Abstract
BRCA1 is a tumor suppressor gene that is mutated in families with breast and ovarian cancer. Several BRCA1 splice variants are found in different tissues, but their subcellular localization and functions are poorly understood at the moment. We previously described BRCA1 splice variant BRCA1a to induce apoptosis and function as a tumor suppressor of triple negative breast, ovarian and prostate cancers. In this study we have analyzed the function of BRCA1 isoforms (BRCA1a and BRCA1b) and compared them to the wild type BRCA1 protein using several criteria like studying expression in normal and tumor cells by RNase protection assays, sub cellular localization/fractionation by immunofluorescence microscopy and western blot analysis, transcription regulation of biological relevant proteins and growth suppression in breast cancer cells. We are demonstrating for the first time that ectopically expressed GFP-tagged BRCA1, BRCA1a, and BRCA1b proteins are localized to the mitochondria, repress ELK-1 transcriptional activity and possess antiproliferative activity on breast cancer cells. These results suggest that the exon 9,10 and 11 sequences (aa 263 – 1365) which contain two nuclear localization signals, p53, Rb, c-Myc, γ- tubulin, Stat, Rad 51, Rad 50 binding domains, angiopoietin-1 repression domain are not absolutely required for mitochondrial localization and growth suppressor function of these proteins. Since mitochondrial dysfunction is a hallmark of cancer, we can speculate that the mitochondrial localization of BRCA1 proteins may be functionally significant in regulating both the mitochondrial DNA damage as well as apoptotic activity of BRCA1 proteins and mislocalization causes cancer.
Keywords: BRCA1/1a/1b proteins, breast cancers, mitochondria, growth suppression, transcriptional regulation, ELK-1
Introduction
The human tumor suppressor gene BRCA1 was identified in 1994 as the breast and ovarian cancer susceptibility gene [Futreal, et al., 1994; Miki, et al., 1994; Rosen, et al., 2006; Boulton, 2006]. This gene codes for a 1863 amino acid (aa) protein that is not only expressed in breast and ovary, but is also abundant in the testis and thymus [Miki, et al., 1994]. The BRCA1 gene consists of only 22 coding exons and codes for a protein with a molecular mass of approximately 220 kilo Daltons (kDa). BRCA1 was shown to be localized mainly to the nucleus; however, its sub cellular localization has been questionable since its discovery. Additional research has shown that BRCA1 is secreted and is localized in the cytoplasm and secretory granules [Chen, et al., 1995; Jensen, et al., 1996; Thakur, et al., 1997; Wang, et al., 1997]. Differences in fixation techniques and confocal and immunofluorescence microscopy have also demonstrated the exclusively nuclear [Scully, et al., 1996], nuclear-cytoplasm tube-like invaginations, Golgi complex, endoplasmic reticulum [Coene, et al., 1997; De Potter, et al., 1998] and combined nuclear and cytoplasmic [Thomas, et al.,1996] localization of BRCA1. BRCA1 has been identified as a protein that actively shuttles between the nucleus and cytoplasm [Wang, et al., 1997; Rodriguez, 2000; Fabbro, 2003; Rodriguez, et al., 2004; Henderson, 2005]. Others have characterized the localization of BRCA1 based on tumor type. Chen et al. concluded that BRCA1 is nuclear in normal breast cells but cytoplasmic in cancerous breast cells [Chen, et al., 1995]. Although mostly located in the nucleus and cytoplasm of normal and malignant breast tissue, one group showed that sporadic tumors exhibited nuclear staining while nuclear staining of BRCA1 was reduced or absent in familial and early onset breast cancer [Jarvis, et al., 1998]. However, Taylor et al. gave similar results and also indicated that the absence of nuclear but not cytoplasmic BRCA1 is prevalent in high grade breast tumors and those with lymph node metastasis [Taylor, et al., 1998]. More recently, phosphorylated BRCA1 has been localized to the nucleus and mitochondria [Coene, et al., 2005]. BRCA1 is known to be involved in the maintenance of genomic stability and DNA repair [Boulton, 2006]. In addition to its growth and tumor suppressor functions, BRCA1 and its isoforms induce apoptosis in human breast cancer cells [Chai, et al., 2007; Shao, et al., 1996]. BRCA1 interacts with p53, c-Myc, Rb, p300, BRCA2, Rad51, STAT1, and many other proteins [Rosen, et al., 2006; Boulton, 2006; Mullan, et al., 2006]. The role of BRCA1 in cell cycle control has been illustrated by its ability to interact with various cyclins and cyclin-dependent kinases (CDK's), activate the CDK inhibitor p21 and the p53 tumor suppressor gene, and regulate several genes that control cell cycle checkpoints [Wang, et al., 1997; Mullan, et al., 2006; Chai, et al., 1999]. DNA damage repair is necessary for genome maintenance as defects in DNA damage repair processes can lead to the development of cancer. BRCA1 plays a role in DNA damage repair and genomic stability by repairing double-strand breaks (DSBs) by homologous recombination (HR) [Boulton, 2006; Gudmundsdottir, 2006]. The association of BRCA1 with the RNA polymerase II holoenzyme complex, which is vital for DNA transcription, via RNA helicase A, has implicated BRCA1 in transcriptional regulation [Rosen, et al., 2006; Boulton, 2006; Mullan, et al., 2006; Monteiro, et al., 1996; Scully, et al., 1997; Anderson, et al., 1998].
The amino-terminal RING finger domain, two nuclear localization signals (NLS), and two BRCA1 C-terminal (BRCT) domains are important determinants of BRCA1 structure and function [Mullan, et al., 2006]. The RING domain of BRCA1 spans amino acids 20-64 and contributes to the many protein interactions of BRCA1 via DNA binding and also exhibits an E3 ubiquitin ligase activity [Rosen, et al., 2006; Boulton, 2006; Monteiro, et al., 1996]. The NLS from aa 500-508 and 609-615 verify the primarily nuclear function of BRCA1 [Thakur, et al., 1997; Monteiro, et al., 1996]. The C-terminal domain of BRCA1, aa 1560-1863, has been shown to function as a transcriptional activation domain when fused with a GAL4 DNA binding domain (DBD) [Rosen, et al., 2006; Boulton, 2006; Mullan, et al., 2006; Chai, et al., 1999; Monteiro, et al., 1996; Anderson, et al., 1998; Chapman, 1996; Cui, et al., 1998]. The C-terminal region is extremely important in the function of BRCA1 as many common mutations that predispose women to breast and ovarian cancer are located within this region [Mullan, et al., 2006; Monteiro, et al., 1996; Anderson, et al., 1998; Chapman, 1996]. The importance of the BRCA1 RING domain and C-terminal region is further illustrated by studies that have shown variations in BRCA1 sub cellular localization due to mutations and/or interactions within these domains. BRCA1 and BARD1 associate via their RING domains [Fabbro, 2002]. Two CRM1/exportin pathway-dependent nuclear export sequences (NES) have been identified in BRCA1 [Rodriguez, 2000; Thompson, et al., 2005] and it is thought that BARD1 blocks the BRCA1 NES to inhibit its cytoplasmic localization [Fabbro, 2002]. Common BRCT mutations affect BRCA1 folding and function due to defects in the nuclear localization of BRCA1 [Rodriguez, et al., 2004; Au, 2005]. These results further contribute to the controversy regarding BRCA1 localization.
We have previously isolated and characterized two BRCA1 splice variants, BRCA1a and BRCA1b, that code for p110 and p100 kDa proteins, respectively. Both BRCA1a and BRCA1b have an in-frame deletion of the majority of exon 11 from aa 263-1365. BRCA1b has an additional deletion of exons 9 and 10 [Wang, et al., 1997; Chai, et al., 1999; Cui, et al., 1998; Shao, et al., 1998]. Full-length BRCA1, BRCA1a and BRCA1b are the most evolutionary conserved of all the other splice variants. A fourth predominant splice variant is missing only exons 9 and 10 and is referred to as BRCA1 Δ (9, 10) [Orban, 2003]. Previously, we have found both BRCA1a and BRCA1b to be multifunctional proteins that are involved in transcriptional activation/repression, cell cycle regulation, apoptosis and growth/tumor suppression of triple negative breast, ovarian and prostate cancers [Wang, et al., 1997; Chai, et al., 2007; Chai, et al., 1999; Cui, 1998; et al., Shao, et al., 1998]. In the present study we have analyzed the structure/function relationship of the BRCA1 splice variants and compared them to full-length BRCA1 using criteria such as in vivo expression in normal and tumor cell lines via RNAse protection assays (RPA) and Western blot analysis, sub cellular localization using FLAG or GFP tagged vectors using immunofluorescence and Western blotting, transcriptional repression using biologically relevant promoters and growth suppression assays in breast cancer cells.
Subcellular fractionation and immuno-fluorescence analysis demonstrate that BRCA1, BRCA1a and BRCA1b are localized within the nucleus, cytoplasm, and mitochondria. Localization of BRCA1 and the splice variants within these three compartments validate the known roles of BRCA1 and confirm the roles of the BRCA1 splice variants in cell cycle regulation and apoptosis. Our results suggest that the BRCA1 isoforms have characteristics that are overlapping with full-length BRCA1. The splice variants possess antiproliferative activity, suggesting that the lack of exon 9,10 and part of exon 11 sequences that interacts with proteins like Rb, STAT1, p53, c-Myc, ZBRK1, Rad51 and two nuclear localization signals are not necessary for the tumor suppressor function of these proteins [Mullan, et al., 2006; Chai, et al., 2007]. Furthermore the presence of BRCA1 proteins in the mitochondria strongly suggests that it may be involved both in apoptosis as well as in maintaining the stability of the mitochondrial genome.
Materials and Methods
Cell Lines
A-431, COLO 320, COS1, HBL-100, HeLa, HL-60, K-562, MCF7, MOLT-4, NIH OVCAR-3 and SK-OV-3 cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA) and maintained according to the manufacturer's suggestions. The HLR-ELK1 Trans-Reporter Cell Line was purchased from Stratagene, La Jolla, CA, and maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 250 μg/ml G418 and 100 μg/ml Hygromycin. The CAL51 cell line was generously provided by J. Gioanni and was grown in MEM supplemented with 10% FBS and 1% penicillin-streptomycin [Gioanni, et al., 1990].
Plasmids
BRCA1a, BRCA1b cDNA subcloned into pFLAG-CMV vector (Eastman Kodak Company) has been described previously [Wang, et al., 1997]. BRCA1a and BRCA1b cDNA was inserted into the pEGFP-C1 vector via directional cloning, after gel purification and ligation. pCDNA3 BRCA1a BRCA1b has been described previously [Wang, et al., 1997]. The pcDNA3 BRCA1 plasmid was kindly provided by Dr.Michael Erdos. Elb luciferase and MEK plasmids were kindly provided by Dr. Roger Davis (Howard Hughes Medical Institute, Worcester, MA).
In vivo Expression of BRCA1a and BRCA1b
COS1 cells were transfected with 12μg of pFLAG-CMV –BRCA1a, BRCA1b or empty vector using FuGENE 6 (Roche, Nutley, NJ) according to manufacturer's instructions. Cellular extracts were prepared 48 hours later using RIPA buffer and 75μg of whole cell lysates were resolved on a 10% SDS-PAGE. The blot was probed with secondary anti-FLAG HRP (Sigma A8592, St Louis MO), developed using an ECL detection kit (GE Healthcare, Piscataway, NJ) and exposed using an LAS-3000 system (Fujifilm). COS1 cells were also transfected with pEGFP-C1-BRCA1a/1b clones using FuGENE 6 (Roche) and cellular extracts were prepared and separated and the blot developed and exposed as discussed above. The blot was probed with mouse anti-GFP antibody (Zymed, Carlsbad CA).
RNAase Protection Assay
RNAase protection assay was performed using Ribonuclease protection Assay kit (Ambion Inc., Austin, TX). The DNA template used for the synthesis of radiolabeled RNA probe was constructed using primers that were located in exon 11 (spanning nucleotides 789 to 1011) and sub cloned into pcDNA3 vector. The DNA template was transcribed in presence of 32P-rUTP to obtain the labeled probe. The radio labeled RNA was then gel purified and hybridized with 20ug of total RNA isolated from various breast, ovarian, prostate, cervical carcinoma, lung, colon cancer cells as indicated in the figure 3 legend. Total RNA isolated from COS-1 cells transfected with pcDNA3 BRCA1a was used as a positive control
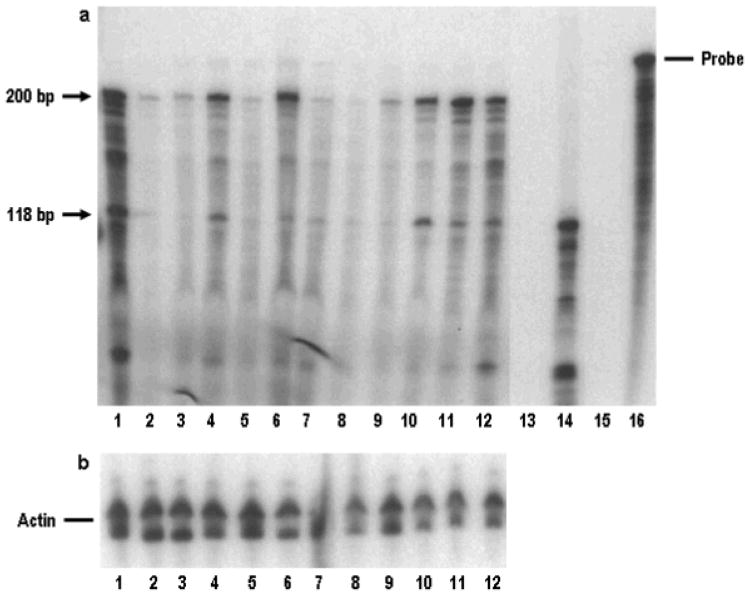
Figure 3.
(a) Analysis of expression BRCA1, BRCA1a and BRCA1b transcripts by RNAase protection assay. RNAase protection assay was carried out on ∼ 30 micrograms of total RNA as described by us previously. Lane1, HBL100; Lane 2, Normal human mammary gland; Lane3, MCF7; Lane 4, CAL-51; Lane 5, SKOV3; Lane 6, NIHOVCAR3; Lane 7, Hela; Lane 8, HL60; Lane 9, A431; Lane 10, COLO320; Lane 11, MOLT-4; Lane 12, K562; Lane 13, COS cells transfected with vector CMV; Lane 14, COS cells transfected with BRCA1a expression plasmid. Protected 200bp (corresponding to BRCA1) and 118bp (corresponding to BRCA1a and BRCAlb) is shown by arrows (b) Represents actin control.
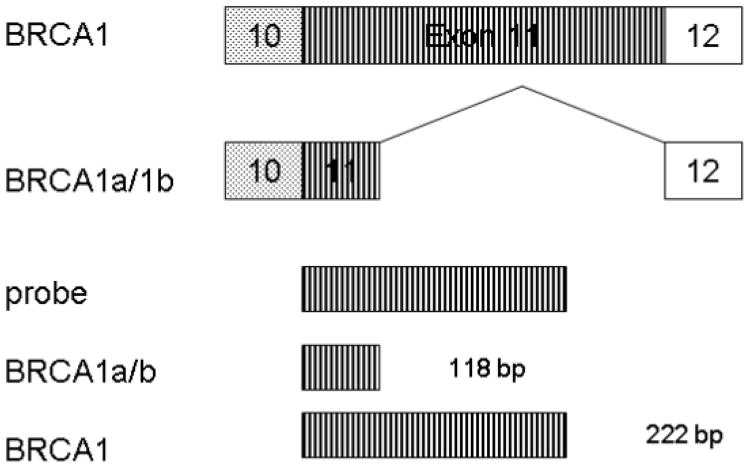
(c) Schematic representation of antisense RNA probe used for RNAase protection assay and also the expected fragments from BRCA1, BRCA1a/1b transcripts are shown.
Luciferase Assays
MCF7 and HLR-ELK1 cells were plated at a density of 6×105 cells per well in 6-well plates. MCF7 cells were cotransfected with 1μg each of GAL4-Elk1 (305-428), MEK, E1bLuc and pCMV β gal. Cells were also transfected with various concentrations of pcDNA3-BRCA1a or BRCA1b or pcDNA3-BRCA1a, BRCA1b Y1853 ter mutants using Lipofectamine™ 2000 (Invitrogen) according to manufacturer's instructions. DNA was held constant at 5μg using Gem3 (Promega). HLR-ELK1 cells contain pFR-Luc and pFA2-Elk1 plasmids stably integrated within their genome. HLR-ELK1 cells were cotransfected with 1μg each of MEK, pCMV β gal, and various concentrations of pcDNA3-BRCA1a, BRCA1b and pcDNA3-BRCA1a and BRCA1b Y1853ter mutants using Lipofectamine™ 2000 (Invitrogen). Forty-eight hours after transfections, cells were harvested and assayed for luciferase activity using a luciferase assay kit (Promega). Transfections were normalized for β-galactosidase activity.
Subcellular Fractionation
COS1 cells were plated at a density of 1×106 cells per 10-cm dish. Cells were transfected with 11μg pEGFPC-BRCA1a or BRCA1b, pEGFPC vector, or pcDNA3-BRCA1 using FuGENE 6 (Roche) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were harvested and nuclear and cytoplasmic extracts were prepared. After trypsinization, the cell pellet was washed in 1× PBS and the pellet was resuspended in 400μl of ice cold buffer A (10mM Hepes, pH 7.9, 50mM NaCl, 1mM EDTA, 0.1mM PMSF, 1mM DTT, Halt™ Protease Inhibitor Cocktail, EDTA-Free (Pierce, Rockford, IL) and incubated on ice for 20 minutes. After incubation, 500μl of ice cold buffer B (buffer A with 0.1% NP-40) was added; the samples were incubated on ice for an additional 20 minutes, and centrifuged at 5000×g for 2 minutes at 4°C. The supernatant containing the cytoplasmic protein was collected. The pellet was resuspended in 400μl buffer A and centrifuged at 5000×g for 2 minutes at 4°C. The supernatant was removed, the pellet resuspended in 100μl of buffer C (20mM Hepes, pH 7.9, 400mM NaCl, 1mM EDTA, 1mM EGTA, 1mM DTT, and 0.1mM PMSF), incubated on ice for 30 minutes, and centrifuged at 5000×g for 2 minutes. The supernatant containing the nuclear protein was collected. Protein concentration was measured using the Bradford method and approximately 80μg of protein was subjected to 8% SDS-PAGE. EGFP-BRCA1a and BRCA1b were detected using mouse anti-GFP (Zymed) and the blot was developed and exposed as described above. Mitochondrial fractions were prepared using a mitochondria isolation kit for cultured cells (Pierce) according to the manufacturer's instructions. Mitochondrial pellets were resuspended in 1× sample buffer and equal volumes were loaded on 4-20% gradient gels. Cytochrome c was detected using anti-cytochrome c H-104 (Santa Cruz).
Immunofluorescence
MCF7 cells were plated at a density of 6×105 cells per well onto coverslips in 6-well plates the day before transfection. Cells were transfected with 4μg of EGFP-BRCA1, BRCA1a, BRCA1b using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. Twenty-four hours post-transfection, the cells were stained with 100nM MitoTracker® for 45 minutes at 37°C and then washed with complete medium. The cells were washed twice with 1× PBS and fixed with 4% paraformaldehyde (pH 7.2) at room temperature for 20 minutes. Cells were washed three times for 5 minutes each and permeabilized in 0.2% Triton-X 100 in 1× PBS at room temperature for 5 minutes. Cells were washed with 1× PBS and blocked in 10% BSA/PBS for 30 minutes at 37°C. After blocking, the cells were probed with anti-GFP mouse monoclonal antibody (Invitrogen) at a 1:200 dilution in 3% BSA/PBS for one hour at room temperature. The primary antibody was removed, and the cells were washed with 1× PBS and probed with Alexa Fluor® 488 goat anti-mouse IgG (H+L) (Molecular Probes) at 10μg/ml in 3% BSA/PBS for 45 minutes at room temperature. Nuclei were stained using 4′, 6-Diamidino-2-phenylindole dihydrochloride (DAPI; Sigma) for 2 minutes. The coverslips were washed with 1× PBS and mounted onto glass slides using UltraCruz™ Mounting Medium (Santa Cruz). Images were taken with an Olympus epifluorescent microscope with a digital camera.
Colony Suppression Assays
MCF-7 cells were plated at a concentration of 1.5 × 105 cells per 100 mm plate in 10ml of complete media and transfected with pcDNA3 or pcDNA3 BRCA1, BRCA1a and BRCA1b plasmids by using the calcium phosphate kit from Promega. Thirty six to forty eight hours later cells were trypsinized and plated directly into complete medium containing 600 μg/ml G418. Cells were fed with fresh medium containing G418 every 3-4 days. Cells were stained for colonies approximately 21 days after transfections using crystal violet blue as described previously [Chai, et al., 2001].
Results
In vivo expression of BRCA1a and BRCA1b Proteins
In order to determine if the BRCA1 splice variants could be expressed in vivo, COS-1 cells were transfected with BRCA1a or BRCA1b using FLAG and GFP vectors. Both BRCA1a (110 kDa) and BRCA1b (100 kDa) were expressed in COS1 cells using an anti-FLAG antibody (Figure 2).
Figure 2.
Western blot analysis of BRCA1a and BRCA1b expression in Cos-1 cells. Cos-1 cells were transfected with 12 μg of CMVFlag BRCA1a/1b and CMVFlag vector. The whole cell lysates (∼75 μg) were resolved on a 10 % SDS-PAGE. The blot was probed with an anti-FLAG antibody as described previously.
Analysis of Expression of BRCA1, BRCA1a and BRCA1b Transcripts by RNAase protection assay
We studied the expression of BRCA1, BRCA1a, BRCA1b transcripts in different human normal and cancer cells using Ribonuclease protection assay (RPA) which is a very sensitive procedure for the detection and quantification of RNA. We observed expression of BRCA1 and BRCA1a/1b transcripts as represented by the predicted 220 bp (BRCA1) and 118 bp (BRCA1a and BRCA1b) protected fragment in normal mammary gland as well as in several cancer cells (Figure 3, HBL-100, CAL-51, NIH OVCAR3, COLO320, MOLT-4 and K562 cells). In some cancer cells like MCF-7 (lane 3), NIH OVCAR3 (lane 6), A431 (lane 9), MOLT4 (lane 11) and K562 (lane 12) the ratio of full length to the isoforms was altered when compared to normal mammary gland. It should be noted that RNAase protection assay does not distinguish between BRCA1a and BRCA1b transcripts.
Effect of BRCA1a and BRCA1b on GAL4-Elk1 Transcriptional Activation
Elk-1 is an ETS-like gene that codes for a DNA binding protein that is involved in transcriptional activation, proliferation, differentiation, an activator of MAP kinases and induces apoptosis [Shao, et al., 1998; Rao, et al., 1989; Hipskind, et al., 1991; Rao, 1994]. Elk-1 forms a ternary complex in conjunction with the serum response element (SRE) and serum response factor (SRF) to activate c-Fos [Rao, 1994]. Previous studies in our laboratory have shown the splice variants BRCA1a and BRCA1b to associate with Elk-1 and to inhibit MAP kinase induced activity of c-Fos [Chai, et al., 2001]. In order to determine any difference between wild type and mutant BRCA1a and BRCA1b proteins on GAL4-Elk1 transcriptional activation, MCF7 breast cancer cells and HLR-ELK1 cells were transfected with various concentrations of BRCA1a and BRCA1b. HLR-ELK1 cells are a double-stable cell line that contains pFR-Luc and pFA2-Elk1 plasmids stably integrated within their genome. Phosphorylation of the transcriptional activation domain of Elk-1 by MAP kinase will activate transcription of a luciferase reporter plasmid in these cells. Luciferase activity levels serve as an indication of the activation or repression of signaling events. Similarly, MCF7 breast cancer cells were cotransfected with GAL4-Elk1 (305-428), MEK, E1bLuciferase reporter and pCMV β– Gal and various concentrations of BRCA1a and BRCA1b. Both BRCA1a and BRCA1b were shown to decrease the relative luciferase activity as compared to the empty vector control (Figure 4a), indicating inhibition of Elk1 transcriptional activity. Because HLR-ELK1 cells already contain Elk1 and a luciferase reporter plasmid integrated in the genome, these cells were cotransfected with MEK, pCMV β–Gal, BRCA1a and BRCA1b. Both BRCA1a and BRCA1b proteins were shown to decrease the relative luciferase activity and therefore Elk-1 activity in HLR-ELK1 cells (Figure 4b). Several germ line mutations in the BRCA1 gene have been identified and are known to be associated with the early onset of breast and ovarian cancers. The change of a tyrosine to a stop codon at position 1853 of BRCA1 (Y1853 ter mutant) leads to a truncated protein eleven amino acids from the C-terminal end [Monteiro, et al., 1996; Friedman, et al., 1994]. Because this mutation is located within the BRCA1 BRCT domain, which is involved in transcriptional activation and protein interactions, it is hypothesized that this mutation will disrupt the ability of BRCA1 to transcriptionally activate or repress Elk-1 [Monteiro, et al., 1996]. MCF7 breast cancer cells and HLR-ELK1 cells were cotransfected with BRCA1a and BRCA1b Y1853 ter mutants. These mutants failed to repress the activation of Elk-1 (Figures 4a and b). These results suggest BRCA1a and BRCA1b proteins to regulate the activity of ELK-1 protein.
Figure 4.
Effect of BRCA1a and BRCA1b on GAL-4-ELK-1 transcriptional activation. (a) MCF7 (b) HLR-ELK-1 cells were plated at a density of 6×105 cells per well in 6-well plates. Cells were cotransfected with 1 μg each of GAL-4-Elk-1, MEK, E1bLuc and pCMV βgal. Cells were also transfected with various concentrations of BRCA1a/BRCA1b, BRCA1a/BRCA1b Y1853 ter mutants and pcDNA3 vector using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. DNA was held constant at 5 μg using Gem3. Forty-eight hours after transfections, cells were harvested and assayed for luciferase activity using a luciferase assay system (Promega). Transfections were normalized for β-galactosidase activity. Values represent relative luciferase activity as percent of pcDNA3 vector control.
Sub cellular Localization of BRCA1, BRCA1a and BRCA1b proteins
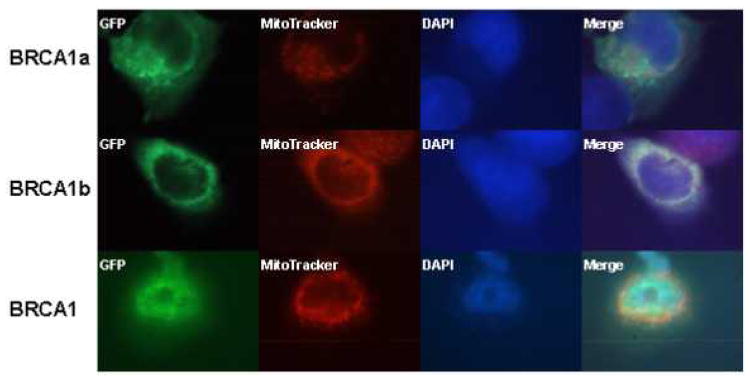
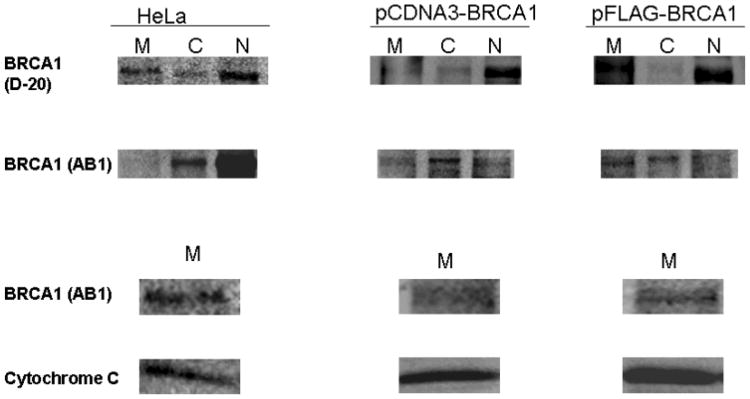
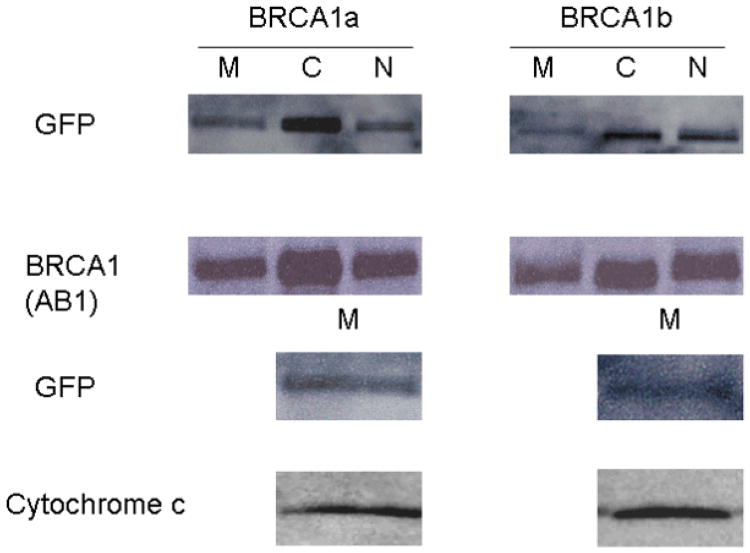
The sub cellular localization of BRCA1 and its splice variants have remained controversial. We have found wild type BRCA1 and BRCA1a to be localized in the nucleus as well as in the cytoplasm [Wang, et al., 1997; Rao, et al., 1996]. Others have shown BRCA1 to be a nuclear protein that resides in secretory granules and also in the cytoplasm [Chen, et al., 1995; Jensen, et al., 1996; Thakur, et al., 1997; Wang, et al., 1997]. Recent studies using immunoelectron microscopy results show endogenous BRCA1 protein to be localized in the mitochondria [Coene, et al., 2005]. To study the localization of BRCA1, BRCA1a and BRCA1b proteins within the cell, we have used GFP -tagged BRCA1 proteins. Immunofluorescence in MCF7 cells that have been transfected with EGFP-BRCA1/BRCA1a/BRCA1b using GFP antibodies showed the localization of BRCA1, BRCA1a and BRCA1b proteins also in the mitochondria (Figure5a). The distribution of BRCA1/BRCA1a/BRCA1b differed between cell to cell as observed previously with BRCA1 using different antibodies (Wang, et al,. 1997; Coene, et al., 2005; Rao, et al., 1996.). COS1 cells were transfected with pCDNA3-BRCA1, EGFP-BRCA1a, EGFP-BRCA1b and nuclear, cytoplasmic and mitochondrial extracts were prepared and subjected to SDS-PAGE and Western blot analysis using BRCA1 and GFP antibodies. Both endogenous as well as ectopically expressed BRCA1 proteins were found in the nucleus; cytoplasm as well as mitochondria using different BRCA1 antibodies (Figure 5b). Coene et al. used confocal and immunoelectron microscopy to illustrate the presence of endogenous BRCA1 in the mitochondria, similar to our results [Coene, et al., 2005]. The EGFP-BRCA1a and EGFP-BRCA1b fusion protein at 137 and 127 kDa, respectively, were also found in mitochondrial, cytoplasmic and nuclear fractions with anti-GFP and anti-BRCA1 antibodies via Western blot analysis and immunofluorescence (Figure 5c). Western blotting of the mitochondrial fractions with cytochrome C further supports the mitochondrial localization of these proteins (Figure 5c). We have found for the first time that epitope-tagged BRCA1, BRCA1a and BRCA1b proteins to be located within mitochondria similar to endogenous BRCA1. Based on these observations we can speculate that the mitochondrial localization of BRCA1 proteins may be linked to their apoptotic function.
Figure 5.
a Mitochondrial localization of BRCA1, BRCA1a, and BRCCA1b. The GFP tagged BRCA1, BRCA1a, or BRCA1b was transiently expressed in MCF-7. The tagged proteins were detected by immunostaining with anti-GFP antibody. MitoTracker staining and DAPI staining are shown in parallel. BRCA1, BRCA1a and BRCA1b proteins are shown to co-localize with the Mitotracker (red color) which specifically stains the mitochondria.
(b) BRCA1 Localization and Expression in COS1 and HeLa Cells. Cells were transfected with pCDNA3-BRCA1 or pFLAG-BRCA1 using Fugene 6 (Roche). Mitochondria, cytoplasmic and nuclear extracts were prepared 48 hours after transfection using a mitochondrial isolation kit for cultured cells (Pierce). Extracts were also prepared from HeLa cells. BRCA1 was detected by Western blot analysis using BRCA1 (D-20; Santa Cruz) or BRCA1 (Ab-1; Calbiochem) Cytochrome c was detected using cytochrome-c (H-104; Santa Cruz). M: mitochondria; C: cytoplasm; N: nucleus. The column labeled Hela represents the endogenous BRCA1 isolated from Hela cells and the columns labeled pCDNA-BRCA1 and pFLAG-BRCA1 refers to the COS-1 transfected BRCA1 proteins.
(c) BRCA1a and BRCA1b Localization and Expression in COS1 Cells. Cells were transfected with pEGFP-C1-BRCA1a/1b using Fugene 6 (Roche). Mitochondria, cytoplasmic and nuclear extracts were prepared 48 hours after transfection using a mitochondrial isolation kit for cultured cells (Pierce). BRCA1 was detected by Western blot analysis using Anti-GFP (Zymed) or BRCA1 (Ab-1; Calbiochem Cytochrome c was detected using cytochrome c (H-104; Santa Cruz). M: mitochondria; C: cytoplasm; N: nucleus.
BRCA1, BRCA1a and BRCA1b proteins suppress growth of human breast cancer cells
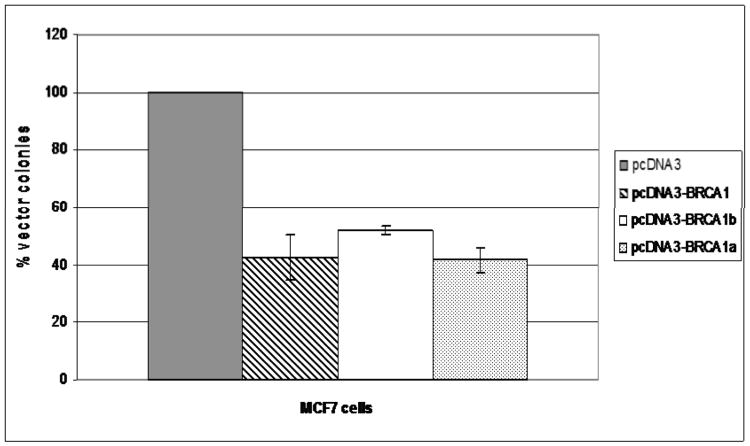
BRCA1, BRCA1a and BRCA1b proteins have been shown to inhibit the growth of breast, ovarian, lung and colon cancer cells [Chai, et al., 2007; Holt, et al., 1996; Aprelikova, et al., 1999]. In an attempt to compare the growth suppressive function of BRCA1, BRCA1a and BRCA1b proteins MCF-7 breast cancer cells were transfected with either a BRCA1 or BRCA1a or BRCA1b or empty vector and selected with G418 and the colonies were counted after 21 days following staining with crystal violet. Over expression of BRCA1 and BRCA1a resulted in 50 – 60% and BRCA1b 50% reduction in the number of G418 – resistant colonies compared to vector alone (Figure 6). These results suggest BRCA1a and BRCA1b to inhibit growth of breast cancer cells similar to BRCA1.
Figure 6.
Effect of BRCA1, BRCA1a and BRCA1b on the growth of MCF7 breast cancer cells. Cells were co-transfected with pcDNA3 or pcDNA BRCA1, BRCA1b, BRCA1a and selected with G418 and the resulting colonies were stained and counted. The number of colonies obtained by pcDNA3 clone was considered as 100%. Each experiment was repeated at least three times and the bars shown represent standard deviation.
Discussion
In the present report, we have presented data concerning the structure/functional relationship of the BRCA1 splice variants BRCA1a and BRCA1b to wild type BRCA1. Our results compare the in vivo expression, sub cellular localization, transcriptional regulation and growth suppression of BRCA1, BRCA1a and BRCA1b proteins. Both variants are missing the majority of the exon 11 sequences (263-1365) while BRCA1b contains an additional deletion of exons 9 and 10. We have studied the expression of BRCA1 and its isoforms using Western blot analysis and RNase protection assays (RPA). BRCA1a/1b was expressed in COS1 cells using FLAG vectors. Using RPA, we were able to illustrate RNA expression of BRCA1 and BRCA1a/BRCA1b in normal human mammary gland and several cancer cells like HBL-100, A-431, CAL-51, COLO 320, HeLa, HL-60, K-562, MCF7, MOLT-4, and NIH: OVCAR-3 and SK-OV-3 (Figure 3). In some of the cancer cells like MCF-7, NIH OVCAR3, A431, MOLT4 and K562 cells the ratio of full length to the isoforms was altered compared to the normal mammary gland. These results suggest that mere down regulation of expression of BRCA1a or BRCA1b can cause cancers. Elk-1 is an ETS-related gene that plays a role in transcriptional activation, proliferation, and differentiation. Elk-1 also induces apoptosis and forms a ternary complex with SRE and SRF to activate the c-Fos gene [Shao, et al., 1998; Rao, et al., 1989; Hipskind, et al., 1991; Rao, 1994]. We have previously shown BRCA1a and BRCA1b proteins to associate with Elk-1 and inhibit c-Fos Oncogene activation mediated by MAP kinases [Chai, et al., 2001]. Fos plays a pivotal role in the regulation of normal cell growth and transformation and is induced by growth factors and estrogen in breast cancer cells. To study the effect of wild type and disease associated Y1853 termination mutant BRCA1a/BRCA1b proteins on Elk-1 transcriptional activation we used a GAL4-Elk-1 fusion vector. Phosphorylation of the transcriptional activation domain of Elk-1 by a MAP kinase activates transcription of a luciferase reporter plasmid and the luciferase activity will serve as an indication of signaling events. Our results indicate BRCA1a and BRCA1b to inhibit Elk-1 transcriptional activation as shown by the decrease in luciferase activity in MCF7 breast cancer cells and the double stable cell line HLR-ELK1. Several germline mutations in the BRCA1 gene are located within the BRCA1 C-terminal region [Mullan, et al., 2006; Monteiro, et al., 1996; Anderson, et al., 1998; Chapman, 1996]. The BRCT domain plays an important role in BRCA1 transcriptional activation and interaction with various other proteins. It has been suggested that mutations within this region predispose women to develop cancer due to the inability of BRCA1 to activate the transcription of genes [Monteiro, et al., 1996]. Therefore, we hypothesized that BRCA1a/BRCA1b Y1853 termination mutants would fail to inhibit Elk-1 activation. The results presented in this study show that the mutants were impaired in their ability to inhibit Elk-1 activation. Previously estrogen (E2) -induced activation (phosphorylation) of Elk-1 protein was shown to be an important downstream target of E2-dependent Ras-MAPK signaling to proliferation in breast cancer cells (Duan, et al., 2001). These results suggest that one of the mechanisms whereby BRCA1 isoforms suppress cell growth is through inhibition of Elk-1 transcriptional activation. Our results also indicate that BRCA1 mutations within the BRCT domain predispose women to breast and/or ovarian cancer development by failing to inhibit Elk-1 activation, a gene involved in cell growth and proliferation.
As mentioned previously, the sub cellular localization of BRCA1 has been controversial. Researchers have shown BRCA1 to be a mainly nuclear localized protein that is also found in the cytoplasm and secretory granules [Chen, et al., 1995; Jensen, et al., 1996; Thakur, et al., 1997; Wang, et al., 1997; Rao, et al., 1996]. More recent studies have illustrated mitochondrial localization of phosphorylated BRCA1 [Coene, et al., 2005]. A variant of BRCA1, BRCA1 delta 672-4095, in which the entire exon 11 sequences is missing, produces a 97 kDa protein that is localized exclusively in the cytoplasm [Thakur, et al., 1997]. This led to the conclusion that there is a nuclear localization signal within exon 11 [Thakur, et al., 1997]. Others have identified a splice variant called BRCA1 delta 11b, which is equivalent to BRCA1a, to be found exclusively in the cytoplasm [Wilson, et al., 1997]. A recently identified BRCA1 locus product, BRCA1-IRIS, whose mRNA contains an uninterrupted open reading frame from codon 1 of full-length BRCA1 and continues 34 triplets into intron 11, has been shown to positively influence DNA replication and is exclusively nuclear and chromatin-associated [Elshamy, 2004; Kvist, et al., 2005; Elshamy, 2005]. We have previously observed BRCA1 and BRCA1a to be localized to both nucleus as well as cytoplasm [Wang, et al., 1997]. Here we have used immunofluorescence and sub cellular fractionation followed by Western blot analysis to determine the location of BRCA1a/1b within the cell. The results of our immunofluorescence and Western blot studies showed BRCA1a and BRCA1b to be localized both in the nucleus and cytoplasm. The fact that BRCA1a and BRCA1b proteins are localized in the nucleus even though they are missing the majority of exon 11 which includes the NLS sequences indicates that the BRCA1 splice variants may contain a cryptic NLS or may use an alternate mechanism for their nuclear import [Fabbro, et al., 2002; Au, 2005]. Our studies demonstrate for the first time the mitochondrial localization of BRCA1 splice variants BRCA1a and BRCA1b using immunofluorescence and Western blot analysis. The presence of BRCA1a and BRCA1b within the mitochondria seems logical due to the role of BRCA1 and the splice variants in apoptosis [Henderson, 2005; Shao, et al., 1996; Fabbro, et al., 2002]. Mitochondria synthesize their own DNA [Coene, et al., 2005] but rely upon the cell nucleus for repair, transcription, translation and replication [Penta, et al., 2001]. Mitochondrial DNA mutations and genome instability have been reported in breast cancer [Penta, et al., 2001; Bianchi, et al., 2001; Parrella, et al., 2001]. Proteins are imported into the mitochondria by a translocation system in the outer and inner membranes [Paschen, 2001]. Although NLS sequences have been identified in BRCA1, no such sequences have been identified that target BRCA1 to mitochondria. Localization of BRCA1, BRCA1a, and BRCA1b in mitochondria leads one to propose that these proteins may also be involved in mitochondrial DNA repair. Therefore, it remains to be determined if patients who develop cancer due to BRCA1 mutations are impaired in apoptotic function due to mislocalization of BRCA1.
In summary, the results in this present study show the BRCA1 proteins to be multifunctional proteins that are associated with proteins involved in transcriptional activation and/or repression, cell cycle regulation, growth suppression and apoptosis [Rosen, et al., 2006; Boulton, 2006; Wang, et al., 1997; Shao, et al., 1996; Mullan, et al., 2006; Shao, et al., 1998; Rao, et al., 1996; Lu, 2000]. We have compared the structure & function of BRCA1a/1b and compared them to wild type BRCA1 using in vivo expression, RNase protection assays, transcriptional regulation using biologically relevant promoters to study their role in transcriptional activation/repression, growth suppression, and subcellular localization studies using immunofluorescence and Western blotting analysis. We demonstrated the anti proliferative function of BRCA1 proteins using growth suppression assays. The results from these studies suggest that the BRCA1 isoforms have overlapping functions and they possess antiproliferative activity in breast cancer cells. The exon 9,10 and 11 sequences possess two nuclear localization signals and binding domains for p53, Rb, c-Myc, STAT1, Rad50 and Rad51, which are absent in the variants BRCA1a and BRCA1b proteins. The absence of this region illustrates that these sequences are not necessary for the growth/tumor suppressor function of the BRCA1 splice variants. Since mitochondrial dysfunction is a hallmark of cancer, we can speculate that the mitochondrial localization of BRCA1 proteins may be functionally significant for the tumor suppressive and apoptotic activity of BRCA1 proteins.
Figure 1.
Schematic representation of the BRCA1, BRCA1a, and BRCA1b isoforms (Not drawn to scale).
Acknowledgments
We thank J. Gioanne and Jean Louis Fischel of Oncopharmcologic laboratories for the CAL-51 cell line and Dr. Michael Erdos (National Human Genome Research Institute, Bethesda, MD, USA) for generously providing the BRCA1 plasmid. We also thank all the other members of Rao and Reddy labs for their help. We thank the Morehouse School of Medicine RCMI Core facilities for their help. This work was funded in part by Georgia Cancer Coalition Distinguished Cancer Scholar Award, NIH-NCRR-RCMI grant G-12-RR03034, U54 and P60 grants to V.N. Rao, Georgia Cancer Coalition Distinguished Cancer Scholar Award to E.S.P. Reddy.
References
- Anderson S, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- Aprelikova O, Fan BS, Meissner EG, Cotter S, Campbell M, Kuthiala A, et al. BRCA1 associated growth arrest is RB-dependant. PNAS. 1999;96:11867–11871. doi: 10.1073/pnas.96.21.11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W, Henderson BR. The BRCA1 RING and BRCT domains cooperate in targeting BRCA1 to ionizing radiation-induced nuclear foci. J Biol Chem. 2005;280:6993–7001. doi: 10.1074/jbc.M408879200. [DOI] [PubMed] [Google Scholar]
- Bianchi N, Bianchi MS, Richard SM. Mitochondrial genome instability in human cancers. Mutant Res. 2001;488:9–23. doi: 10.1016/s1383-5742(00)00063-6. [DOI] [PubMed] [Google Scholar]
- Boulton S. Cellular functions of the BRCA1 tumor-suppressor proteins. Biochem SOC Trans. 2006;34:633–645. doi: 10.1042/BST0340633. [DOI] [PubMed] [Google Scholar]
- Chai Y, Chipitsyna G, Cui J, et al. C-Fos oncogene regulator Elk-1 interacts with BRCA1 splice variants BRCA1a/1b and enhances BRCA1a/1b-mediated growth suppression in breast cancer cells. Oncogene. 2001;20:1357–1367. doi: 10.1038/sj.onc.1204256. [DOI] [PubMed] [Google Scholar]
- Chai Y, Cui J, Shao N, Reddy ESP, Rao VN. The second BRCT domain of BRCA1 proteins interacts with p53 and stimulates transcription from the p21WAF1/CIP1 promoter. Oncogene. 1999;18:263–268. doi: 10.1038/sj.onc.1202323. [DOI] [PubMed] [Google Scholar]
- Chai Y, Shao N, Rao R, Aysola P, Reddy V, Oprea-llies G, et al. BRCA1a has antitumor activity in TN breast, ovarian and prostate cancers. Oncogene. 2007;26:6031–6037. doi: 10.1038/sj.onc.1210420. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chapman M, Verma IM. Transcriptional activation by BRCA1. Nature. 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen CF, Riley DJ, et al. Aberrant subcellular localization of BRCA1 in breast cancer. Science. 1995;270:789–791. doi: 10.1126/science.270.5237.789. [DOI] [PubMed] [Google Scholar]
- Coene E, Hollinshead MS, Waeytens AA, et al. Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol Biol Cell. 2005;16:997–1010. doi: 10.1091/mbc.E04-10-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coene E, Van OP, Willems K, van EJ, De Potter CR. BRCA1 is localized in cytoplasmic tube-like invaginations in the nucleus. Nat Genet. 1997;16:122–124. doi: 10.1038/ng0697-122. [DOI] [PubMed] [Google Scholar]
- Cui J, Wang H, Reddy ES, Rao VN. Differential transcriptional activation by the N-terminal region of BRCA1 splice variants BRCA1a and BRCA1b. Oncol Rep. 1998;5:585–589. doi: 10.3892/or.5.3.585. [DOI] [PubMed] [Google Scholar]
- De Potter C, Coene ED, Schelfhout VR. Localization of BRCA1 protein at the cellular level. J Mammary Gland Biol Neoplasia. 1998;3:423–429. doi: 10.1023/a:1018740216630. [DOI] [PubMed] [Google Scholar]
- Duan R, Xie W, Burghardt RC, Safe S. Estrogen receptor-mediated activation of the serum response element in MCF-7 cells through MAPK-dependent phosphorylation of Elk-1. J Biol Chem. 2001;276:11590–11598. doi: 10.1074/jbc.M005492200. [DOI] [PubMed] [Google Scholar]
- Elshamy W, Livingston DM. Identification of BRCA1-IRIS, a BRCA1 locus product. Nat Cell Biol. 2004;6:954–967. doi: 10.1038/ncb1171. [DOI] [PubMed] [Google Scholar]
- Elshamy W, Livingston DM. Promoter usage of BRCA1-IRIS. Nat Cell Biol. 2005;7:326. doi: 10.1038/ncb0405-326a. [DOI] [PubMed] [Google Scholar]
- Fabbro M, Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp Cell Res. 2003;282:59–69. doi: 10.1016/s0014-4827(02)00019-8. [DOI] [PubMed] [Google Scholar]
- Fabbro M, Rodriguez JA, Baer R, Henderson BR. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J Biol Chem. 2002;277:21315–21324. doi: 10.1074/jbc.M200769200. [DOI] [PubMed] [Google Scholar]
- Friedman L, Ostermeyer EA, Szabo CI, et al. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- Futreal P, Liu Q, Shattuck-Eidens D, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- Gioanni J, Le FD, Zanghellini E, et al. Establishment and characterisation of a new tumorigenic cell line with a normal karyotype derived from a human breast adenocarcinoma. Br J Cancer. 1990;62:8–13. doi: 10.1038/bjc.1990.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- Henderson B. Regulation of BRCA1, BRCA2 and BARD1 intracellular trafficking. Bioessays. 2005;27:884–893. doi: 10.1002/bies.20277. [DOI] [PubMed] [Google Scholar]
- Hipskind R, Rao VN, Mueller CG, Reddy ES, Nordheim A. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature. 1991;354:531–534. doi: 10.1038/354531a0. [DOI] [PubMed] [Google Scholar]
- Holt J, Thompson ME, Szabo C, Robinson-Benion C, Arteaga CL, King MC, et al. Growth retardation and tumor inhibition by BRCA1. Nat Genetics. 1996;12:298–302. doi: 10.1038/ng0396-298. [DOI] [PubMed] [Google Scholar]
- Jarvis E, Kirk JA, Clarke CL. Loss of nuclear BRCA1 expression in breast cancers is associated with a highly proliferative tumor phenotype. Cancer Genet Cytogenet. 1998;101:109–115. doi: 10.1016/s0165-4608(97)00267-7. [DOI] [PubMed] [Google Scholar]
- Jensen R, Thompson ME, Jetton TL, et al. BRCA1 is secreted and exhibits properties of a granin. Nat Genet. 1996;12:303–308. doi: 10.1038/ng0396-303. [DOI] [PubMed] [Google Scholar]
- Kvist A, Rovira C, Borg A, Medstrand P. Promoter usage of BRCA1-IRIS. Nat Cell Biol. 2005;7:325–326. doi: 10.1038/ncb0405-325. [DOI] [PubMed] [Google Scholar]
- Lu M, Arrick BA. Transactivation of the p21 promoter by BRCA1 splice variants in mammary epithelial cells: evidence for both common and distinct activities of wildtype and mutant forms. Oncogene. 2000;19:6351–6360. doi: 10.1038/sj.onc.1204025. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Monteiro A, August A, Hanafusa H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci USA. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan P, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- Orban T, Olah E. Emerging roles of BRCA1 alternative splicing. Mol Pathol. 2003;56:191–197. doi: 10.1136/mp.56.4.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrella P, Xiao Y, Fliss M, et al. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001;61:7623–7626. [PubMed] [Google Scholar]
- Paschen S, Neupert W. Protein import into mitochondria. IUBMB Life. 2001;52:101–112. doi: 10.1080/15216540152845894. [DOI] [PubMed] [Google Scholar]
- Penta J, Johnson FM, Wachsman JT, Copeland WC. Mitochondrial DNA in human malignancy. Mutant Res. 2001;488:119–133. doi: 10.1016/s1383-5742(01)00053-9. [DOI] [PubMed] [Google Scholar]
- Prasher D, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- Rao V, Huebner K, Isobe M, ar-Rushdi A, Croce CM, Reddy ES. Elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science. 1989;244:66–70. doi: 10.1126/science.2539641. [DOI] [PubMed] [Google Scholar]
- Rao V, Reddy ES. Elk-1 proteins interact with MAP kinases. Oncogene. 1994;9:1855–1860. [PubMed] [Google Scholar]
- Rao V, Shao N, Ahmad M, Reddy ES. Antisense RNA to the putative tumor suppressor gene BRCA1 transforms mouse fibroblasts. Oncogene. 1996;12:523–528. [PubMed] [Google Scholar]
- Rodriguez J, Au WW, Henderson BR. Cytoplasmic mislocalization of BRCA1 caused by cancer-associated mutations in the BRCT domain. Exp Cell Res. 2004;293:14–21. doi: 10.1016/j.yexcr.2003.09.027. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Henderson BR. Identification of a functional nuclear export sequence in BRCA1. J Biol Chem. 2000;275:38589–38596. doi: 10.1074/jbc.M003851200. [DOI] [PubMed] [Google Scholar]
- Rosen E, Fan S, Ma Y. BRCA1 regulation of transcription. Cancer Letters. 2006;236:175–185. doi: 10.1016/j.canlet.2005.04.037. [DOI] [PubMed] [Google Scholar]
- Scully R, Anderson SF, Chao DM, et al. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Ganesan S, Brown M, et al. Location of BRCA1 in human breast and ovarian cancer cells. Science. 1996;272:123–126. doi: 10.1126/science.272.5258.123. [DOI] [PubMed] [Google Scholar]
- Shao N, Chai Y, Cui JQ, et al. Induction of apoptosis by Elk-1 and deltaElk-1 proteins. Oncogene. 1998;17:527–532. doi: 10.1038/sj.onc.1201931. [DOI] [PubMed] [Google Scholar]
- Shao N, Chai YL, Reddy ES, Rao VN. Induction of apoptosis by the tumor suppressor protein BRCA1. Oncogene. 1996;13:1–7. [PubMed] [Google Scholar]
- Taylor J, Lymboura M, Pace PE, et al. An important role for BRCA1 in breast cancer progression is indicated by its loss in a large proportion of non-familial breast cancers. Int J Cancer. 1998;79:334–342. doi: 10.1002/(sici)1097-0215(19980821)79:4<334::aid-ijc5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Thakur S, Zhang HB, Peng Y, et al. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol Cell Biol. 1997;17:444–452. doi: 10.1128/mcb.17.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Smith M, Rubinfeld B, Gutowski M, Beckmann RP, Polakis P. Subcellular localization and analysis of apparent 180-kDa and 220-kDa proteins of the breast cancer susceptibility gene, BRCA1. J Biol Chem. 1996;271:28630–28635. doi: 10.1074/jbc.271.45.28630. [DOI] [PubMed] [Google Scholar]
- Thompson M, Robinson-Benion CL, Holt JT. An amino-terminal motif functions as a second nuclear export sequence in BRCA1. J Biol Chem. 2005;280:21854–21857. doi: 10.1074/jbc.M502676200. [DOI] [PubMed] [Google Scholar]
- Wang H, Shao N, Ding QM, Cui J, Reddy ES, Rao VN. BRCA1 proteins are transported to the nucleus in the absence of serum and splice variants BRCA1a, BRCA1b are tyrosine phosphoproteins that associate with E2F, cyclins and cyclin dependent kinases. Oncogene. 1997;15:143–157. doi: 10.1038/sj.onc.1201252. [DOI] [PubMed] [Google Scholar]
- Wilson C, Payton MN, Elliott GS, et al. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene. 1997;14:1–16. doi: 10.1038/sj.onc.1200924. [DOI] [PubMed] [Google Scholar]