Abstract
Background
High levels of cardiac troponin T measured by a highly sensitive assay (hs-cTnT) are strongly associated with incident coronary heart disease (CHD) and heart failure (HF). No large-scale genome-wide association study (GWAS) of hs-cTnT has been reported to date. We sought to identify novel genetic variants that are associated with hs-cTnT levels.
Methods and Results
We performed a GWAS in 9,491 European-Americans and 2,053 African-Americans free of CHD and HF from 2 prospective cohorts: the Atherosclerosis Risk in Communities Study (ARIC) and the Cardiovascular Health Study (CHS). GWASs were conducted in each study and race stratum. Fixed-effect meta-analyses combined the results of linear regression from 2 cohorts within each race stratum, and then across race strata to produce overall estimates and p-values. The meta-analysis identified a significant association at chromosome 8q13 (rs10091374, p = 9.06 × 10−9) near the nuclear receptor coactivator 2 (NCOA2) gene. Over-expression of NCOA2 can be detected in myoblasts An additional analysis using logistic regression and the clinically motivated 99th percentile cut-point detected a significant association at 1q32 (rs10091374, p = 9.06 × 10−8) in the gene TNNT2, which encodes the cardiac troponin T protein itself. The hs-cTnT-associated SNPs were not associated with CHD in a large case-control study, but rs12564445 was significantly associated with incident HF in ARIC European-Americans (HR = 1.16, p-value = 0.004).
Conclusions
We identified 2 loci, near NCOA2 and in the TNNT2 gene, at which variation was significantly associated with hs-cTnT levels. Further use of the new assay should enable replication of these results.
Keywords: genetics, genome-wide association study, troponin
Cardiac troponin T (cTnT) is a thin filament protein that participates in cardiac muscle contraction. Detection of cTnT in peripheral blood indicates cardiomyocyte injury, and cTnT is one of the preferred biochemical markers for diagnosis and prognosis of acute coronary syndromes.1, 2 This low molecular weight protein can be released into the circulation not only after damage to myocytes, but also after inflammation and trauma.3, 4 The detection limit of the conventional assay for cTnT is approximately 0.01 μg per liter, but studies have shown that cTnT levels below this limit may further differentiate individuals at high or low risk for future cardiovascular events or death.5, 6 A recently developed highly sensitive assay for cardiac troponin T (hs-cTnT) can detect hs-cTnT levels 10 times lower than conventional assays.7 A population-based study of older adults reported that increased levels when compared to undetectable levels of hs-cTnT are associated with incident heart failure (HF) and cardiovascular mortality.8 Moreover, in a population-based sample without coronary heart disease (CHD), hs-cTnT is a significant predictor of incident CHD, as well as overall mortality and HF.9
Cardiac troponin T is encoded by the TNNT2 gene, and mutations in this gene account for approximately 15% of familial hypertrophic cardiomyopathy, which is associated with high risk of sudden cardiac death.10, 11 It is plausible that serum cTnT levels may be influenced by genetic variation in the TNNT2 and other genes, but to date no genome-wide association study of cTnT levels has been published. In this study, we sought to identify novel genetic variants that contribute to cTnT levels, which were measured by the new highly sensitive assay.
Methods
Study populations
ARIC is a prospective cohort study designed to ascertain the etiology and predictors of cardiovascular disease (CVD), which enrolled 15,792 middle-aged adults from four U.S. communities in 1987–1989. CHS is a prospective observational cohort study designed to investigate CVD in older adults, which enrolled 5,201 individuals in the original cohort in 1989–1990, with further enrollment of a minority sample of 687 African-Americans in 1992–1993. Their detailed designs have been published elsewhere.12, 13 Participants were excluded if they had prevalent CHD or HF when hs-cTnT levels were measured, if they were first degree relatives of someone else in the study, if there were sample handling errors or race or sex discrepancies between reported data and genotype data, or if they did not give consent for use of DNA information.
hs-cTnT Measurement
In ARIC, hs-cTnT levels were measured using visit 4 plasma samples in 1996–1998. In CHS, hs-cTnT levels were measured at baseline in 1989–90 and in 1992–93 for European Americans and African Americans recruited in the original cohort, and in 1992–93 and 1994–95 for African Americans recruited in the minority cohort. Plasma samples were stored at −70 to −80°C and thawed before testing. Hs-cTnT levels were measured with the Elecsys Troponin T high sensitive assay [Roche Diagnostics, Indianapolis, IN]. The range of detection of this assay is from 0.003 – 10 μg/L. For CHS participants with more than one hs-cTnT measurement, we selected the first measurement.
Genotyping and Imputation
Autosomal SNPs were genotyped using the Affymetrix 6.0 chip for ARIC and the Illumina 370CNV chip for CHS. Each study imputed their genotype data to the ~2.5 million SNPs identified in HapMap CEU samples for European Americans. For African-Americans, SNP data were imputed based on a panel of reference haplotypes using HapMap CEU and YRI samples. MACH v1.0 in ARIC and BIMBAM v0.99 in CHS were used to do the imputation, and allele dosage information was summarized in the imputation results. SNPs were excluded if they had no chromosomal location, were monomorphic, had a call rate < 95%, or had a Hardy-Weinberg equilibrium p-value <10−6 for ARIC or p-value <10−5 for CHS. For each eligible SNP in both cohorts, the ratio of the observed versus expected variance of the dosage served as a measure of imputation quality. A ratio < 0.3 was considered to be poor imputation quality, and these SNPs were removed from further analyses.
Statistical Analyses
Hs-cTnT levels were treated as a continuous and a dichotomous variable, in two distinct regression analyses. In addition to genotype, covariates used in the regression were obtained at the time when hs-cTnT levels were measured. When analyzing continuous hs-cTnT levels, standard linear regressions were used. In European-Americans, the adjustment covariates were age, sex and study site. In African-Americans, additional covariates included the first 10 principal components to account for population stratification.14 Individuals whose cTnT levels were below the lowest detectable limit were assigned a value of 0.003 ug/L (the known detection limit of the assay7). All cTnT values were natural log-transformed prior to analysis. When analyzing dichotomized hs-cTnT levels, we used the recently-recommend value of greater than the 99th percentile of hs-cTnT concentration to diagnose non-ST- elevation myocardial infarction (NSTEMI).7, 15 Therefore, we grouped hs-cTnT levels into 2 categories: < 99th percentile vs. ≥ 99th percentile and used logistic regression with this binary outcome. Because of the small number of African-Americans in the two cohorts with hs-cTnT levels ≥ 99th percentile, the logistic regression was applied only in the European-Americans. The study-specific 99th percentile of hs-cTnT for ARIC and CHS is 0.027 ug/L and 0.036 ug/L, respectively.
In all analyses, the effects of SNPs were estimated under an additive genetic model; a locus with two copies of the coded alleles was coded as 2, the heterozygote was coded as 1 and two copies of the non-coded alleles was coded as 0. For each model and after genomic control adjustment, inverse variance fixed-effect meta-analyses were used to combine the results of the 2 cohorts within race strata, and then again to meta-analyze the race-specific results to obtain an overall β coefficient, standard error and p-value. Because of the relative small sample size of African Americans, analyses with low MAF would be underpowered or lead to spurious associations. Thus, SNPs with a minor allele frequency (MAF) < 1% for European Americans and < 10% for African Americans, or absolute value of β coefficient > 5 were excluded before the meta-analyses. Only SNPs present in both cohorts and race strata were included in the meta-analysis results. Quantile-quantile (QQ) plots of the observed and expected p-values for all eligible SNPs were generated for each analysis to illustrate the behavior of the test statistics. Genome-wide significance was defined as an overall p-value < 5 × 10−8 in this study, and a pvalue < 1 × 10−5 was regarded as suggestive evidence for association. If more than one suggestive or significant SNP clustered at a genetic locus, the SNP with the smallest p-value was reported as the locus’s sentinel marker. Forest plots showing cohort/race specific findings and regional plots showing linkage disequilibrium and gene information were generated for genome-wide significant SNPs in each analytic model. All analyses in CHS were performed by R (www.rproject.org), and by ProbABEL16 in ARIC. Meta-analyses were performed by METAL (http://genome.sph.umich.edu/wiki/METAL_Program). The estimated post-hoc power based on the parameter estimates presented here are: 1) For the linear model: 85% power with 12,000 individuals, MAF = 0.48, alpha = 5 × 10−8 and 0.35% variation explained by the SNP. 2) Binary model: 70% power with 10,000 individuals, MAF = 0.2, alpha = 5 × 10−8 and OR = 2.5.
Since hs-cTnT levels are known to have a skewed distribution, we analyzed the association of the top ranking SNP in the linear model with categorical hs-cTnT levels using proportional odds logistic regression8,9. We grouped the undetectable cTnT levels as the reference group, and the remaining participants were divided into approximate fourths (0.003–0.0055 ug/L, 0.0056–0.0085 ug/L, 0.0086–0.0135 ug/L and > 0.0135 ug/L). The proportional odds assumption was tested using the Brant test and the same strategy was applied for the meta-analysis as described above. These analyses were performed using Stata (StataCorp, www.stata.com) and R (www.r-project.org).
Association with HF and CHD
The association between the genome-wide significant SNPs in each model and incident HF were tested in ARIC using a Cox proportional hazards model adjusted for age, sex and study site. The analyses were performed by R (www.r-project.org). In ARIC, the diagnosis of HF was based on International Classification of Diseases, Ninth Revision (ICD-9) code 428.3, while incident HF was defined as the first hospitalization or death from HF for those without a prior HF hospitalization. Individuals were followed up for events through December 31, 2008; those who were lost to follow-up were censored at the date of last contact. The association between the genome-wide significant SNPs and CHD were examined from results in 22,233 cases and 64,762 controls from the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study.17, 18 Although details differed among the contributing studies in CARDIoGRAM, the definition of CHD included clinically defined myocardial infarction or angiographically accessed coronary artery disease. Information about the CARDIoGRAM study is provided in Supplemental A.
Results
A total of 9,491 European-Americans and 2,053 African-Americans were included in these genome-wide association analyses. The characteristics of 11,544 study participants free of CHD and HF at the time hs-cTnT levels were measured are shown in Table 1. The average age at baseline varied from 61.68 to 72.82 and females were the majority in each cohort. By design, CHS study participants were older than those in ARIC. The mean hs-cTnT levels and the proportion of hs-cTnT levels below the detectable limit were similar across race strata and cohorts. Both European-Americans and African-Americans had average body mass indices greater than 25, and African Americans tended to have higher prevalence of hypertension and diabetes.
Table 1.
Characteristics of Participants by Cohort and Race
| Characteristics | ARIC
|
CHS
|
||
|---|---|---|---|---|
| EA | AA | EA | AA | |
| Participants, n | 6460 | 1539 | 3031 | 514 |
| Age, y | 62.86 ± 5.61 | 61.68 ± 5.70 | 72.74 ± 5.37 | 72.82 ± 5.55 |
| Age range, y | (53.00, 75.00) | (53.00, 75.00) | (65.00, 95.00) | (65.00, 92.00) |
| Female, % | 56.11 | 64.85 | 62.22 | 64.98 |
| hs-cTnT, μg/L* | 0.004 (0.003, 0.007) | 0.005 (0.003, 0.008) | 0.005 (0.003, 0.009) | 0.005 (0.003, 0.01) |
| Participants with hs-cTnT below the detectable limit, % | 34.0 | 32.6 | 37.1 | 40.3 |
| Body mass index, kg/m2 | 28.11 ± 5.19 | 30.47 ± 6.18 | 26.29 ± 4.45 | 28.41 ± 5.39 |
| Systolic blood pressure, mmHg | 125.56 ± 18.34 | 133.16 ± 19.74 | 135.23 ± 21.13 | 141.75 ± 22.04 |
| Diastolic blood pressure, mmHg | 70.04 ± 9.76 | 75.84 ± 10.46 | 70.57 ± 11.15 | 76.43 ± 11.50 |
| Total cholesterol, mg/dl | 202.39 ± 35.6 | 199.48 ± 37.73 | 212.34 ± 38.48 | 207.98 ± 38.60 |
| Triglyceride, mg/dl | 148.9 ± 83.95 | 113.59 ± 64.74 | 162.27 ± 146.04 | 112.92 ± 55.72 |
| Low-density, lipoprotein, mg/dl | 123.25 ± 32.35 | 123.65 ± 35.37 | 129.72 ± 34.84 | 127.25 ± 34.72 |
| High-density lipoprotein, mg/dl | 49.88 ± 16.31 | 53.25 ± 16.53 | 55.21 ± 15.65 | 58.99 ± 15.70 |
| Current smoker, % | 14.23 | 17.8 | 10.22 | 16.54 |
| Prevalent hypertension, % | 39.36 | 64.32 | 52.89 | 56.61 |
| Prevalent diabetes, % | 11.72 | 24.98 | 12.44 | 20.23 |
hs-cTnT levels, median (quartile 1, quartile 3). For continuous variables, mean values ± standard errors are shown. Categorical variables are given as percentage. EA indicates European Americans; AA, African Americans.
In ARIC, prevalent hypertension included use of antihypertensive medication or measured blood pressure (systolic blood pressure ≥ 140 mmHg, diastolic blood pressure ≥ 90 mmHg); prevalent diabetes was defined as fasting glucose levels ≥ 126 mg/dl, nonfasting glucose levels ≥ 200 mg/dl, or self-reported diagnosis of diabetes or use of diabetic medication. In CHS, prevalent hypertension included measured blood pressure (systolic blood pressure ≥140% mmHg, diastolic blood pressure ≥90 mmHg) or use of antihypertensive drug and reports of physician diagnosis of hypertension; prevalent diabetes was defined as fasting glucose levels ≥ 126 mg/dl, or use of insulin or an oral hypoglycemic drug.
Linear Model and Proportional odds Logistic Model
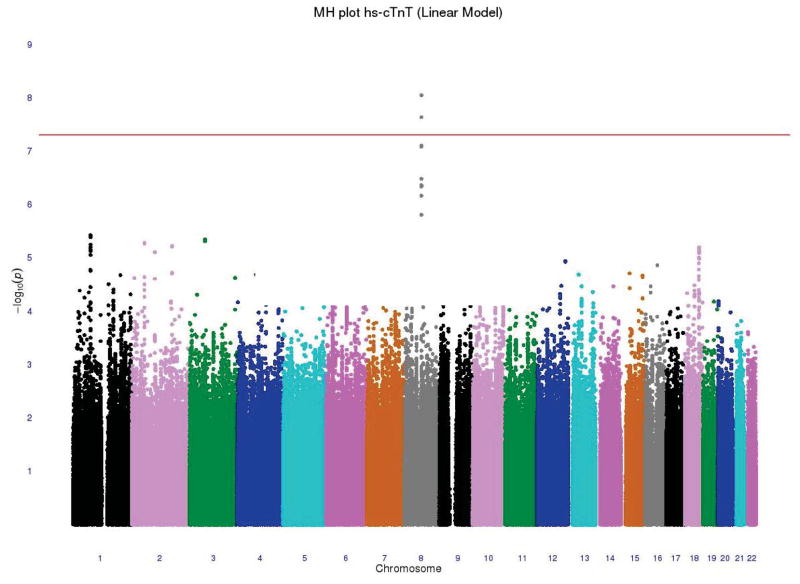
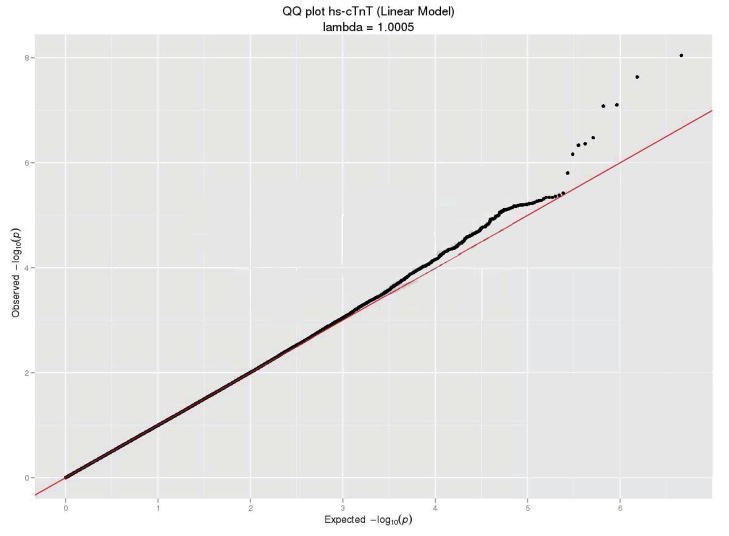
For the analyses of hs-cTnT as a continuous variable, the race-and-cohort-specific genomic control parameters were 1.03 for ARIC EAs, 1.03 for ARIC AAs, 1.02 for CHS EAs and 1.05 for CHS AAs. After doing the two-stage meta-analyses, two SNPs exceeded the genome-wide significance threshold (p-value < 5 × 10−8). The overall genomic control parameter of 1.0 for this model suggested negligible population stratification. The Manhattan plot and QQ plot of the two-stage meta-analyzed p-values are shown in Figure 1. Twelve genetic loci were identified with genome-wide suggestive evidence for association (p-value < 1 × 10−5). These suggestive loci, presented in Supplemental B, are not discussed further.
Figure 1.
(A) −log10 p-value of an additive genetic model for each SNP according to its location in the 22 autosomal chromosomes from the two-stage meta-analysis (linear analysis). Horizontal line indicates the a priori 5 × 10−8 threshold of genome-wide significance. (B) Plot of the expected and observed −log p-values from the two-stage meta-analysis
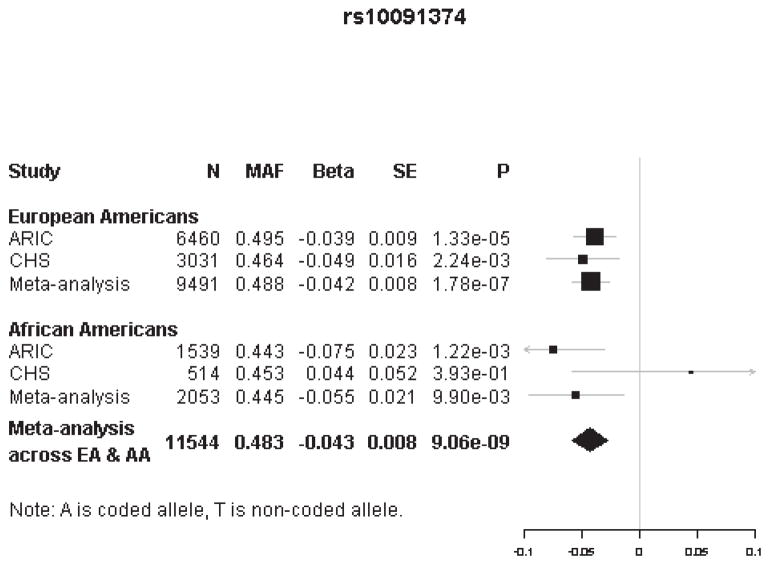
The two genome-wide significant SNPs were rs10091374 (p-value = 9.06 × 10−9) and rs6989313 (p-value = 2.33 × 10−8) located at chromosome 8q13. These two SNPs were in strong linkage disequilibrium (LD), r2 ≥ 0.8. The top SNP, rs10091374, was an imputed SNP with MAF = 0.483 and estimated effect size β = −0.04 (T→A), corresponding to a ~4% reduction in hscTnT level per additional A allele. This SNP lies between two genes; it is 70.9 kb from NCOA2 (nuclear receptor coactivator 2) and 98.5 kb from TRAM1 (translocation associated membrane protein 1). A forest plot of the two-stage meta-analysis result for rs10091374 is shown in Figure 2, and information about LD and other SNPs in this region are presented in Supplemental B.
Figure 2.
Forest plot shows the two-stage meta-analysis (linear analysis) of the association between rs10091374 and hs-cTnT levels (EA indicates European Americans; AA, African Americans; N, number of sample; MAF, minor allele frequency; BETA, β coefficients; SE, standard error; P, p-value).
We further conducted proportional odds logistic regression for rs10091374, and per additional A allele for rs10091374, the odds of being in the four higher hs-cTnT categories versus the undetectable hs-cTnT category was reduced by 11% (OR = 0.89, p-value = 4.5 × 10−6).
Logistic Model
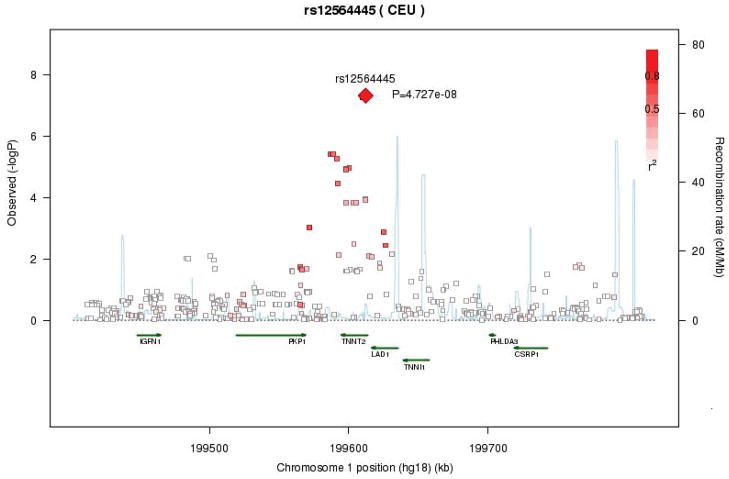
In the dichotomized analyses of hs-cTnT for European Americans, the genomic control parameters were 1.01 for ARIC, 1.06 for CHS, and 1.0 after the meta-analysis. One SNP, rs12564445, reached the genome-wide significance threshold (OR = 2.33, G→A p-value = 4.73 × 10−8, MAF = 0.197). This SNP was imputed in ARIC and CHS EA, but genotyped in CHS AA. Rs12564445 is located in an intron of the gene that codes for cardiac troponin T type 2 (TNNT2), and is responsible for 133% greater odds of being in the 99th percentile group for each additional A allele. Information about LD and other SNPs in this region is presented in Figure 3. In addition to this region, eight more regions were identified with suggestive evidence for genome-wide association (p-value < 1 × 10−5). Details about these suggestive regions are provided in Supplemental B. The effect of rs12564445 on the continuous measure of hs-cTnT was not statistically significant (β = −0.0017, p-value = 0.86), corresponding to a 0.2% reduction in hs-cTnT level per additional A allele. The effect of rs10091374, which was significant in the linear model meta-analysis of the continuous measure of hs-cTnT, was not statistically significant for the categorical measure (OR=0.79, p-value = 0.10).
Figure 3.
Regional association plot of hs-cTnT for the genome-wide significant marker rs12564445 in European Americans (linear analysis).
Association with HF and CHD
We next analyzed the association of rs10091374 and rs12564445 with 1119 incident HF events in ARIC European Americans and with 22,233 CHD cases from the CARDIoGRAM consortium.17, 18 In all cases except one, the results were not statistically significant (data not shown; p-value > 0.05). Rs12564445 was significantly associated with incident HF among 8,894 ARIC European Americans with an average 18 years follow-up time (HR = 1.16, 95%CI: 1.05–1.28, p-value = 0.004). We repeated the analyses with a competing risk model developed by Fine and Grey19 and the results were very similar to the Cox regression reported here, subhazard ratio was 1.17 (95%CI: 1.06–1.30, p = 0.002).
Discussion
This study evaluated the association between genetic variants and hs-cTnT levels among participants free of CHD and HF. The meta-analysis, which included 9,491 European Americans and 2,053 African Americans, identified one locus on chromosome 8q13 that was significantly associated with hs-cTnT levels and 12 additional suggestive loci. Furthermore, one locus, TNNT2, was associated with high hs-cTnT levels (≥ 99th percentile) in 9,491 European Americans together with eight additional loci showing suggestive association.
The effects of both SNPs on quantitative and categorical hs-cTnT levels are generally consistent. For rs10091374, the direction of effect is the same in all four analyses (two race groups × two phenotype definitions). There is ~ 21% lower odds of being in the 99th percentile of hs-cTnT per additional A allele, which is consistent with the A allele lowering hs-cTnT levels. For rs12564445, the effect is consistent in African Americans but not in European Americans. It is possible that the significant association of rs12564445 with elevated hs-cTnT levels represents the biologic effect of a low frequency variant private to European Americans.
The gene most likely responsible for the association between hs-cTnT levels and chromosome 8q13 is NCOA2, which encodes a transcriptional coregulatory protein that aids in the function of nuclear hormone receptors. Our observation that an additional A allele at this locus decreases hs-cTnT levels was confirmed by proportional odds logistic regression. Overexpression of NCOA2 has been detected in both proliferating and confluent myoblasts and Western blot analysis shown that it increases during myogenesis,20 and reduced expression has been observed in pulmonary arterial hypertension (PAH).21 Thus, NCOA2 may play a role in promoting muscle cells maintenance and growth, eventually influencing cTnT levels.
In addition, we hypothesize that specific genes may contribute to high hs-cTnT levels ≥ 99th percentile in the population). Under such model, rs12564445 in the TNNT2 gene reached the genome-wide significance threshold in our analysis of. TNNT2 encodes cardiac troponin T, and it is well-known that mutations in this gene can cause familial hypertrophic cardiomyopathy (CMH),22–24 familial dilated cardiomyopathy (CMD),25–27 and left ventricular noncompaction.28 The specificity of hs-cTnT assay for categorizing NSTEMI is ~80%.7 Given the function of TNNT2, it would be interesting to explore if variation in TNNT2 contributes to the remaining ~20% false-positives.
Several recent epidemiological studies have reported that hs-cTnT levels predict incident HF and CHD in multiple populations.8, 9, 29 In this GWAS, two loci reached genome-wide significance, but only rs12564445 was significantly associated with incident HF in European Americans and neither of them was significantly associated with CHD. In previous studies, mutations in TNNT2 have been associated with hypertrophic cardiomyopathy 21–23 and in ARIC this variant is associated with ECG-determined left ventricular hypertrophy (data not shown). In this study, TNNT2 gene variation is related to both hs-cTnT levels and incident HF, and it has been repeatedly reported that hs-cTnT levels are associated with incident HF. It is likely that TNNT2 alters the levels of hs-cTnT, which subsequently influences the onset of HF. But such relationships were not found for incident CHD. Given these results, we suggest that hs-cTnT codes may not be in the causal pathway for CHD, but may in some cases be in the causal pathway for HF.
Strengths and Limitations
This study is the first GWAS to reveal genetic risk variants for hs-cTnT levels. The effect size and MAF of the genetic variant detected in the linear model were consistent across European Americans and African Americans, which provides some evidence that the genetic mechanism for hs-cTnT may be similar between the two groups. Our study also has several limitations. A study in CHS indicated that the changes of hs-cTnT levels over time were associated with HF,8 but hs-cTnT levels were only measured at one visit in ARIC. Thus, we were unable to comment on the genetic contribution to the variability of hs-cTnT levels. Different thresholds for hs-cTnT based on 99th study specific percentiles were used in the present study. We hypothesized that genetic variation contributes to very high hs-cTnT levels, but discovered that the 99th percentile of 0.014 ug/L provided by the manufacturer is based on healthy people who are approximately 10 and 20 years younger than ARIC and CHS, respectively.30 Because of the increased age of CHS and ARIC participants, the distribution of hs-cTnT was shifted up in both studies. Thus, we analyzed a study-specific 99th percentile threshold, which could complicate future replication studies. For African Americans, we could not examine genetic association with extremely high hs-cTnT levels due to limitations of the available sample size. Even though the sensitivity of this new cTnT assay is improved, there still is a lowest detectable limit, and those with missing data at baseline were imputed with this low value limit as a proxy for an undetectably low true value. If this low value were due to genetic variation, the limitations of the assay may impact the statistical power of these analyses. Finally, the effect of the significant SNP observed under the linear model was interpreted in the context of the nearest gene, but it is possible that the true casual variant may located in another gene or in an intergenic region.
Conclusions
We identified two loci that reached genome-wide significance from a GWAS of hs-cTnT in two prospective cohort studies: ARIC and CHS. One SNP, rs10091374, near NCOA2 on chromosome 8q13 was significantly associated with quantitative hs-cTnT levels in European Americans and African Americans. The other SNP, rs12564445, in TNNT2 on chromosome 1q32 was significantly related to high levels of hs-cTnT in European Americans. These results contribute to the knowledge-base of cTnT physiology and to our understanding of its role as a biomarker of cardiac injury. Further use of the new assay should enable replication of these results.
Supplementary Material
Acknowledgments
We acknowledge the essential role of the CHARGE Consortium in developing and support for this article. CHARGE members include ARIC, CHS, FHS, RS and Age, Gene/Environment Susceptibility. The authors also thank the staff and participants of the ARIC and CHS study for their important contributions. B.Y. is supported in part by the Burroughs Wellcome Fund.
Funding Sources: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
This CHS research was supported by NHLBI contracts N01-HC-85239, N01-HC-85079 through N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133 and NHLBI grants HL080295, HL075366, HL085251, HL087652, HL105756 with additional contribution from NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the NIA. See also http://www.chsnhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center for Research Resources CTSI grant UL 1RR033176 and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center and the Cedars-Sinai Board of Governors’ Chair in Medical Genetics (JIR).
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Lindahl B, Toss H, Siegbahn A, Venge P, Wallentin L. Markers of myocardial damage and inflammation in relation to long-term mortality in unstable coronary artery disease. Frisc study group. Fragmin during instability in coronary artery disease. The New England journal of medicine. 2000;343:1139–1147. doi: 10.1056/NEJM200010193431602. [DOI] [PubMed] [Google Scholar]
- 2.Gupta S, de Lemos JA. Use and misuse of cardiac troponins in clinical practice. Prog Cardiovasc Dis. 2007;50:151–165. doi: 10.1016/j.pcad.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Spies C, Haude V, Fitzner R, Schroder K, Overbeck M, Runkel N, et al. Serum cardiac troponin t as a prognostic marker in early sepsis. Chest. 1998;113:1055–1063. doi: 10.1378/chest.113.4.1055. [DOI] [PubMed] [Google Scholar]
- 4.Fulda GJ, Giberson F, Hailstone D, Law A, Stillabower M. An evaluation of serum troponin t and signal-averaged electrocardiography in predicting electrocardiographic abnormalities after blunt chest trauma. J Trauma. 1997;43:304–310. doi: 10.1097/00005373-199708000-00016. [DOI] [PubMed] [Google Scholar]
- 5.de Lemos JA, Drazner MH, Omland T, Ayers CR, Khera A, Rohatgi A, et al. Association of troponin t detected with a highly sensitive assay and cardiac structure and mortality risk in the general population. JAMA. 2010;304:2503–2512. doi: 10.1001/jama.2010.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochholzer W, Reichlin T, Twerenbold R, Stelzig C, Hochholzer K, Meissner J, et al. Incremental value of high-sensitivity cardiac troponin t for risk prediction in patients with suspected acute myocardial infarction. Clin Chem. 2011;57:1318–1326. doi: 10.1373/clinchem.2011.162073. [DOI] [PubMed] [Google Scholar]
- 7.Giannitsis E, Kurz K, Hallermayer K, Jarausch J, Jaffe AS, Katus HA. Analytical validation of a high-sensitivity cardiac troponin t assay. Clin Chem. 2010;56:254–261. doi: 10.1373/clinchem.2009.132654. [DOI] [PubMed] [Google Scholar]
- 8.deFilippi CR, de Lemos JA, Christenson RH, Gottdiener JS, Kop WJ, Zhan M, et al. Association of serial measures of cardiac troponin t using a sensitive assay with incident heart failure and cardiovascular mortality in older adults. JAMA. 2010;304:2494–2502. doi: 10.1001/jama.2010.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saunders JT, Nambi V, de Lemos JA, Chambless LE, Virani SS, Boerwinkle E, et al. Cardiac troponin t measured by a highly sensitive assay predicts coronary heart disease, heart failure, and mortality in the atherosclerosis risk in communities study. Circulation. 2011;123:1367–1376. doi: 10.1161/CIRCULATIONAHA.110.005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watkins H, McKenna WJ, Thierfelder L, Suk HJ, Anan R, O’Donoghue A, et al. Mutations in the genes for cardiac troponin t and alpha-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med. 1995;332:1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 11.Pasquale F, Syrris P, Kaski JP, Mogensen J, McKenna WJ, Elliott P. Long-term outcomes in hypertrophic cardiomyopathy caused by mutations in the cardiac troponin t gene. Circ Cardiovasc Genet. 2012;5:10–17. doi: 10.1161/CIRCGENETICS.111.959973. [DOI] [PubMed] [Google Scholar]
- 12.The atherosclerosis risk in communities (aric) study: Design and objectives. The aric investigators. American journal of epidemiology. 1989;129:687–702. [PubMed] [Google Scholar]
- 13.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, et al. The cardiovascular health study: Design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 14.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.Giannitsis E, Becker M, Kurz K, Hess G, Zdunek D, Katus HA. High-sensitivity cardiac troponin t for early prediction of evolving non-st-segment elevation myocardial infarction in patients with suspected acute coronary syndrome and negative troponin results on admission. Clin Chem. 2010;56:642–650. doi: 10.1373/clinchem.2009.134460. [DOI] [PubMed] [Google Scholar]
- 16.Aulchenko YS, Struchalin MV, van Duijn CM. Probabel package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preuss M, Konig IR, Thompson JR, Erdmann J, Absher D, Assimes TL, et al. Design of the coronary artery disease genome-wide replication and meta-analysis (cardiogram) study: A genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet. 2010;3:475–483. doi: 10.1161/CIRCGENETICS.109.899443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 20.Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, grip-1, is necessary for mef-2c-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- 21.Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, Kaminski N, et al. Genomewide rna expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol. 2010;298:H1235–1248. doi: 10.1152/ajpheart.00254.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, et al. Alphatropomyosin and cardiac troponin t mutations cause familial hypertrophic cardiomyopathy: A disease of the sarcomere. Cell. 1994;77:701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 23.Tardiff JC, Factor SM, Tompkins BD, Hewett TE, Palmer BM, Moore RL, et al. A truncated cardiac troponin t molecule in transgenic mice suggests multiple cellular mechanisms for familial hypertrophic cardiomyopathy. J Clin Invest. 1998;101:2800–2811. doi: 10.1172/JCI2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menon SC, Michels VV, Pellikka PA, Ballew JD, Karst ML, Herron KJ, et al. Cardiac troponin t mutation in familial cardiomyopathy with variable remodeling and restrictive physiology. Clin Genet. 2008;74:445–454. doi: 10.1111/j.1399-0004.2008.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 26.Mogensen J, Murphy RT, Shaw T, Bahl A, Redwood C, Watkins H, et al. Severe disease expression of cardiac troponin c and t mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2033–2040. doi: 10.1016/j.jacc.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 27.Mirza M, Marston S, Willott R, Ashley C, Mogensen J, McKenna W, et al. Dilated cardiomyopathy mutations in three thin filament regulatory proteins result in a common functional phenotype. J Biol Chem. 2005;280:28498–28506. doi: 10.1074/jbc.M412281200. [DOI] [PubMed] [Google Scholar]
- 28.Klaassen S, Probst S, Oechslin E, Gerull B, Krings G, Schuler P, et al. Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation. 2008;117:2893–2901. doi: 10.1161/CIRCULATIONAHA.107.746164. [DOI] [PubMed] [Google Scholar]
- 29.Everett BM, Cook NR, Magnone MC, Bobadilla M, Kim E, Rifai N, et al. Sensitive cardiac troponin t assay and the risk of incident cardiovascular disease in women with and without diabetes mellitus: The women’s health study. Circulation. 2011;123:2811–2818. doi: 10.1161/CIRCULATIONAHA.110.009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mingels A, Jacobs L, Michielsen E, Swaanenburg J, Wodzig W, van Dieijen-Visser M. Reference population and marathon runner sera assessed by highly sensitive cardiac troponin t and commercial cardiac troponin t and i assays. Clinical chemistry. 2009;55:101–108. doi: 10.1373/clinchem.2008.106427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.