eur J Hum Genet. 2005;13:970–977. doi: 10.1038/sj.ejhg.5201445. [DOI] [PubMed] [Google Scholar]
- 38.Huang K, Yanai A, Kang R, Arstikaitis P, Singaraja RR, Metzler M, Mullard A, Haigh B, Gauthier-Campbell C, Gutekunst C-A, Hayden MR, El-Husseini A. Huntingtin-Interacting Protein HIP14 Is a Palmitoyl Transferase Involved in Palmitoylation and Trafficking of Multiple Neuronal Proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 39.Oyama T, Miyoshi Y, Koyama K, Nakagawa H, Yamori T, Ito T, Matsuda H, Arakawa H, Nakamura Y. Isolation of a novel gene on 8p21.3–22 whose expression is reduced significantly in human colorectal cancers with liver metastasis. Genes, Chromosomes and Cancer. 2000;29:9–15. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1001>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 40.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto Y, Chochi Y, Matsuyama H, Eguchi S, Kawauchi S, Furuya T, Oga A, Kang JJ, Naito K, Sasaki K. Gain of 5p15.33 is associated with progression of bladder cancer. Oncology. 2007;72:132–138. doi: 10.1159/000111132. [DOI] [PubMed] [Google Scholar]
- 42.Yu L, Reader JC, Chen C, Zhao XF, Ha JS, Lee C, York T, Gojo I, Baer MR, Ning Y. Activation of a novel palmitoyltransferase ZDHHC14 in acute biphenotypic leukemia and subsets of acute myeloid leukemia. Leukemia. 2011;25:367–371. doi: 10.1038/leu.2010.271. [DOI] [PubMed] [Google Scholar]
- 43.Draper JM, Smith CD. DHHC20: a human palmitoyl acyltransferase that causes cellular transformation. Mol Membr Biol. 2010;27:123–136. doi: 10.3109/09687681003616854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 Constitute a Human Protein Fatty Acyltransferase with Specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 45.Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, Linder ME. 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J Lipid Res. 2009;50:233–242. doi: 10.1194/jlr.M800270-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Hu J, Hofer K, Wong AM, Cooper JD, Birnbaum SG, Hammer RE, Hofmann SL. DHHC5 interacts with PDZ domain 3 of post-synaptic density-95 (PSD-95) protein and plays a role in learning and memory. J Biol Chem. 2010;285:13022–13031. doi: 10.1074/jbc.M109.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas GM, Hayashi T, Chiu SL, Chen CM, Huganir RL. Palmitoylation by DHHC5/8 targets GRIP1 to dendritic endosomes to regulate AMPA-R trafficking. Neuron. 2012;73:482–496. doi: 10.1016/j.neuron.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Martin BR, Cravatt BF, Hofmann SL. DHHC5 protein palmitoylates flotillin-2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. J Biol Chem. 2012;287:523–530. doi: 10.1074/jbc.M111.306183. [DOI] [PMC free article] [PubMed] [Google Scholar]