Abstract
The family Polyomaviridae is comprised of circular double-stranded DNA viruses, several of which are associated with diseases, including cancer, in immunocompromised patients. Here we describe a novel polyomavirus recovered from the fecal microbiota of a child in Malawi, provisionally named STL polyomavirus (STLPyV). We detected STLPyV in clinical stool specimens from USA and The Gambia at up to 1% frequency. Complete genome comparisons of two STLPyV strains demonstrated 5.2% nucleotide divergence. Alternative splicing of the STLPyV early region yielded a unique form of T antigen, which we named 229T, in addition to the expected large and small T antigens. STLPyV has a mosaic genome and shares an ancestral recombinant origin with MWPyV. The discovery of STLPyV highlights a novel alternative splicing strategy and advances our understanding of the complex evolutionary history of polyomaviruses.
Keywords: Polyomavirus, Virus discovery, Tumor antigen, Alternative splicing, Recombination
Introduction
Polyomaviruses are small circular double stranded DNA viruses. Members of the family Polyomaviridae have been isolated from a variety of human specimen types, as well as from other hosts, including primates, rodents and birds. The genomes of polyomaviruses range in size from 4,754–5387 bp and can be divided by transcriptional criteria into an early region, a late region and a non-coding control region (Van Ghelue et al., 2012). The early region specifies the large tumor antigen (LTAg) and small tumor antigen (STAg). Rodent polyomaviruses encode an additional middle tumor antigen (MTAg) (Gottlieb and Villarreal, 2001). The late region encodes the structural proteins VP1, VP2 and VP3. Additionally, avian polyomaviruses harbor a unique VP4 upstream of VP1, VP2 and VP3, that is absent from mammalian polyomaviruses (Johne and Müller, 2007). The VP4 of avian polyomaviruses differs from the similarly named SV40 VP4, which is an opening reading frame within the VP2 transcript (Daniels et al., 2007). Finally, a limited subset of polyomaviruses encode an agnoprotein 5’ of the late region (Van Ghelue et al., 2012).
Alternative splicing is a critical mechanism for regulating expression of different gene products from the early region of polyomaviruses (Huang and Carmichael, 2009). Three major forms of the T antigen (LTAg, MTAg and STAg) have been described, as have multiple alternative forms of the LTAg. While most polyomaviruses express STAg from an unspliced mRNA transcript, STAg from rodent polyomaviruses is encoded from an alternatively spliced transcript. Alternative splicing also results in LTAg and STAg sharing approximately 80 amino acid (aa) residues at their N-terminus. Additionally, SV40 encodes a 17KT protein that shares the first 131 aa residues with LTAg, followed by 4 aa residues due to differential splicing (Zerrahn et al., 1993). JCPyV encodes 3 additional proteins (T-135, T-136 and T-165), which also similarly share 132 N-terminal aa residues with LTAg, but differ at the C-terminus due to alternative splicing patterns (Trowbridge and Frisque, 1995). Similarly, MCPyV encodes a 57kT antigen by alternative splicing of the LTAg transcript (Shuda et al., 2008). Moreover, a truncated form of LTAg expressed by BKPyV results from alternative splicing of its LTAg transcript (Abend et al., 2009). Furthermore, alternative splicing can also result in the translation of a middle tumor antigen (MTAg), although this is thought to be a feature unique to rodent polyomaviruses (Huang and Carmichael, 2009). While the 17KT, 57kT, T’- and truncated T antigens share their splice donor site with their cognate LTAg transcripts, the splice donor site of MTAg is shared by STAg instead. Importantly, many of these proteins expressed from alternatively spliced early products have been shown to have transforming potential (Bollag et al., 2000; Boyapati et al., 2003).
Eight new polyomaviruses have been discovered in human clinical specimens within the last five years (Allander et al., 2007; Feng et al., 2008; Gaynor et al., 2007; Schowalter et al., 2010; Scuda et al., 2011; Siebrasse et al., 2012b; van der Meijden et al., 2010), leading to new insights in the fundamental biology and pathogenesis of polyomavirus. These discoveries, along with the discoveries of additional animal polyomaviruses have led to a recent taxonomic proposal for classification of polyomaviruses (Johne et al., 2011). According to this proposal, polyomaviruses are broadly classified into three genera, Avipolyomavirus, Wukipolyomavirus, and Orthpolyomavirus, primarily based on phylogenetic analyses on full-length polyomavirus genomes (Johne et al., 2011). Polyomaviruses have co-evolved with their hosts over long evolutionary timescale (Pérez-Losada et al., 2006; Sharp and Simmonds, 2011). While several intra-strain recombinations have been observed (Chen et al., 2004; Hatwell and Sharp, 2000), large scale recombination of polyomaviruses is generally thought to be rare (Bhattacharjee, 2010; Crandall et al., 2006). However, emerging studies highlight several lineages with inconsistency amongst topologies constructed from different regions of the genome, indicative of recombination events (Sauvage et al., 2011; Schowalter et al., 2010; Siebrasse et al., 2012b).
Here, we describe a novel polyomavirus provisionally named STL polyomavirus (STLPyV). We sequenced the complete genomes of STLPyV strains from Malawi and Saint Louis and found that they differed by 5.2% at nucleotide level. We show that the early region of STLPyV, in addition to the unspliced STAg, undergoes alternative splicing that would encode for LTAg and a unique 229 amino acid T antigen (229T) unlike that of previously characterized rodent MTAg or various modified forms of LTAg, such as 17KT, T’ or truncated T antigens. STLPyV is most closely related to MW polyomavirus (MWPyV) and we establish that their common ancestor had a recombinant origin. We also developed a sensitive degenerate PCR assay that detects both STLPyV and MWPyV and used this assay to screen for STLPyV and MWPyV in clinical specimens from Saint Louis, USA and Gambia. Both viruses were detected in pediatric stool specimens, but there was no statistically significant evidence of association between either virus with diarrheal cases. These results contribute to a deeper understanding of polyomavirus diversity, evolution and gene content.
Results
Discovery of the novel STL polyomavirus
As part of a broader effort to characterize the human gut virome in health and disease, we performed shotgun 454 pyrosequencing of DNA amplified by multiple displacement amplification (MDA) from the stool of a healthy 15 month child in Malawi. Prior to DNA extraction, the stool was processed by CsCl ultracentrifugation to generically enrich for viral particles as described previously (Reyes et al., 2010). Two reads from this sample shared limited sequence identity to known polyomaviruses (Figure 1A). At the time the sequences were generated, the first read (308 nt) shared only 46% amino acid identity to the LTAg of Squirrel monkey polyomavirus, its top scoring hit after tBlastx search to the Genbank nt database. The second read (87 nt) mapped to a separate region of the LTAg in California sea lion polyomavirus based (81% amino acid identity). We recently reported the discovery of another novel polyomavirus, MWPyV (Siebrasse et al., 2012b); sequence analyses indicated that these two reads were most closely related to, but clearly distinct from, MWPyV. From these two initial reads, we designed primers to PCR amplify products that span the two initial pyrosequencer reads in either direction. The resulting complete circular genome of 4,776 bp was sequenced to more than 3X coverage (Figure 1A). Whole genome comparison to all known polyomaviruses indicated that this virus shared the highest nucleotide sequence similarity (64.2%) to MWPyV, which is less than the 81% criterion outlined in species classification guidelines from the International Committee on Taxonomy of Viruses (ICTV) (Johne et al., 2011). We gave this new species of polyomavirus the provision name of STLPyV.
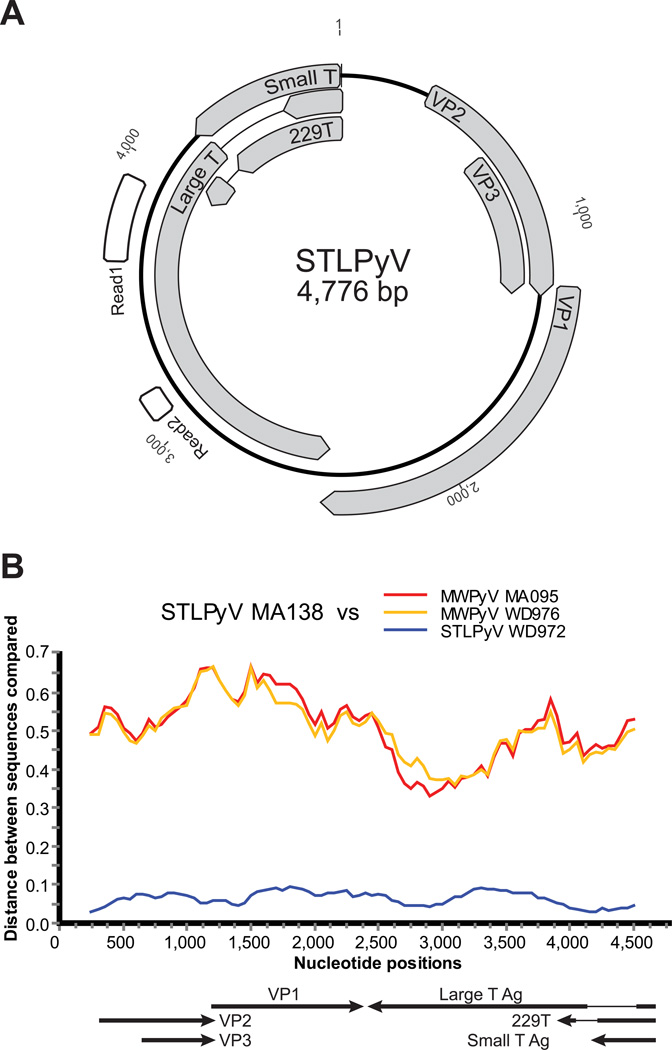
Figure 1. STLPyV is a novel polyomavirus.
(A) Diagram of STLPyV genome based on strain GA138. Positions of the two reads obtained from the initial 454 pyrosequencing of DNA generated by MDA of purified virus particles from the index Malawian case are indicated in white. Predicted open reading frames of STLPyV are shown in grey. (B) Diversity plots of nucleotide sequences are shown between STLPyV GA138 and STLPyV WD972 (blue). Their closest relatives MWPyV MA095 (red) and MWPyV WD976 (orange) are included as reference. The proportion of DNA sequence difference is indicated (0.1 = 10%).
We also amplified, cloned, and sequenced to greater than 3X coverage a second full-length STLPyV strain (WD972). This strain was amplified, using a similar PCR strategy as the index strain, from a fecal specimen obtained from a child who had been enrolled a study of diarrheal diseases conducted at Saint Louis Children’s hospital (described in detail below). The complete genome of STLPyV strain WD972 was 4,775 bp, which included a one nucleotide deletion in the non-coding control region compared to the index Malawi strain. Whole genome sequence analyses indicated that the overall nucleotide sequence identity between the two STLPyV strains was 94.8% with divergence of up to 13% within regions of VP1 and LTAg (Figure 1B).
Genome annotation and alternative splicing of a unique T antigen mRNA transcript
The STLPyV genome organization and sizes of its predicted open reading frames were characteristic of known polyomaviruses (Fig 1A and Table 1). Further, the non-coding control region of STLPyV (nucleotide position 1 – 352) contained features typical of polyomaviruses (Van Ghelue et al., 2012). The ori region encoded five consensus T antigen binding pentanucleotide sequence G(A/G)GGC (nucleotide position 29, 35, 44, 51 and 249) and one non-canonical GTGGC pentanucleotide sequence (nucleotide position 147) (Cantalupo et al., 2005). In addition, there was a 17 nucleotide stretch of AT-rich region including the putative TATA box (nucleotide position 59 – 75).
Table 1.
Putative proteins encoded by STLPyV (strain MA138)
| Protein | Putative coding region(s) |
Predicted size (aa) |
Calculated mass (kDa) |
Range (aa) in other polyomaviruses |
|---|---|---|---|---|
| STAg | 4776-4189 | 195 | 23.0 | 124-199 |
| 230T | 4776-4206, 4090-3972 | 230 | 27.2 | N/A |
| LTAg | 4776-4534, 4188-2452 | 660 | 75.9 | 599-817 |
| VP1 | 1242-2447 | 401 | 43.4 | 343-497 |
| VP2 | 353-1264 | 303 | 33.2 | 241-415 |
| VP3 | 677-1264 | 195 | 21.9 | 190-272 |
The early proteins encoded by polyomaviruses are translated from alternatively spliced transcripts. Typically, gene predictions for LTAg are made based upon the presence of conserved splice donor and splice acceptor sites defined by alignment of other polyomaviruses. However, STLPyV lacked the consensus splice donor sites commonly utilized by most polyomaviruses. Moreover, the rarer splice donor sites identified were incongruent with the predicted LTAg open reading frame due to an out-of-frame arrangement when paired with the corresponding splice acceptor sites. Therefore, we experimentally determined the splice sites used by STLPyV early mRNA transcripts. To do this, we cloned the genomic region encoding the putative LTAg of STLPyV (nucleotide position 4776 – 2452) into an expression vector, transiently transfected the plasmids into 293T cells, and performed RT-PCR on total cell RNA extracts. Primers were designed to span a 666 bp fragment of the early region judged most likely to harbor the splice donor and acceptor sites. As expected, PCR using a plasmid template containing STLPyV genomic DNA yielded the predicted band, while RNA extracted from mock-transfected cells did not amplify any products (Figure 2A). Surprisingly, we detected three bands following RT-PCR of STLPyV transfected extracts (Figure 2A, STLPyV). Each of the three bands was cloned and sequenced and subsequently determined to be a unique product. The largest band represented the unspliced mRNA transcript that yielded a 195 aa open reading frame expected of STAg. The smallest band represented an mRNA transcript generated by excision of a 345 bp intron, yielding an open reading frame with features consistent with those expected for a LTAg (660 aa). The LTAg of STLPyV contained a putative pRb-binding motif (LSCNE beginning at aa residues 105), an N-terminal DnaJ domain (HPDKGG commencing at aa residue 42), and a predicted Bub1 binding site (WDQWW beginning at aa residue 90) – features that are highly conserved in polyomaviruses (Van Ghelue et al., 2012).
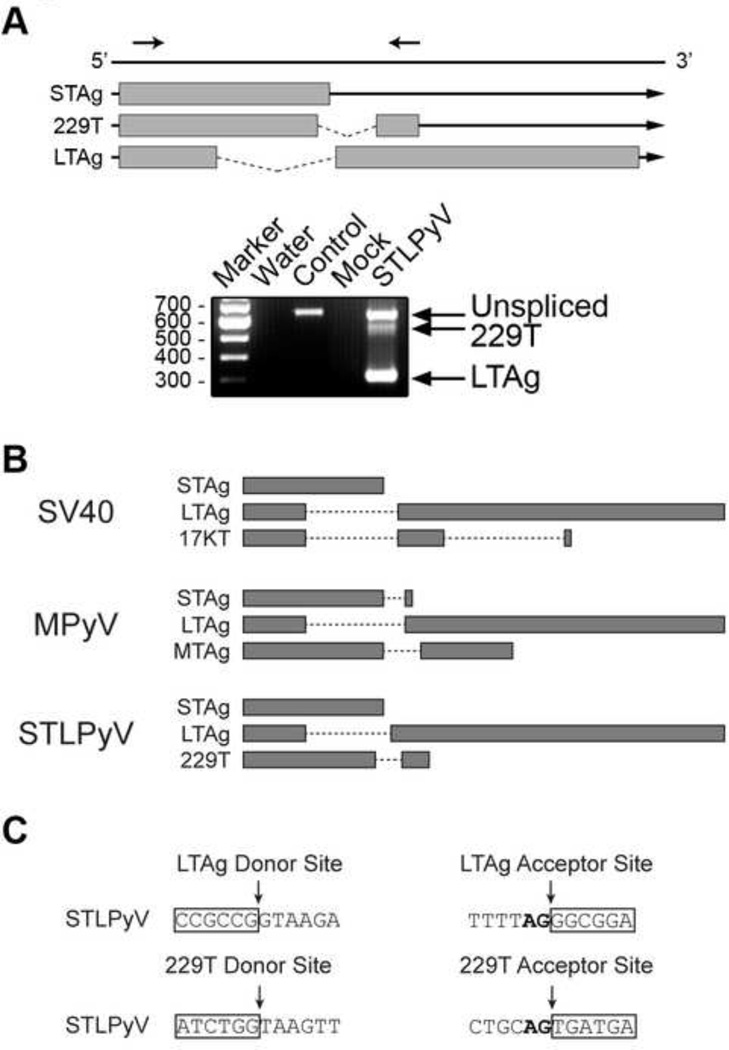
Figure 2. Alternative splicing of STLPyV early region.
(A) Schematic shows mRNA transcripts expressed from the early region of STLPyV. Exonic regions are indicated by grey boxes, intronic regions by lines. Arrows above the diagram indicate the position of primers. 293T cells were transfected with a plasmid expressing STLPyV LTAg for 48 hr. Cells were harvested and lysed for RT-PCR analysis. RT-PCR reactions were performed with water (Water), STLPyV LTAg plasmid (Control), RNA harvested from mock-transfected (Mock), STLPyV LTAg transfected 293T cells (STLPyV). Data are representative of at least three independent experiments. The three alternatively splice mRNA transcripts were confirmed with a different set of primers (data not shown) and verified in HeLa cells (data not shown). (B) Diagram showing the coding region of T antigens, separated by intron regions in dashed lines. The T’- and truncated T antigens of JCPyV and BKPyV are spliced from LTAg transcripts in a similar manner as SV40 17KT. (C) Sequences of the splice donor and acceptor sites for STLPyV LTAg and 229T transcripts (strain MA138). Exon sequences are highlighted in boxes. The nucleotide sequences of the splice donor and acceptor sites of STLPyV shown are conserved in the three STLPyV strains characterized.
The intermediate band corresponded to an mRNA transcript derived by splicing of a 115 bp intron that yields a unique open reading frame of 229 aa; the first 190 aa were shared with the STAg, the 191st aa encompassed the splice junction and the remaining 38 aa were derived from the second exon. Based on its predicted amino acid length, we named this putative protein 229T. The splice sites that generated 229T were unique from the splice sites utilized by the STLPyV LTAg. Although there is precedence in rodent polyomaviruses for splicing of STAg, in those instances the splice acceptor site is identical to the LTAg splice acceptor, which was not the case for STLPyV. 229T also differed from the 17KT and T’ antigens of SV40 or BKPyV which share common splice sites with their LTAg transcripts (Fig 2B). To verify the alternatively spliced transcripts, we designed a second set of primers for RT-PCR. Cloning and sequencing independently confirmed the splice junctions, demonstrating that the results were not due to an artifact of the initial primers (data not shown). The splicing patterns were also confirmed with both primer pairs in HeLa cells indicating that the splice variants were not cell line specific (data not shown).
We next sequenced a fragment of the STLPyV genome in the region spanning the splice sites (nt 4731 – 4066) from an additional sample from another child in the Saint Louis study found to be positive for the presence of STLPyV (using a PCR assay described below) so that we could perform a sequence comparison with the two STLPyV strain. While we observed up to a 3.2% difference in the nucleotide sequence across the 666 bp region between the three strains, both 5’ splice and 3’ sites for LTAg and 229T were entirely conserved. Thus, we conclude that the early region of STLPyV has unique alternative splicing features.
Phylogenetic analysis indicates ancient recombinant origin of STL polyomavirus
To estimate the phylogenetic relationship of the new STLPyV to other polyomaviruses, we used the LTAg, VP1 and VP2 sequences to construct phylogenies using both Bayesian (BI) and maximum likelihood (ML) methods. Both methods yielded trees with similar topologies. Midpoint rooting of the large T antigen phylogeny, which was the most alignable region across all polyomaviruses, positioned the avian polyomaviruses (Avipolyomavirus genera) basal to mammalian polyomaviruses (Figure 3A). This is consistent with other studies (Pérez-Losada et al., 2006; Sauvage et al., 2011). Furthermore, the outgroup positioning of the Avipolyomavirus is consistent with marked differences in the genomic organization, pathogenesis and biology of the avian polyomaviruses (Johne et al., 2011). Therefore, the avian polyomaviruses were used as an outgroup to root the phylogenies.
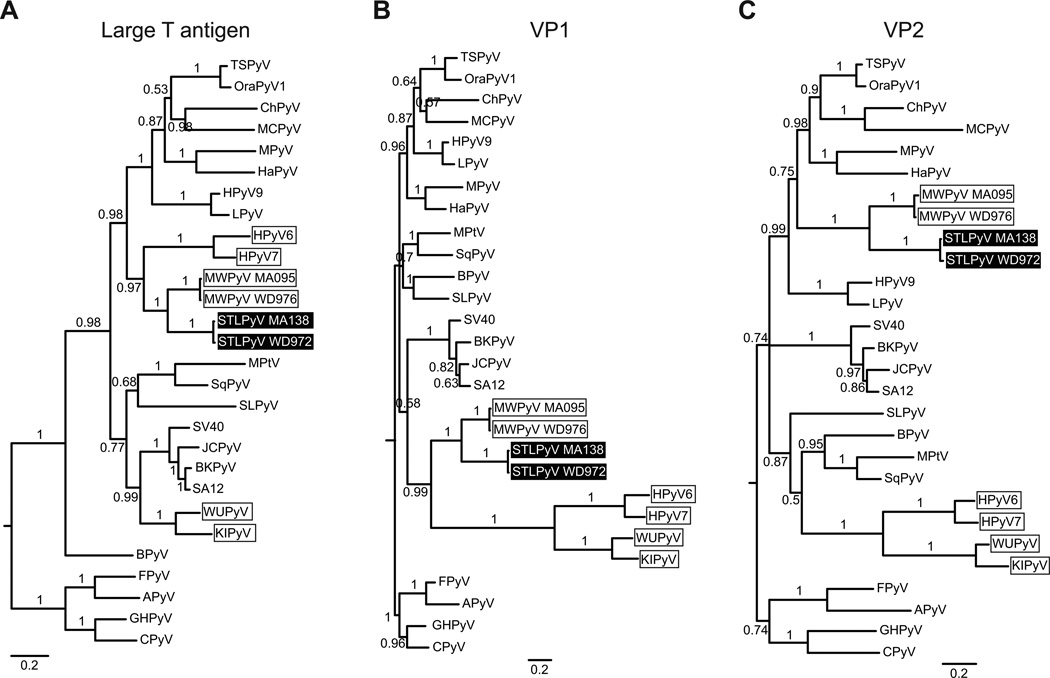
Figure 3. Phylogenetic analysis of STLPyV.
Phylogenetic relationships of 28 diverse polyomavirus sequences were inferred from alignment of protein sequences from LTAg (A), VP1 (B) and VP2 (C). Avian polyomaviruses were used as an outgroup to root the phylogenies. STLPyV strains are highlighted in black; MWPyV strains and Wukipolyomavirus whose members show discordant phylogenetic relationships are indicated by boxes. Internal branch labels indicate Bayesian posterior probabilities. The ML method yielded trees with similar topologies.
The phylogenies of LTAg, VP1 and VP2 showed that both STLPyV strains are closely related to MWPyV. STLPyV and MWPyV strains formed a monophyletic clade with high confidence (Figure 3). However, the topologies of the LTAg and VP1 derived trees were different. Within the LTAg region, STLPyV and MWPyV were closely related to HPyV6 and HPy7 and formed a clade with several Orthopolyomavirus species including TSPyV, MCPyV, MPyV and HPyV9 (Figure 3A). Strikingly, in the VP1 region, STLPyV and MWPyV were most closely related to HPyV6, HPyV7, WUPyV and KIPyV, but clustered with different Orthopolyomavirus species (SV40, BKPyV, JCPyV and SA12) (Figure 3B). Finally, the VP2-derived tree indicated that STLPyV and MWPyV cluster with the similar Orthopolyomavirus species as in the LTAg derived phylogeny, whereas HPyV6 and HPyV7 are closely related to WUPyV and KIPyV. The VP2 region of polyomaviruses has many insertions and deletions; therefore, it is important to be cautious about over-interpreting the different branching orders. Nonetheless, the discordant phylogenetic relationship of the STLPyV and MWPyV clade in LTAg and VP1 derived trees are indicative of recombinant events in their common ancestor. HPyV6 and HPyV7 are also likely recombinant lineages, although the phylogenies suggest that the evolutionary histories of HPyV6 and HPyV7 as compared to that of STLPyV and MWPyV are likely distinct because the relationships of all four viruses cannot be adequately explained by the same recombination events. Thus, our results indicate that STLPyV and MWPyV share an ancestral recombinant origin.
Molecular characterization demonstrates STLPyV presence in Saint Louis and Gambia
In order to determine the prevalence of STLPyV, we developed a PCR assay targeted against the large T antigen region. Since STLPyV is closely-related to the newly-identified MWPyV (Figure 3A), we designed an assay capable of detecting both STLPyV and MWPyV, thus allowing us to characterize the prevalence of both novel polyomaviruses as well as to identify additional variants of these two viruses. We confirmed that the assay amplified a specific 481bp (STLPyV) or 484bp (MWPyV) PCR product using plasmids encoding the large T antigen of STLPyV or MWPyV (Figure 4A). To validate our PCR assay, we examined pediatric fecal specimens sent for bacterial culture to the clinical microbiology laboratory at the Saint Louis Children’s Hospital. These samples were collected from patients, primarily with diarrhea, who were examined between July 2009 and June 2010. This cohort was previously screened for the presence of MWPyV by a real-time PCR assay and 12 specimens were found to be positive (Siebrasse et al., 2012b). In addition to the 12 samples, the new PCR assay described in this report identified two additional samples that were positive for MWPyV, for a total of 14 samples (Figure 4B, right column). In the previous study, these two samples yielded detectable Ct values in the real time assay, but were below the cutoff value used to define positive samples (data not shown).
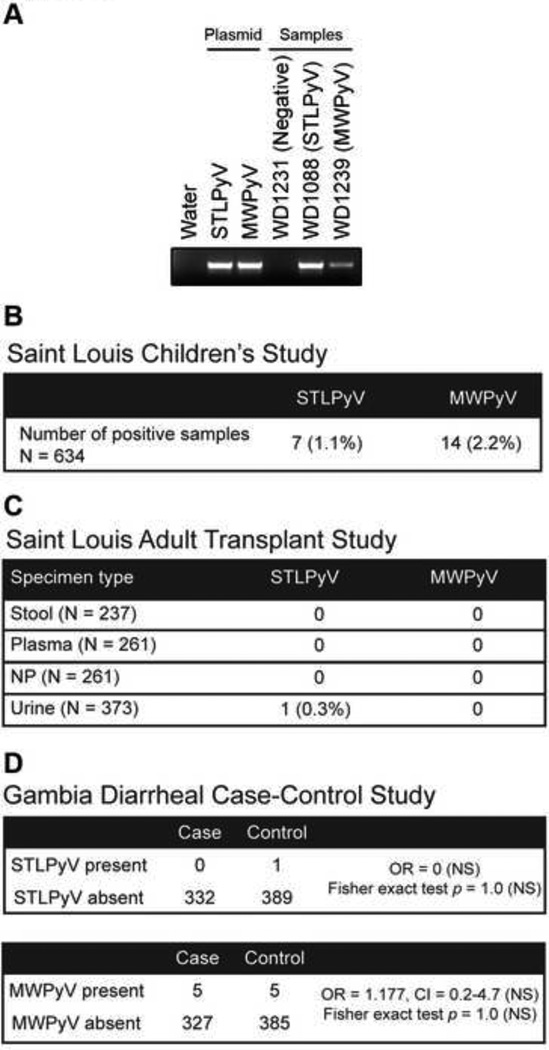
Figure 4. Prevalance of STLPyV and MWPyV.
(A) PCR analysis of STLPyV and MWPyV is shown for control plasmids of the respective LTAg, or representative samples found to be negative,and positive for STLPyV or MWPyV. Bands corresponds to a STLPyV (481 bp) or MWPyV (484 bp) PCR product. PCR products for all positive samples were cloned and sequenced verified.(B) Prevalence of STLPyV and MWPyV in 634 stool specimens collected from a study of children from Saint Louis , as defined by the direct PCR assay described in panel A. Numbers in parenthesis indicate frequency. (C) Results of PCR screening for STLPyV and MWPyV in feces, plasma, nasopharyngeal swabs (NP), and urine specimens collected from a cohort of adult USA kidney transplant recipients. Numbers in parenthesis beside each specimen type indicate the number of specimens screened.(D) Prevalence of STLPyV and MWPyV in a Gambian diarrheal case control study of children is shown, and is based on the PCR screening assay. Odds ratio (OR), 95% confidence intervals (CI) and Fisher exact test indicate that there is no statistically evidence that STLPyV or MWPyV are significantly associated with diarrheal cases (NS = not significant).
Seven samples from the Saint Louis children’s study were found to be positive for STLPyV (Figure 4B, left column) demonstrating that STLPyV can also be found in a geographically separate region from the initial strain identified in Malawi. Interestingly, three samples contained both STLPyV and MWPyV (samples WD972, WD976 and WD1226). These three samples were collected from the same individual, a five year-old lung transplant recipient who presented with persistent, recurring diarrhea. The first two samples were taken on consecutive days while the third sample was collected four months later. The other four STLPyV samples came from four different individuals.
Previous reports indicate that the prevalence of polyomaviruses is elevated in immunocompromised patients. Hence, we sought to understand the prevalence of STLPyV and MWPyV in adult transplant recipients by screening specimens collected from kidney transplant patients at Washington University in Saint Louis. Different types of biospecimens (fecal, urine, nasopharyngeal swab and sera) from individuals were examined with our PCR assay. We did not detect STLPyV in fecal samples (n=237), plasma (n=261) or nasopharyngeal swabs (n=261) (Figure 4C). We found that one urine sample was positive for STLPyV. Interestingly, we did not detect MWPyV in any of the specimens. The absence of STLPyV and MWPyV in fecal samples from the adult study was unexpected since the prevalence of STLPyV and MWPyV in the children we surveyed from St. Louis was about 1% and 2.2% respectively (Figure 4B). To verify the integrity of the specimens, we screened urine samples for the presence of JCPyV using a published real time PCR assay (Siebrasse et al., 2012a). Forty-five samples were positive for JCPyV (data not shown); the 12% prevalence of JCPyV in this study is consistent with estimates from other reports suggesting that the low/lack of STLPyV and MWPyV was not due to compromised specimen conditions. Thus, the prevalence of STLPyV and MWPyV in stool samples from our study of adult kidney recipients is lower than that observed in the children’s study.
Since STLPyV and MWPyV were readily detected in the stool samples from children, we examined whether STLPyV and MWPyV are associated with childhood diarrhea by analyzing fecal samples collected from Gambia as part of the ongoing Global Enteric Multi-center Study (GEMS) (Kotloff et al., submitted for publication). For each enrolled case with diarrhea, one healthy control without diarrhea (matched for age, gender and time of presentation) was randomly selected from the community. We screened 332 cases and 389 controls for the presence of STLPyV and MWPy, finding one sample from the controls and none from the cases to be positive for STLPyV, and five samples from cases and five from controls to be positive for MWPyV. Based on these results, we concluded that neither STLPyV nor MWPyV had a statistically significant association with diarrheal cases (Figure 4D, right column).
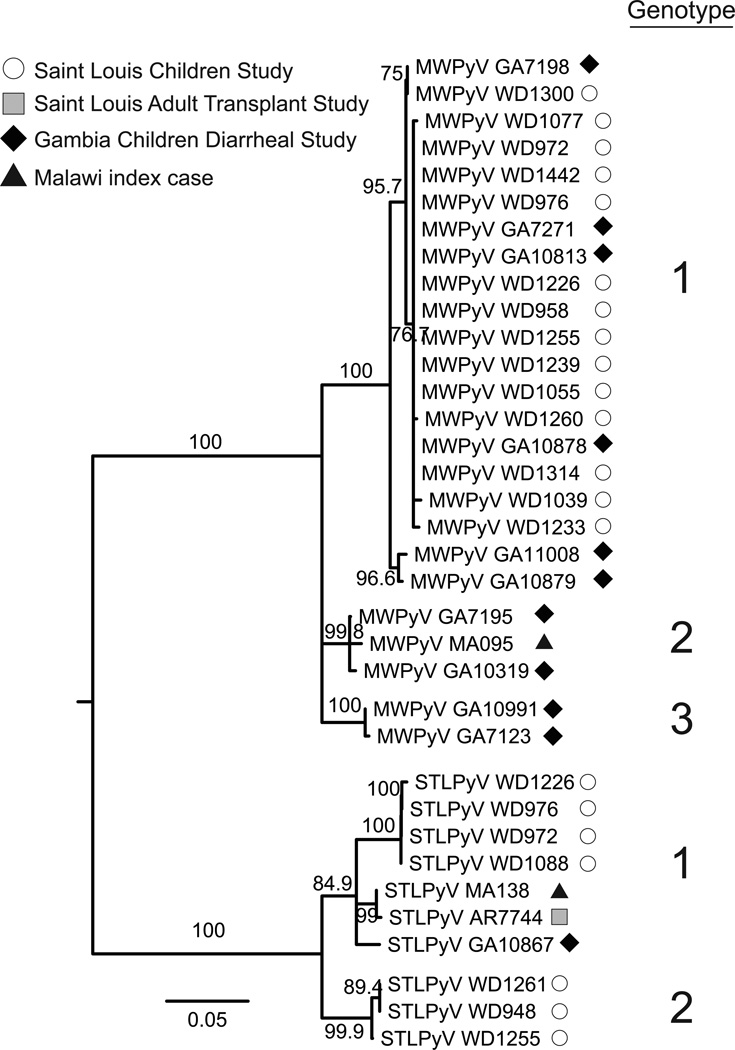
We next examined the diversity of STLPyV and MWPyV strains based on the amplicon sequences generated from the positive samples from the GEMS and St. Louis studies described above. Phylogenetic analyses indicated that STLPyV and MWPyV sequences formed distinct clades with high confidence (Figure 5). The STLPyV strains formed two monophyletic clades that differed by at least 5% nucleotide sequence identity between clades, hence we tentatively designated them as two genotypes. MWPyV strains formed three monophyletic clades with high confidence, with at least 5% difference in inter-clade nucleotide sequence identity, which we designated as three genotypes. A previous study of MWPyV identified members of two of these clades (Siebrasse et al., 2012b); in the present study, additional members of those two clades were identified as well as the existence of a third clade. By comparison, the global variation of WUPyV strains is approximately 1.2%, while the BKPyV strains vary by up to 5.3% at the nucleotide sequence (Bialasiewicz et al., 2010; Krumbholz et al., 2009). Thus, this indicates that there is diverse strain variation in STLPyV and MWPyV.
Figure 5. STLPyV and MWPyV strains are diverse.
Midpoint-rooted neighbor-joining phylogeny inferred from the nucleotide sequences of 9 STLPyV strains and 25 MWPyV strains identified in the three cohorts (Figure 4) and the Malawi index case. Primer sequences were removed to yield a 437 nt alignment. The two genotypes of STLPyV and three genotypes of MWPyV, with strong bootstrap support and > 5% inter-clade sequence difference, are indicated on the right. The ML method yielded a tree with similar topologies.
Discussion
Novel T antigen alternative splicing
Alternative splicing of the early mRNA transcripts of a few canonical polyomaviruses is well documented. In recent years, as application of molecular methods dramatically increased the rate of discovery of new polyomaviruses, many of the genome annotations have been based on strictly computational analyses and predictions; there have been few studies that have experimentally defined early region splicing patterns in these newly identified viruses. Because of a lack of consensus splice donor and acceptor sites, we experimentally determined the splice junctions in the early transcript of STLPyV, leading us to identify a unique splicing pattern. As the predicted protein generated by this splicing event consists of 229 aa, we have termed this protein 229T. The splicing pattern that yields STLPyV 229T is distinct from the one that yields rodent MTAgs and modified versions of LTAg described in other polyomavirus species (Figure 2B). STLPyV 229T does not arise from secondary splicing of the LTAg transcript, as seen in 17KT/T’ antigens. Furthermore, STLPyV 229T splicing is different from the splicing pattern of rodent polyomaviruses, which rely on the same splice donor site for STAg and MTAg; instead, STLPyV 229T uses a distinct downstream splice donor and acceptor site from its LTAg alternative splicing. The putative open reading frame of STLPyV 229T included the N-terminal DnaJ domain (HPDKGG) motif, which recruits and activates ATPase activity of DnaKs in studies of other polyomaviruses (Campbell et al., 1997; Srinivasan et al., 1997; Sullivan et al., 2000). No additional conserved protein motifs were detected using Motif scan and Prosite. Interestingly, MTAg of MPyV has been shown to be the major transforming protein (Rassoulzadegan et al., 1982; Treisman et al., 1981). While we do not have functional evidence for the role of STLPyV 229T, this raises provocative questions about the role of STLPyV 229T and whether an analogous T antigen might be present but unrecognized in other polyomaviruses.
Complex evolutionary history of polyomaviruses
The evolutionary history of polyomaviruses is complex. Phylogenetic analysis of individual genes yielded distinct topologies depending on the locus we analyzed. One constant was the consistent placement of STLPyV with MWPyV at all loci, suggesting that STLPyV shares an ancestral recombinant origin with MWPyV. The recombinant nature of these two viruses poses a conundrum for the current taxonomic scheme for polyomaviruses, which cannot accommodate the complex phylogenetic relationships of polyomaviruses that are becoming apparent. For example, in addition to the clear recombinant nature of STLPyV and MWPyV, HPyV6 and HPyV7 (currently members of Wukipolyomavirus genera) are evidently recombinant species as well, as shown by their discordant phylogenies; they do not form a monophyletic clade with WUPyV and KIPyV when the LTAg is analyzed (Figure 3A and 3B, compare HPyV6, HPyV7 to WUPyV, KIPyV). Second, species of the Orthpolyomavirus taxa are paraphyletic regardless of designating either Avipolyomavirus or Wukipolyomavirus as the outgroup to root the phylogenies.
Given these issues, the current taxonomic system may need further refinement with the broad goal of making genus level taxonomic assignments concordant with phylogenetic structure. More broadly, these results underscore the importance of assessing phylogeny using multiple loci. Finally, unlike RNA viruses and ssDNA viruses, dsDNA viruses like polyomaviruses have a low mutation rate as they rely on the host polymerase for replication (Duffy et al., 2008). Hence, the clear evidence for recombination within the family Polyomaviridae is significant because it suggests that polyomaviruses can compensate for their limited mutational changes by recombination to gain new, and possibly more advantageous, genetic information. Indeed, from the multiple sequence alignments, we observed many clade-specific insertions and deletions in the late region proteins (data not shown). Moreover, a more extreme recombination between two viruses of Polyomaviridae and Papillomaviridae has been previously described (Woolford et al., 2007), corroborating that recombination in Polyomaviridae can occur. Thus, recombination events are important milestones in the evolutionary history of polyomaviruses.
Insight into STLPyV epidemiology
In addition to the index case from Malawi, we detected STLPyV in fecal specimens collected from Saint Louis and Gambia, demonstrating that it is geographically widespread. Through the use of a degenerate PCR assay, we were able to define simultaneously the prevalence of both STLPyV and MWPyV in multiple patient cohorts. In each of the cohorts, STLPyV was found in fewer fecal samples compared to MWPyV (Figure 4). Strikingly, both STLPyV and MWPyV were detected in three serial fecal samples collected from the same patient. This patient had received a lung transplant three years previously; however it is unclear whether immunosuppression contributed to this observation. Further screening to determine whether this observation is a statistical anomaly or whether it reflects some underlying biological linkage between the two viruses is warranted.
Although both viruses were detected in fecal specimens collected from pediatric patients, we did not detect either virus in 237 fecal specimens collected from adult renal transplant patients. Screening of nasopharygeal specimens, serum and urine from these patients yielded a single urine sample positive for STLPyV. By contrast we detected JCPyV in 45 (12%) of the urine samples tested. At this point in time, it is still not known whether either STLPyV or MWPyV causes bona fide infection in humans. If we assume that STLPyV does in fact infect humans, its lower prevalence than JCPyV in adult renal transplant patients may be the result of one or more of the following factors: (1) STLPyV infection may be relatively rare; (2) acute STLPyV infection may occur primarily in children; (3) STLPyV may have a distinct tropism that is not reflected by the samples tested; (4) the life cycle of STLPyV may not involve persistence and then reactivation in the context of immunosuppression.
While there are many outstanding questions regarding the potential role of STLPyV in human infection, STLPyV and the recently described MWPyV are the first two polyomaviruses to be discovered in human stool samples, raising the question as to whether they may have a primary gastrointestinal tropism. Together with the frequent detection of WUPyV and KIPyV in respiratory specimens (Babakir-Mina et al., 2011), and the apparent primary tropism of HPyV6 and HpyV7 for the skin (Schowalter et al., 2010), these findings demonstrate the ubiquity of polyomaviruses in different human specimens. As disease associations have been established for neurotropic polyomaviruses (JCPyV), renal-tropic polyomaviruses (BKPyV) and skin-tropic polyomaviruses (Merkel, TSPyV), it remains to be seen whether disease associations for the other novel polyomaviruses will emerge.
Materials and methods
Clinical Specimens
The index stool specimen was obtained in January 2009 from a healthy, 15 month-old male living in Malawi as part of a human gut microbiome survey of healthy and malnourished children (Yatsunenko et al., 2012). The sample was processed by CsCl ultracentrifugation as described previously (Reyes et al., 2010).
This study was approved by the College of Medicine Research Ethics Committee of the University of Malawi; the Human Research Protection Office (HRPO) of Washington University in St. Louis, Missouri, USA; the Institutional Review Board of the University of Maryland Baltimore, Baltimore, Maryland, USA; the Joint MRC/Gambia Government Ethics Committee and by the Ethics committee of the London School of Hygiene and Tropical Medicine, UK.
A panel of 514 stool samples (Saint Louis Children’s cohort) were collected previously from children age 0 to 18 years old, primarily with diarrheal diseases from July, 2009 through June, 2010 (Siebrasse et al., 2012b). A total of 237 fecal samples, 261 plasma samples, 261 nasopharyngeal swabs and 373 urine specimens were obtained from adult kidney transplant recipients at Washington University’s O’Brien Center Kidney Translational Research Core using a protocol approved by the University’s HRPO. Patients enrolled in this study were defined as ‘new’ transplant recipients if the samples were obtained within 12 months after they received a kidney or as ‘pre-existing’ transplant recipients if sampling occurred more than 12 months after the transplant. In addition, we surveyed 722 fecal samples (332 cases and 390 controls) that had been collected, using a protocol approved by the Ethics committee, from children aged 0 to 5 years old who living in The Gambia as part of the Global Enterics Multi-Center Study (GEMS), a study of diarrhea etiologies (Kotloff et al., submitted for publication). For each enrolled child with diarrhea, one healthy control child without diarrhea matched to the case by age, gender, and time of presentation is randomly selected from the censused community in which the case resides, matched to the case by age, gender, and time of presentation.
Sample preparation for high throughput sequencing
From the index Malawi case, DNA was purified from the fecal sample, amplified by rolling circle amplification and subjected to FLX Titanium pyrosequencing as previously described (Siebrasse et al., 2012b). Unique high quality reads with no detectable similarity to the reference human genome or NCBI nt database by BLASTn were analyzed by BLASTx alignment against the NCBI non-redundant (nr) protein database.
Amplification of complete genomes and early region splice sites
The complete genomes of the STLPyV MA138 and WD972 strains were PCR amplified in three overlapping fragments, cloned and sequenced. DNA from the samples was initially subjected to rolling circle amplification using the Illustra GenomiPhi V2 kit (GE Healthcare) prior to PCR amplification. The following primers were used: (i) STLPyVMA138-P1F (5’–GCATTCATAGGGTTTCAGAC–3’) with STLPyVMA138-P5r (5’–GTAGCAGCTGCTATATTAG–3’); (ii) STLPyVMA138-Q2F (5’–CCTTCAGGCCTGGATTGTTTTGTTACAGC–3’) with STLPyVMA138-P5r; and (iii) STLPyVMA138-P2r (5’–GGCTGGAAAACATAGAGTG–3’) with STLPyVMA138-P3F (5’–CTAATATAGCAGCTGCTAC–3’). WD972 strain was amplified using the following PCR primers: (i) STLPyVWD972-P3F (5’–CTAATATAGCAGCTGCTACAGTTG–3’) with STLPyVWD972-P5r (5’–CAACTGTAGCAGCTGCTATATTAG–3’), (ii) and STLPyVWD972-P3F with STLPyVMA138-P2r.
Amplicons were cloned into pCR4 using TOPO TA cloning kit (Invitrogen) and sequenced; for the STLPyV MA138 strain primers included, STLPyVMA138-Seq1F (5’–CTGATGTTGTTGATGTGGTGGCAACCTG–3’), STLPyVMA138-Seq1r (5’–GTACATCACATGTTTCCAAGCATGAAGGC–3’), STLPyVMA138-Seq2F (5’–CTCCAGGTAAGGGTTCTGAGCCATCTG–3’), STLPyVMA138-Seq2r (5’–GCTTTCAAATTGTGGACCAAGCAATTCACC–3’), STLPyVMA138-Seq3F (5’–GGTAAAGTTGGAGCAGCAGGAGTTGC–3’), STLPyVMA138-Seq3r (5’–TGAGAGTAGTAACTTCGGCAGCGAGC–3’), STLPyVMA138-Seq4F (5’–ACTGCATCAGGGCCTACTTGAATGTC–3’), and STLPyVMA138-Seq4r (5’–AGTTTCAGGTGATGCTGCTGCCATTG–3’), while primers for the STLPyV WD972 strain consisted of STLPyVWD972-Seq1F (5’–GTACATCACATGTTTCCAAGCATGAAG–3’), STLPyVWD972-Seq1r (5’–CTTCATGCTTGGAAACATGTGATGTAC–3’), STLPyVWD972-Seq2F (5’–GGTTGTCCTGATACTCTGGGCATTTGG–3’), STLPyVWD972-Seq2r (5’–GGAGGACAGTTAATATTTAAGGTTCTGCC–3’), STLPyVWD972-Seq3F (5’–GTAGTCCTGCATTTTCAGCTGCCTGTAG–3’), and STLPyVWD972-Seq3r (5’–CAGTGACAGCAACTCCTGCTGCTCCAAC–3’).
A segment of STLPyV viral genome that encompasses the early region splice sites was verified from an additional sample (WD1226) found to be positive by PCR screening in the Saint Louis Children’s study. PCR amplification was performed on the rolling circle amplified sample, using primers STLPyVConSpliceF (5’–CTTCTRGGGCTTCCAGAAGAYTCCTG–3’) in combination with STLPyVConSpliceR (5’–TGGGATGCAGAGGTCCCTTCATCATC–3’).
Cells and plasmids
293T cells were maintained at 37°C in Dulbecco's modified Eagle's medium, supplemented with 10% bovine growth serum, and 1% penicillin-streptomycin, under an atmosphere of 5% CO2/95% air. The large T antigen region of STLPyV MA138 from nucleotide positions 4776 – 2452 was cloned and ligated into a pCDNA3.1 expression vector.
RT-PCR
293T cells were seeded at 1.7 × 105 cells/ml. Plasmid DNA (500 ng) was transfected with TransIT-LT1 reagent (Mirius) according to the manufacturer’s recommendations. Forty-eight hours post-transfection, cells were washed with Dulbecco's phosphate-buffered saline and removed with 0.05% trypsin. RNA was purified from the cells using a RNeasy Mini Kit (Qiagen). Reverse transcription was performed with the OneStep RT-PCR kit (Qiagen) according to the manufacturer’s instructions. STLPyV mRNA was amplified with “forward primer” STLSplice1F (5’–CTTCTGGGGCTTCCAGAAGATTCCTG–3’) in combination with “reverse primer” STLSplice1r (5’–GGATGCAGAGGTCCCTTCATCATCAC–3’). A second set of primers was used to independently verify the splice junction [STLSplice2F (5’–GATCAAGCTCTCTCTAGGCAAGAAGC–3’) in combination with STLSplice2r (5’–GGTGTTGAATTCTGAGTAGAAGAATCAGG–3’)]. Bands corresponding to the unspliced, middle T antigen spliced, and large T antigen spliced transcripts were purified by QIAquick gel extraction kit (Qiagen), cloned into pCR4 vector using TOPO TA cloning kit (Invitrogen) and clones from multiple transformed bacterial colonies were sequenced.
Diversity plots and phylogenetic analyses
Nucleotide sequences of the full-length genome from the STLPyV MA138, STLPyV WD972, MWPyV MA095 and MWPyV WD976 strains were aligned by MUSCLE (Edgar, 2004), and minor editing was done manually. Diversity plots were generated with Simplot (Lole et al., 1999), employing sliding windows of 300 nt in 50 nt steps, with Kimura (2-parameter) correction.
Phylogenetic trees were constructed from alignments of the LTAg, VP1 and VP2 protein sequences from 28 polyomaviruses: avian polyomavirus (NC_004764, APyV), crow polyomavirus (NC_007922, CPyV), finch polyomavirus (NC_007923, FPyV), goose hemorrhagic polyomavirus (NC_004800, GHPyV), trichodysplasia spinulosa-associated polyomavirus (NC_014361, TSPyV), bornean orangutan polyomavirus (NC_013439, OraPyV1), chimpanzee polyomavirus (NC_014743, ChPyV), Merkel cell polyomavirus (HM011557, MCPyV), murine polyomavirus (NC_001515, MPyV), hamster polyomavirus (NC_001663, HaPyV), human polyomavirus 9 (NC_015150, HPyV9), B-lymphotropic polyomavirus (NC_004763, LPyV), simian virus 40 (NC_001669, SV40), BK polyomavirus (NC_001538, BKPyV), JC polyomavirus (NC_001699, JCPyV), baboon polyomavirus (NC_007611, SA12), California sea lion polyomavirus (NC_013796, SLPyV), bovine polyomavirus (NC_001442, BPyV), murine pneumotropic virus (NC_001505, MPtV), squirrel monkey polyomavirus (NC_009951, SqPyV), human polyomavirus 6 (NC_014406, HPyV6), human polyomavirus 7 (NC_014407, HPyV7), KI polyomavirus (NC_009238, KIPyV), WU polyomavirus (NC_009539, WUPyV), MW polyomavirus MA095 strain, (JQ898291, MWPyV MA095), MW polyomavirus WD976 strain (JQ898292, MWPyV WD976), and the two strains of STL polyomaviruses (STLPyV MA138 and STLPyV WD972). Alignments were performed with a probabilistic, multiple sequence alignment algorithm Fast Statistical Alignment (FSA) (Bradley et al., 2009). Phylogenies were constructed with MrBayes v3.2.1 (Huelsenbeck and Ronquist, 2001) using a Bayesian MCMC inference (BI), and PhyML v3.0 (Guindon and Gascuel, 2003) by the maximum likelihood (ML) method. MrBayes analyses (RtREV+I+G+F) were run for 4,000,000 – 6,000,000 steps with a sample frequency set to 500 and a 25% burn-in period. Convergence and mixing were assessed with Tracer v1.5 (Drummond and Andrew, 2009). Analyses were performed at least twice. Support for ML trees (LG+I+G+F) was assessed by 1,000 nonparametric bootstraps. The two methods yielded trees with similar topologies.
For the phylogenetic analysis of STLPyV and MWPyV sequences obtained from screening, nucleotide sequences were aligned by Muscle (Edgar, 2004) and primer sequences were trimmed from the alignment. A phylogeny was constructed by the neighbor-joining method using the Jukes Cantor method of correction (Drummond et al., 2011). Consistent results were obtained with the ML method.
Diagnostic PCR amplification
Standard precautions to avoid end product contamination were taken for all PCR assays, including the use of PCR hoods and maintaining separate areas for PCR set up and analysis. For every 88 samples tested, seven no-template negative controls were interspersed between the actual samples. Accuprime hot start Taq (Invitrogen) was used to amplify 5 µl of extracted samples using the following PCR program: 95°C for 5 min, 40 cycles of 95°C for 30 sec, 55°C for 30 sec, 72°C for 29 sec, followed by 72°C for 10 min. STLPyV and MWPyV were detected with the “forward primer” STLMWScreenF (5’–GRATGAAAYRCWWTTACAGGTTGCCACC–3’) in combination with the “reverse primer” STLMWScreenr (5’– GTGGWAAAACAACTGTAGCWGCTGC–3’) that together generate a 481 bp (STLPyV) or 484 bp (MWPyV) amplicon from the 3’-end of the large T antigen coding region. Products were visualized following electrophoresis on 1.25% agarose gels. Amplicons were purified by QIAquick gel extraction kit (Qiagen), cloned into pCR4 using TOPO TA cloning kit (Invitrogen) and clones from multiple transformed bacterial colonies were sequenced to verify their identity.
Highlights.
Identification of a new polyomavirus, STL polyomavirus, from a child in Malawi
Novel alternative splicing strategy generates a unique T antigen
STLPyV has a mosaic genome of ancestral recombinant origin
We detectedSTLPyV in USA and The Gambia
Acknowledgements
This work was supported in part by NIH grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research, NIH grant K24 DK002886 and NIH DK079333 (DCB), a grant from the Roche Organ Transplantation Foundation, The Medical Research Council (UK) and the Bill & Melinda Gates Foundation (OPP1016839). DW holds an investigator in the pathogenesis of infectious disease award from the Burroughs Wellcome Fund. We thank Irma Bauer for performing the JCPyV screening.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers. The sequences of the two complete genomes of STLPyV have been entered into the GenBank database under accession numbers JX463183 (strain MA138) and JX463184 (strain WD972). Amplicon sequences from the STLPyV and MWPyV strains have been deposited under accession numbers JX463185 – JX463215.
References
- Abend JR, Joseph AE, Das D, Campbell-Cecen DB, Imperiale MJ. A truncated T antigen expressed from an alternatively spliced BK virus early mRNA. J Gen Virol. 2009;90:1238–1245. doi: 10.1099/vir.0.009159-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. Identification of a third human polyomavirus. J Virol. 2007;81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babakir-Mina M, Ciccozzi M, Perno CF, Ciotti M. The novel KI, WU, MC polyomaviruses: possible human pathogens? New Microbiol. 2011;34:1–8. [PubMed] [Google Scholar]
- Bhattacharjee S. Evolutionary interrelationships among polyomaviruses based on nucleotide and amino acid variations. Indian Journal of Biotechnology. 2010;9:252–264. [Google Scholar]
- Bialasiewicz S, Rockett R, Whiley DW, Abed Y, Allander T, Binks M, Boivin G, Cheng AC, Chung JY, Ferguson PE, Gilroy NM, Leach AJ, Lindau C, Rossen JW, Sorrell TC, Nissen MD, Sloots TP. Whole-genome characterization and genotyping of global WU polyomavirus strains. J Virol. 2010;84:6229–6234. doi: 10.1128/JVI.02658-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag B, Prins C, Snyder EL, Frisque RJ. Purified JC virus T and T' proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology. 2000;274:165–178. doi: 10.1006/viro.2000.0451. [DOI] [PubMed] [Google Scholar]
- Boyapati A, Wilson M, Yu J, Rundell K. SV40 17KT antigen complements dnaj mutations in large T antigen to restore transformation of primary human fibroblasts. Virology. 2003;315:148–158. doi: 10.1016/s0042-6822(03)00524-5. [DOI] [PubMed] [Google Scholar]
- Bradley RK, Roberts A, Smoot M, Juvekar S, Do J, Dewey C, Holmes I, Pachter L. Fast statistical alignment. PLoS Comput Biol. 2009;5:e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, Mullane KP, Aksoy IA, Stubdal H, Zalvide J, Pipas JM, Silver PA, Roberts TM, Schaffhausen BS, DeCaprio JA. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KW, Lewis AM, Pipas JM. Complete nucleotide sequence of polyomavirus SA12. J Virol. 2005;79:13094–13104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sharp PM, Fowkes M, Kocher O, Joseph JT, Koralnik IJ. Analysis of 15 novel full-length BK virus sequences from three individuals: evidence of a high intra-strain genetic diversity. J Gen Virol. 2004;85:2651–2663. doi: 10.1099/vir.0.79920-0. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Pérez-Losada M, Christensen RG, McClellan DA, Viscidi RP. Phylogenomics and molecular evolution of polyomaviruses. Adv Exp Med Biol. 2006;577:46–59. doi: 10.1007/0-387-32957-9_3. [DOI] [PubMed] [Google Scholar]
- Daniels R, Sadowicz D, Hebert DN. A very late viral protein triggers the lytic release of SV40. PLoS Pathog. 2007;3:e98. doi: 10.1371/journal.ppat.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious v5.4. 2011 Available from http://www.geneious.com/
- Drummond AJ, Andrew R. Tracer v1.5. 2009. [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3:e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb KA, Villarreal LP. Natural biology of polyomavirus middle T antigen. Microbiol Mol Biol Rev. 2001;65:288–318. doi: 10.1128/MMBR.65.2.288-318.2001. second and third pages, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hatwell JN, Sharp PM. Evolution of human polyomavirus JC. J Gen Virol. 2000;81:1191–1200. doi: 10.1099/0022-1317-81-5-1191. [DOI] [PubMed] [Google Scholar]
- Huang Y, Carmichael GG. RNA processing in the polyoma virus life cycle. Front Biosci. 2009;14:4968–4977. doi: 10.2741/3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Johne R, Buck CB, Allander T, Atwood WJ, Garcea RL, Imperiale MJ, Major EO, Ramqvist T, Norkin LC. Taxonomical developments in the family Polyomaviridae. Arch Virol. 2011;156:1627–1634. doi: 10.1007/s00705-011-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R, Müller H. Polyomaviruses of birds: etiologic agents of inflammatory diseases in a tumor virus family. J Virol. 2007;81:11554–11559. doi: 10.1128/JVI.01178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumbholz A, Bininda-Emonds OR, Wutzler P, Zell R. Phylogenetics, evolution, and medical importance of polyomaviruses. Infect Genet Evol. 2009;9:784–799. doi: 10.1016/j.meegid.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Losada M, Christensen RG, McClellan DA, Adams BJ, Viscidi RP, Demma JC, Crandall KA. Comparing phylogenetic codivergence between polyomaviruses and their hosts. J Virol. 2006;80:5663–5669. doi: 10.1128/JVI.00056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Cowie A, Carr A, Glaichenhaus N, Kamen R, Cuzin F. The roles of individual polyoma virus early proteins in oncogenic transformation. Nature. 1982;300:713–718. doi: 10.1038/300713a0. [DOI] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage V, Foulongne V, Cheval J, Ar Gouilh M, Pariente K, Dereure O, Manuguerra JC, Richardson J, Lecuit M, Burguière A, Caro V, Eloit M. Human polyomavirus related to African green monkey lymphotropic polyomavirus. Emerg Infect Dis. 2011;17:1364–1370. doi: 10.3201/eid1708.110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kühn J, Hengel H, Ehlers B. A novel human polyomavirus closely related to the african green monkey-derived lymphotropic polyomavirus. J Virol. 2011;85:4586–4590. doi: 10.1128/JVI.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Simmonds P. Evaluating the evidence for virus/host co-evolution. Curr Opin Virol. 2011;1:436–441. doi: 10.1016/j.coviro.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebrasse E, Bauer I, Holtz L, Le B-m, Lassa-Claxton S, Hmiel P, Shenoy S, Sweet S, Turmelle Y, Shepherd R, Wang D. Prevalence of human polyomaviruses in a pediatric transplant cohort. Emerging Infectious Diseases. 2012a In Press. [Google Scholar]
- Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D. Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol. 2012b doi: 10.1128/JVI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, McClellan AJ, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky JL, Pipas JM. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Cantalupo P, Pipas JM. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol Cell Biol. 2000;20:6233–6243. doi: 10.1128/mcb.20.17.6233-6243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R, Cowie A, Favaloro J, Jat P, Kamen R. The structures of the spliced mRNAs encoding polyoma virus early region proteins. J Mol Appl Genet. 1981;1:83–92. [PubMed] [Google Scholar]
- Trowbridge PW, Frisque RJ. Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. J Neurovirol. 1995;1:195–206. doi: 10.3109/13550289509113966. [DOI] [PubMed] [Google Scholar]
- van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6:e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ghelue M, Khan MT, Ehlers B, Moens U. Genome analysis of the new human polyomaviruses. Rev Med Virol. 2012 doi: 10.1002/rmv.1711. [DOI] [PubMed] [Google Scholar]
- Woolford L, Rector A, Van Ranst M, Ducki A, Bennett MD, Nicholls PK, Warren KS, Swan RA, Wilcox GE, O'Hara AJ. A novel virus detected in papillomas and carcinomas of the endangered western barred bandicoot (Perameles bougainville) exhibits genomic features of both the Papillomaviridae and Polyomaviridae. J Virol. 2007;81:13280–13290. doi: 10.1128/JVI.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerrahn J, Knippschild U, Winkler T, Deppert W. Independent expression of the transforming amino-terminal domain of SV40 large I antigen from an alternatively spliced third SV40 early mRNA. EMBO J. 1993;12:4739–4746. doi: 10.1002/j.1460-2075.1993.tb06162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]