SUMMARY
The tuberous sclerosis complex (TSC) tumor suppressors form the TSC1-TSC2 complex, which limits cell growth in response to poor growth conditions. Through its GTPase-activating protein (GAP) activity toward Rheb, this complex inhibits the mechanistic target of rapamycin (mTOR) complex 1 (mTORC1), a key promoter of cell growth. Here, we identify and biochemically characterize TBC1D7 as a stably-associated and ubiquitous third core subunit of the TSC1-TSC2 complex. We demonstrate that the TSC1-TSC2-TBC1D7 (TSC-TBC) complex is the functional complex that senses specific cellular growth conditions and possesses Rheb-GAP activity. Sequencing analyses of samples from TSC patients suggest that TBC1D7 is unlikely to represent TSC3. TBC1D7 knockdown decreases the association of TSC1 and TSC2 leading to decreased Rheb-GAP activity, without effects on the localization of TSC2 to the lysosome. Like the other TSC-TBC components, TBC1D7 knockdown results in increased mTORC1 signaling, delayed induction of autophagy, and enhanced cell growth under poor growth conditions.
INTRODUCTION
The mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) is a protein kinase complex that plays a key evolutionarily conserved role in promoting cell growth (i.e., an increase in cell size) through the inhibition of catabolic processes, such as autophagy, and stimulation of anabolic processes, including protein and lipid synthesis (Laplante and Sabatini, 2012). Due to the substantial energy and nutrient demands of such anabolic processes, cells have evolved an exquisite network of signaling pathways that sense and relay the status of cellular growth conditions to mTORC1. Two classes of small G-proteins, the Rag and Rheb GTPases, lie directly upstream of mTORC1 to control its activation state in response to specific growth signals. Recent evidence suggests that the Rag proteins, in complex with the Ragulator, specifically mediate the ability of mTORC1 to sense amino acids (Kim et al., 2008; Sancak et al., 2010; Sancak et al., 2008; Zoncu et al., 2011), which constitute an essential signal for mTORC1 activation (Hara et al., 1998). On the other hand, Rheb is controlled by numerous stimuli affecting mTORC1, including growth factors, hormones and cytokines, cellular energy levels, and stress (Huang and Manning, 2008; Laplante and Sabatini, 2012). Due to perturbations in the signaling network upstream of Rheb, mTORC1 is aberrantly regulated in a variety of disease settings, including genetic tumor syndromes, the majority of sporadic cancers, common neurological disorders, such as autism and Alzheimer’s, and metabolic diseases, such as obesity and type-2 diabetes (Ehninger and Silva, 2011; Laplante and Sabatini, 2012; Menon and Manning, 2009). Therefore, a detailed understanding of the regulation of Rheb and mTORC1 will provide mechanistic insights into both normal growth control and the molecular events contributing to the pathology of these diverse diseases.
TSC1 and TSC2 are the tumor suppressor genes mutated in the tumor syndromes tuberous sclerosis complex (TSC) and lymphangioleiomyomatosis (LAM), and their gene products form a protein complex that integrates signals upstream of Rheb and mTORC1. TSC1 and TSC2 (also referred to as hamartin and tuberin) are large proteins with limited similarity to other proteins, with the exception of an approximately 200 amino acid stretch at the C-terminus of TSC2 that resembles the GTPase-activating protein (GAP) domain of Rap1Gap. This domain within TSC2 acts as a GAP for Rheb, and complex formation with TSC1 stabilizes TSC2 and enhances its GAP activity (Garami et al., 2003; Inoki et al., 2003a; Tee et al., 2003; Zhang et al., 2003b). Through stimulation of the intrinsic GTPase activity of Rheb, the TSC1-TSC2 complex switches Rheb from its mTORC1-activating, GTP-bound state to its inactive GDP-bound state. Interestingly, most of the signals that regulate Rheb and mTORC1 impinge on the TSC1-TSC2 complex, such that poor growth conditions activate the complex while growth-promoting conditions inhibit the complex to, respectively, inhibit or activate Rheb and mTORC1 (Huang and Manning, 2008). For instance, many growth factors and cytokines activate mTORC1 via an Akt-mediated inhibitory phosphorylation of TSC2 within the complex (Inoki et al., 2002; Manning et al., 2002; Potter et al., 2002), while energy stress inhibits mTORC1, at least in part, through an AMPK-dependent activating phosphorylation on TSC2 (Inoki et al., 2003b; Shaw et al., 2004). Consistent with these signaling mechanisms, loss of function of the TSC1-TSC2 complex leads to constitutive mTORC1 activation that is largely insensitive to perturbations in cellular growth conditions (Jaeschke et al., 2002; Kwiatkowski et al., 2002). It is now clear that the TSC1-TSC2 complex is a point of convergence for a network of signaling pathways that convey information regarding cellular growth conditions to Rheb and mTORC1 to properly control cell growth. However, much remains to be understood regarding the molecular characteristics of this key signal-integrating node that is commonly misregulated in human diseases.
The TSC1-TSC2 complex is believed to function as a heterodimer (van Slegtenhorst et al., 1998). While dozens of interacting proteins have been described in the literature (Guo et al., 2010; Rosner et al., 2008), the functional significance of these associations remains unknown. Importantly, none of the proteins found to bind to the TSC1-TSC2 complex, in either hypothesis-driven or unbiased experiments, have been characterized as additional subunits of the complex. Here, we describe the identification of Tre2-Bub2-Cdc16 (TBC) 1 domain family, member 7 (TBC1D7) in a stringent search for proteins that associate with the TSC1-TSC2 complex. The biochemical characterization presented here demonstrates that TBC1D7 is a third resident component of the TSC1-TSC2 complex, and like the other members of the complex, it is required for the proper regulation of Rheb and mTORC1 by cellular growth conditions.
RESULTS
TBC1D7 is a stably-associated and ubiquitous binding partner of the TSC1-TSC2 complex
To better understand the molecular function of the TSC1-TSC2 complex, we sought to identify proteins that interact with the complex at high stoichiometry. Co-expressed flag-tagged TSC1 and TSC2 were immunopurified from cells and flag-peptide eluates were processed for protein identification using mass spectrometry. The most abundant protein in two independent purifications was a 293 amino acid (~34kDa) protein designated TBC1D7 (Figures 1A and S1A). While there are very few publications on this protein, two previous studies reported an association with TSC1 (Nakashima et al., 2007; Sato et al., 2010); see Discussion). Sequence alignment searches revealed that TBC1D7 is broadly conserved across multiple phyla (Figure S1B), with a conservation pattern that largely overlaps with that of TSC1 and TSC2 orthologs (Serfontein et al., 2010). TBC1D7 predominantly consists of a putative TBC domain (Figure S1C), a structure represented in the genomes of all eukaryotes. The TBC domains of several proteins have been shown to posses GAP activity towards specific members of the Rab family of small GTPases, and this is thought to be the conserved function of this domain family (Frasa et al., 2012). However, the central region of the TBC1D7 TBC domain does not align well with other TBC domains and, importantly, lacks two out of three signature motifs that contain residues shown to be essential for the Rab-GAP activity of other TBC proteins (Figures S1C, D; Frasa et al., 2012).
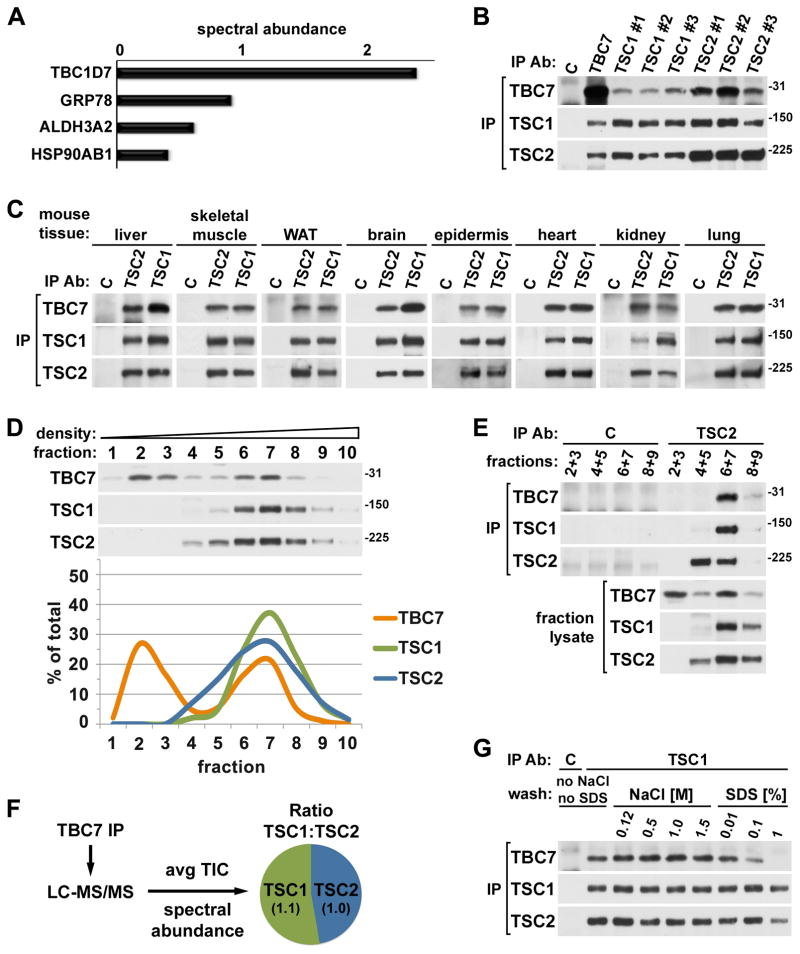
Figure 1. Endogenous TBC1D7 binds tightly and ubiquitously to the TSC1-TSC2 heterodimer.
(A) Relative abundance of proteins identified in TSC1-TSC2 purifications. Co-expressed Flag-TSC1 and -TSC2 were immunoprecipitated from lysates of HEK-293 cells, and tryptic peptides from 3xFlag-peptide eluates were identified using LC-MS/MS. Proteins with three or more peptides identified in both of two independent purifications, but absent in anti-Flag immunoprecipitates from cells expressing empty vector, are listed. The spectral abundance of a protein is calculated from the number of peptides identified (spectral count) and the amino acid length of the protein. See supporting data in Figure S1A.
(B) Reciprocal co-immunoprecipitations of endogenous TBC1D7, TSC1, and TSC2. Endogenous proteins were immunoprecipitated from HeLa cell lysates using the indicated antibodies (listed in Experimental Procedures). See supporting data in Figure S1E.
(C) Co-immunoprecipitation of TBC1D7 with TSC1 and TSC2 from mouse tissues. TSC1 and TSC2 were immunoprecipitated from lysates of the indicated mouse tissues. WAT (white adipose tissue).
(D) One of two cellular pools of TBC1D7 co-fractionates with TSC1 and TSC2 in a density gradient. HeLa cell lysate was ultracentrifuged over a sucrose gradient and ten sequential fractions of increasing density were collected. Immunoblot band intensities were quantified using ImageJ software and graphed as a percentage of the summed band intensities in all fractions for each protein.
(E) TBC1D7 and TSC1 co-immunoprecipitate with TSC2 from high-density fractions. TSC2 was immunoprecipitated from the indicated combined fractions from (D).
(F) Stoichiometry of TBC1D7-bound TSC1 and TSC2. Endogenous TBC1D7 (or control IgG) was immunoprecipitated from a HeLa cell lysate and tryptic peptides were detected using LC-MS/MS. The ratio of TSC1 and TSC2 in the TBC1D7 immunoprecipitate (the control lacked these proteins) was estimated using two established quantitative methods, spectral abundance and average total ion current (TIC). Both methods yielded the same ratio as indicated in the schematic summary.
(G) TBC1D7 is tightly bound to the TSC1-TSC2 complex. HeLa cells were lysed with buffer lacking NaCl and SDS. A single TSC1 immunoprecipitate from this lysate was split and washed with lysis buffer containing the indicated concentrations of NaCl or SDS.
The endogenous interaction of TBC1D7 with the TSC1-TSC2 complex was confirmed in reciprocal immunoprecipitations with a panel of six unique antibodies to TSC1 or TSC2, along with an antibody raised against TBC1D7 (Figure 1B), the specificity of which was confirmed through siRNA-mediated knockdown of TBC1D7 (Figure S1E). TBC1D7 was present in both TSC1 and TSC2, but not control IgG, immunoprecipitations from a diverse series of mouse tissues, including metabolic tissues and those most commonly affected in the TSC disease (Figure 1C), suggesting that the expression and interaction of TBC1D7 with the TSC proteins is ubiquitous. To determine what proportion of total cellular TBC1D7 was associated with the TSC1-TSC2 complex, cell lysates were fractionated over a sucrose density gradient. Interestingly, TBC1D7, which is considerably smaller than TSC1 (~130 kDa) and TSC2 (~200 kDa), fractionated into two pools, one in low-density fractions and a second in high-density fractions that also contained TSC1 and TSC2 (Figure 1D). Importantly, like TSC1, TBC1D7 co-immunoprecipitated with TSC2 only from these denser fractions, indicating that their co-fractionation reflects their association (Figure 1E). From multiple such experiments, it appears that 40–50% of total cellular TBC1D7 is associated with the TSC1-TSC2 complex. We were surprised to also find a pool of TSC2 not associated with stoichiometric levels of TSC1 in fractions of intermediate density (Figures 1D, E), indicating that both TSC2 and TBC1D7 exist as TSC1-free and -associated entities in cells. The co-fractionation of TBC1D7 with both TSC1 and TSC2 suggests that it primarily binds to the TSC1-TSC2 complex, as opposed to either protein individually. TSC1 and TSC2 are thought to interact at a 1:1 ratio (Zhang et al., 2003a) and to function as a heterodimer in all organisms in which they are expressed (Gao and Pan, 2001; Plank et al., 1998; Potter et al., 2001; van Slegtenhorst et al., 1998). Indeed, label-free quantitative mass spectrometry-based analyses to estimate the relative stoichiometry of TSC1 and TSC2 that co-immunoprecipitated with endogenous TBC1D7 yielded a near 1:1 ratio of TSC1 and TSC2 (Figure 1F), providing further evidence that TBC1D7 primarily binds to the TSC1-TSC2 complex.
TSC1 and TSC2 strongly associate with one another, as the purified complex is resistant to various detergents, high salt concentrations, and pH extremes (Gao and Pan, 2001; Nellist et al., 1999; Plank et al., 1998; Potter et al., 2001; van Slegtenhorst et al., 1998), whereas the binding of more loosely associated proteins is readily disrupted under such conditions (Huang et al., 2008). To assess the strength of the TSC1-TSC2-TBC1D7 interaction, endogenous complexes were immunoprecipitated with a TSC1 antibody and then washed extensively with buffers containing increasing concentrations of NaCl or SDS (Figure 1G). Like TSC2, the association of TBC1D7 with the complex was found to be resistant to washes containing ten times the physiological concentration of salt, whereas higher concentrations of SDS could separate TBC1D7 from the complex. Collectively, these findings identify TBC1D7 as a strongly-associated, ubiquitous binding partner of the TSC1-TSC2 heterodimer, distinguishing it from previously characterized TSC1- or TSC2-associated proteins.
TBC1D7 binds to and is stabilized by TSC1 within the TSC1-TSC2 complex
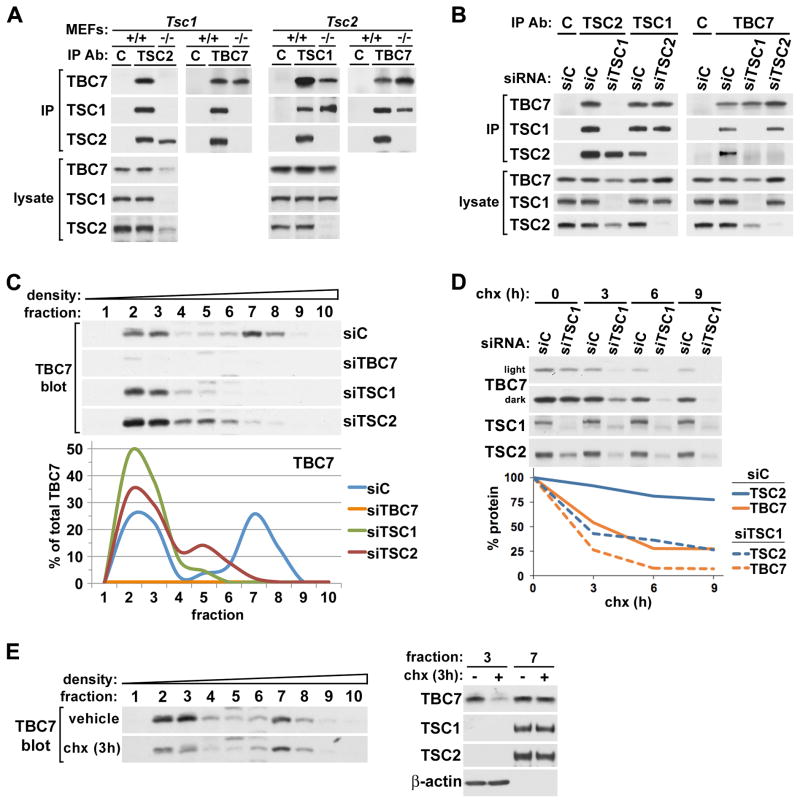
Having established that TBC1D7 strongly and ubiquitously interacts with the TSC1-TSC2 complex, we sought to determine the interdependency of the three proteins for their association within the complex. Through reciprocal immunoprecipitations, we found that all three proteins associate in wild-type mouse embryonic fibroblasts (MEFs) but that TBC1D7 and TSC2 do not interact in littermate-derived Tsc1−/− MEFs (Figure 2A). On the other hand, TBC1D7 and TSC1 can associate in the absence of TSC2, as observed in Tsc2−/− MEFs, although their association was reduced compared to wild-type cells (Figure 2A). To confirm these results under isogenic conditions, we performed the same immunoprecipitations from HeLa cells following siRNA-mediated knockdown of TSC1 or TSC2. Again, TBC1D7 bound to TSC1 in the absence of TSC2, but not TSC2 in the absence of TSC1 (Figure 2B), confirming that TBC1D7 binds to the complex through TSC1. To establish whether the separation of TBC1D7 into high-density fractions (Figure 1D) reflects binding to the TSC1-TSC2 heterodimer through TSC1, we repeated the sucrose density fractionations following siRNA-mediated knockdown of the three complex components. Indeed, knockdown of TSC1 had a dramatic effect on fractionation of the remaining TBC1D7, which completely shifted to low-density fractions (Figure 2C). TSC2 knockdown also eliminated TBC1D7 from high-density fractions, increasing its levels in low-density fractions. However, more TBC1D7 was also found in intermediate-density fractions upon loss of TSC2, consistent with its ability to still bind TSC1 in the absence of TSC2. Nearly identical results were obtained in cell lines with stable shRNA-mediated knockdowns of TBC1D7, TSC1, and TSC2 (Figure S2A). These data demonstrate that TBC1D7 resides in high-density fractions due solely to its association with the TSC1-TSC2 complex. It is important to note that although TBC1D7 can bind TSC1 in the absence of TSC2, our collective data indicate that TBC1D7-TSC1 complexes lacking TSC2 are rare, or nonexistent, in wild-type cells and that TBC1D7 primarily binds to the TSC1-TSC2 complex.
Figure 2. TBC1D7 binds to and is stabilized by TSC1 within the TSC1-TSC2 complex.
(A) TSC1-dependent association of TBC1D7 with TSC2 in MEFs. Immunoprecipitations with TBC1D7, TSC1, TSC2, or control IgG (C) antibodies were performed using lysates of littermate-derived wild-type (+/+) and either Tsc1−/− (left) or Tsc2−/− (right) MEFs.
(B) TSC1-dependent association of TBC1D7 with TSC2 in HeLa cells. Immunoprecipitations as in (A) were performed using lysates of HeLa cells with siRNA-mediated knockdown of TSC1 or TSC2, compared to control (siC).
(C) High-density fractionation of TBC1D7 is dependent on TSC1 and TSC2. Lysates of HeLa cells with siRNA-mediated knockdown of TBC1D7, TSC1, and TSC2 were ultracentrifuged over a sucrose gradient and ten sequential fractions of increasing density were collected. TBC1D7 immunoblot band intensities were quantified using ImageJ software and graphed as a percentage of the summed band intensities in all fractions for each protein. See supporting data in Figure S2A.
(D) Loss of TSC1 decreases the stability of both TBC1D7 and TSC2. HeLa cells with siRNA-mediated knockdown of TSC1 were incubated with 100 μM cyclohexamide (chx) for the indicated duration. Immunoblot band intensities were quantified as in (C) and graphed as a percentage of the untreated (0 h) samples.
(E) The “free” pool of TBC1D7 is unstable. HeLa cells were incubated with cyclohexamide or water (vehicle) for 3 h and fractionated as in (C). Fractions 3 and 7 from both treatments are also compared on the same immunoblot (right).
Consistent with previous findings (Benvenuto et al., 2000; Kwiatkowski et al., 2002), loss of TSC1 in these experiments led to a decrease in the levels of TSC2 (Figures 2A, B). Interestingly, like TSC2, TBC1D7 protein levels were diminished upon loss of TSC1 (Figure 2A–C), and this decrease in both TSC2 and TBC1D7 was indistinguishable under various cell culture conditions (Figure S2B). Transient knockdown of TSC2 had the opposite effect on TBC1D7, modestly increasing its levels (Figure 2B and S2B). To clarify the effects of TSC1-deficiency on TBC1D7 and TSC2, we monitored their stability in TSC1-knockdown cells over a time course of cyclohexamide treatment to block new protein synthesis (Figure 2D). In control cells, TSC2 levels remained relatively stable over time, while TBC1D7 was found to be more labile. However, in TSC1-knockdown cells, the rate and degree of degradation of both TBC1D7 and TSC2 were substantially increased, demonstrating that TSC1 stabilizes both proteins. Given that approximately 50% of TBC1D7 was rapidly degraded, even in the presence of TSC1, while the remainder was more stable, we hypothesized that the two pools of TBC1D7 vary in their stability. To test this, we compared levels of TBC1D7 in these two pools in vehicle- versus cyclohexamide-treated cells (Figure 2E). Indeed, only TBC1D7 in low-density fractions, representing the free pool, was degraded over the 3-hour treatment. This is consistent with TSC1 loss leading to decreased TBC1D7 stability and indicates that the free pool of TBC1D7 present in cells turns over rapidly compared to that tightly bound to the TSC1-TSC2 complex.
Knockdown of TBC1D7 decreases the association of TSC1 and TSC2 without effects on the lysosomal localization of TSC2
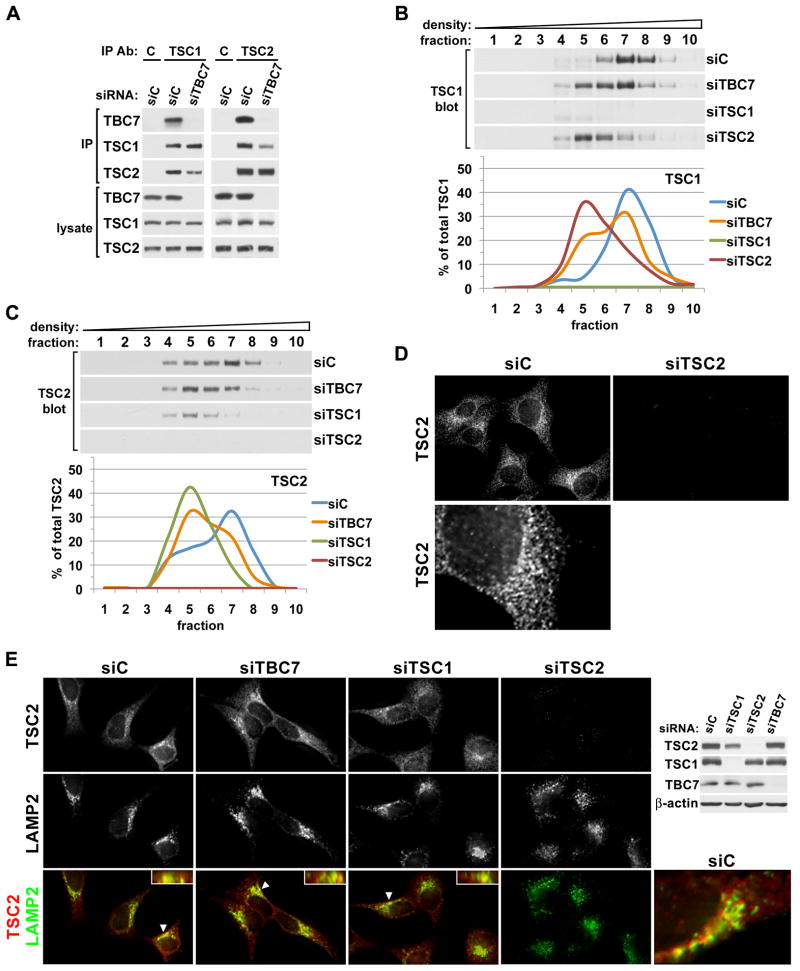
In order to determine whether TBC1D7, as a component of the TSC1-TSC2 complex, affects the association of TSC1 and TSC2, these proteins were immunoprecipitated from cells in which TBC1D7 was knocked down (Figure 3A). In the absence of TBC1D7, a complex between TSC1 and TSC2 was still detected, but a reproducible reduction in their association was observed (Figure 3A). As expected, in sucrose density fractions, both transient (Figure 3B,C) and stable (Figure S3A,B) knockdown of TSC2 caused a complete shift of TSC1 to fractions of intermediate density (Figures 3B and S3A), and TSC1 knockdown had the same effect on the fractionation of TSC2 (Figures 3C and S3B). Consistent with the partial dissociation of TSC1 and TSC2 observed in immunoprecipitation experiments, both transient and stable knockdown of TBC1D7 resulted in the shifting of some TSC1 (Figures 3B and S3A) and TSC2 (Figures 3C and S3B) to intermediate-density fractions containing the uncomplexed proteins. As a control, none of these knockdowns affected the fractionation pattern of the mTORC2 component Rictor (Figure S3C), which weakly interacts with the TSC1-TSC2 complex (Huang et al., 2008). These data further demonstrate that TBC1D7 represents a third core subunit of the TSC1-TSC2 complex.
Figure 3. Knockdown of TBC1D7 leads to decreased association of TSC1 and TSC2 without effects on TSC2 localization.
(A) Knockdown of TBC1D7 leads to a decrease in the co-immunoprecipitation of TSC1 and TSC2. Immunoprecipitations with TSC1, TSC2, or control IgG (C) antibodies were performed using lysates of HeLa cells with siRNA-mediated knockdown of TBC1D7, compared to control (siC).
(B,C) Knockdown of TBC1D7 alters the fractionation patterns of both TSC1 and TSC2. Lysates from HeLa cells with siRNA-mediated knockdown of TBC1D7, TSC1, or TSC2 were ultracentrifuged over a sucrose gradient and ten sequential fractions of increasing density were collected. Immunoblot band intensities for TSC1 (B) and TSC2 (C) were quantified using ImageJ software and graphed as a percentage of the summed band intensities in all fractions. See supporting data in Figure S3A–C.
(D) Localization of endogenous TSC2. HeLa cells with siRNA-mediated knockdown of TSC2 compared to a control (siC) were serum starved for 16 h and immunofluorescently labeled with a TSC2 antibody. An enlarged view of a single cell is shown below.
(E) Knockdowns of TBC1D7 and TSC1 do not have gross effects on the localization of TSC2, including its co-localization with LAMP2 at the lysosome. HeLa cells with the indicated siRNA-mediated knockdowns were prepared as in (D), but co-immunolabeled for TSC2 (red) and LAMP2 (green). In merged images, yellow and orange regions indicate co-localization, and inset boxes show planes perpendicular to the main image that were constructed from z-stacks (arrows indicate cell shown in inset). Corresponding immunoblots and an enlarged view of a single cell are shown to the right. See supporting data in Figure S3D,E.
In order to begin to understand the role of TBC1D7 within the TSC1-TSC2-TBC1D7 (TSC-TBC) complex, we determined its effects on the subcellular localization of TSC2. While several studies have reported various localization patterns of TSC2 by immunofluorescence, most have done so with over-expressed proteins or have failed to validate the specificity of the antibodies used. We have tested most commercially available TSC2 antibodies for specific localization patterns that are eliminated upon knockdown of endogenous TSC2 expression. A single highly specific TSC2 antibody was identified that demonstrates a punctate localization pattern throughout the cell with a more concentrated pattern at a perinuclear region (Figure 3D). To date, we have been unable to identify antibodies that reveal specific localization patterns for endogenous TSC1 or TBC1D7 using this same approach. Through co-localization studies with multiple markers of membranous cellular compartments, we found that the perinuclear region to which TSC2 localization concentrates represents, in part, late endosomes or lysosomes, as there is substantial overlap (~50%, Figure S3D) with LAMP2 staining under serum starvation conditions (Figure 3E, perpendicular images are inset). Knockdown of TBC1D7 had no significant effect on TSC2 localization (Figures 3E and S3D). Surprisingly, this was also true of TSC1 knockdown, although the overall levels of TSC2 were reduced. Importantly, we found that TSC2 localization to this compartment does not reflect its degradation in the lysosome, as lysosomal inhibitors did not increase the levels of TSC2 in the presence or absence of TSC1 (Figure S3E). It is worth noting that the partial co-localization of endogenous TSC2 with LAMP2 is similar to that reported for mTOR (Sancak et al., 2010; Sancak et al., 2008; Zoncu et al., 2011; see below).
TBC1D7 does not appear to be a third TSC gene
Germline mutations in the TSC1 or TSC2 genes that disrupt or functionally inactivate the TSC1-TSC2 complex give rise to tuberous sclerosis complex (TSC), an autosomal dominant tumor syndrome characterized by the widespread occurrence of benign tumors, classified as hamartomas, and a high incidence of various neurological manifestations, including epilepsy and autism spectrum disorders (Crino et al., 2006). While mutations in TSC1 or TSC2 account for up to 85% of TSC cases, approximately 15% of patients clinically diagnosed with TSC do not have apparent mutations in either of these two genes, leading to speculation that a third TSC gene (i.e., TSC3) might exist (Camposano et al., 2009; Kwiatkowski, 2005). We considered the possibility that TBC1D7, as a third resident component of the TSC1-TSC2 complex, might be mutated in this subset of TSC patients. Therefore, we sequenced the seven coding exons of the TBC1D7 locus (Figure S4A) from twelve TSC patients with no mutation identified in TSC1 or TSC2 after extensive genetic analyses (Qin et al., 2010). However, aside from known single nucleotide polymorphisms (SNPs), we found no point mutations in exons 2 through 8 of the TBC1D7 gene, indicating that such mutations do not occur at appreciable frequency in the germline of this population of TSC patients.
The TSC1-TSC2-TBC1D7 (TSC-TBC) complex receives growth signals and is the functional GAP for Rheb
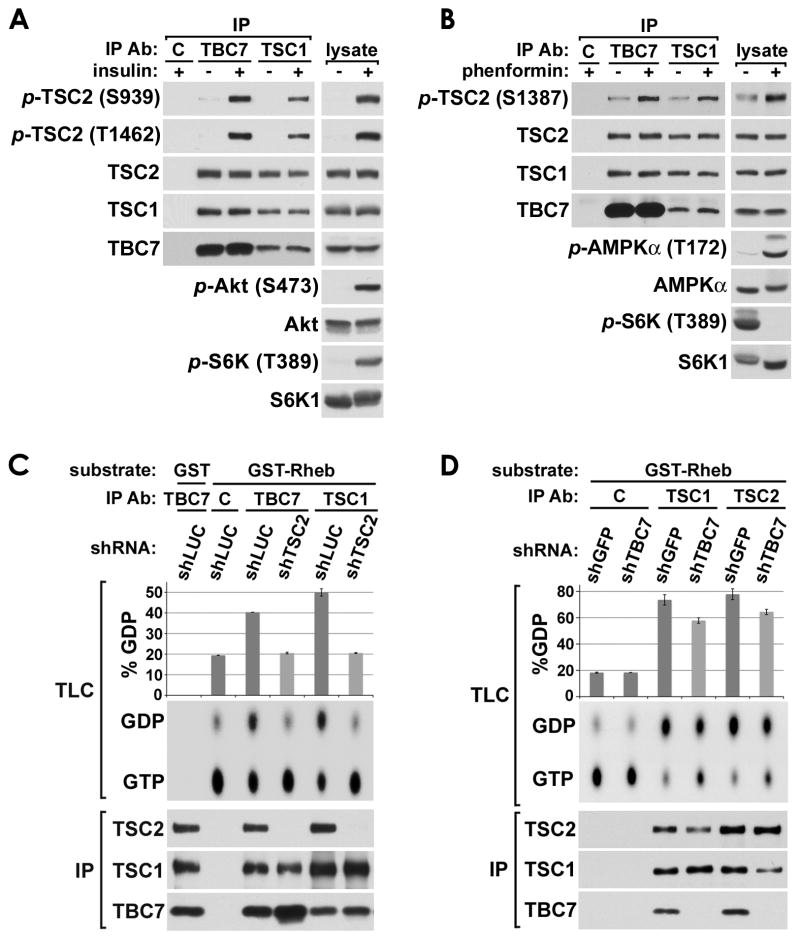
As the only well-established role of the TSC1-TSC2 complex is the regulation of Rheb and mTORC1, we determined whether the TSC-TBC complex is the functional unit in this capacity. The ability of the TSC1-TSC2 complex to act as a GAP for Rheb in cells is controlled by upstream pathways leading to inhibitory or activating phosphorylation of various sites on TSC2. To determine whether the TSC-TBC complex is controlled in this manner, the regulated phosphorylation of TSC2 was assessed in TBC1D7 versus TSC1 immunoprecipitations of the complex. Interestingly, both the insulin-stimulated, Akt-mediated (S939 and T1462; Figure 4A) and phenformin-stimulated, AMPK-mediated (S1387; Figure 4B) phosphorylation of TSC2 was found at similar levels in TBC1D7 and TSC1 immunoprecipitations. Therefore, it is primarily the TSC-TBC complex that is regulated by these well-characterized pathways upstream of Rheb.
Figure 4. The TSC-TBC complex responds to growth cues and has Rheb-GAP activity.
(A) TBC1D7 is associated with TSC2 that is phosphorylated by Akt in response to insulin. Serum starved (18 h) HeLa cells were stimulated with insulin (100 nM) for 10 min prior to lysis and immunoprecipitation (IP) with TBC1D7, TSC1, or control IgG (C) antibodies.
(B) TBC1D7 is associated with TSC2 that is phosphorylated by AMPK in response to energy stress. MEFs were grown in the presence of fresh serum (10%) with or without phenformin (1 mM) for 2 h prior to lysis and IP as in (A).
(C) TBC1D7 is associated with TSC2 that has Rheb-GAP activity. HeLa cells with stable shRNA-mediated knockdown of TSC2 (shTSC2) or a firefly luciferase control (shLUC) were serum starved for 6 h prior to lysis and IP as in (A). Immunopurified complexes were subjected to Rheb-GAP assays using recombinant GST or GST-Rheb pre-loaded with GTP[α-32P]. Rheb-bound GTP and GDP were separated by thin layer chromatography (TLC) and quantified using a phosphoimager in duplicate experiments. Mean GDP levels were graphed as percent of total GTP plus GDP (%GDP), with error bars for standard error of the mean.
(D) Knockdown of TBC1D7 leads to a net decrease in Rheb-GAP activity of TSC2. Rheb-GAP assays were performed, analyzed, and graphed as in (C) except using HeLa cells with stable shRNA-mediated knockdown of TBC1D7 (shTBC7) or a GFP control (shGFP), and twice the amount of cells. See supporting data in Figure S4B.
The TSC2-GAP domain is essential for the inhibition of Rheb by the TSC1-TSC2 complex (Inoki et al., 2003a; Tee et al., 2003; Zhang et al., 2003b). To establish whether the TSC-TBC complex is functional as a Rheb-GAP, we immunopurified endogenous complexes with either TBC1D7 or TSC1 antibodies and performed GAP assays on recombinant, purified Rheb (Figure 4C). TBC1D7 immunoprecipitates exhibited comparable levels of Rheb-GAP activity to TSC1 immunoprecipitates, especially considering that more TSC2 was present in the TSC1 immunoprecipitates. Importantly, this GAP activity was dependent on TSC2, as endogenous complexes from cells with stable knockdown of TSC2 failed to stimulate the GTPase activity of Rheb above the intrinsic levels observed with control IgG. Therefore, most, if not all, of the Rheb-GAP activity of the TSC1-TSC2 complex is in association with TBC1D7. In order to determine whether TBC1D7 knockdown affects the biochemical activity of the complex, we immunoprecipitated endogenous complexes from cells with control or TBC1D7 knockdown and measured Rheb-GAP activity. Interestingly, this activity was partially reduced in cells with stable shRNA knockdown of TBC1D7, and this was evident in Rheb-GAP assays with both TSC1 and TSC2 immunoprecipitations (Figure 4D). The effect of TBC1D7 knockdown on the GAP activity of the complex was not due to an increase in the inhibitory phosphorylation of TSC2 by Akt (Figure S4B). In fact, TBC1D7 knockdown led to decreased phosphorylation of TSC2-T1462 and Akt-S473 in response to insulin, reminiscent of the effects of TSC1 or TSC2 loss on mTORC2 and Akt signaling (reviewed in Huang and Manning, 2009). The reduction in endogenous GAP activity upon loss of TBC1D7 is consistent with the resulting decreased association of TSC1 and TSC2 described above (Figures 3A–C and S3A, B) and detected again here (Figure 4D immunoblot). The fact that reduced GAP activity is also evident in TSC2 immunoprecipitates, despite identical levels of TSC2 being present in the control and TBC1D7 knockdown assays, reflects the importance of complex formation with TSC1 for the full GAP activity of TSC2 towards Rheb (Tee et al., 2003).
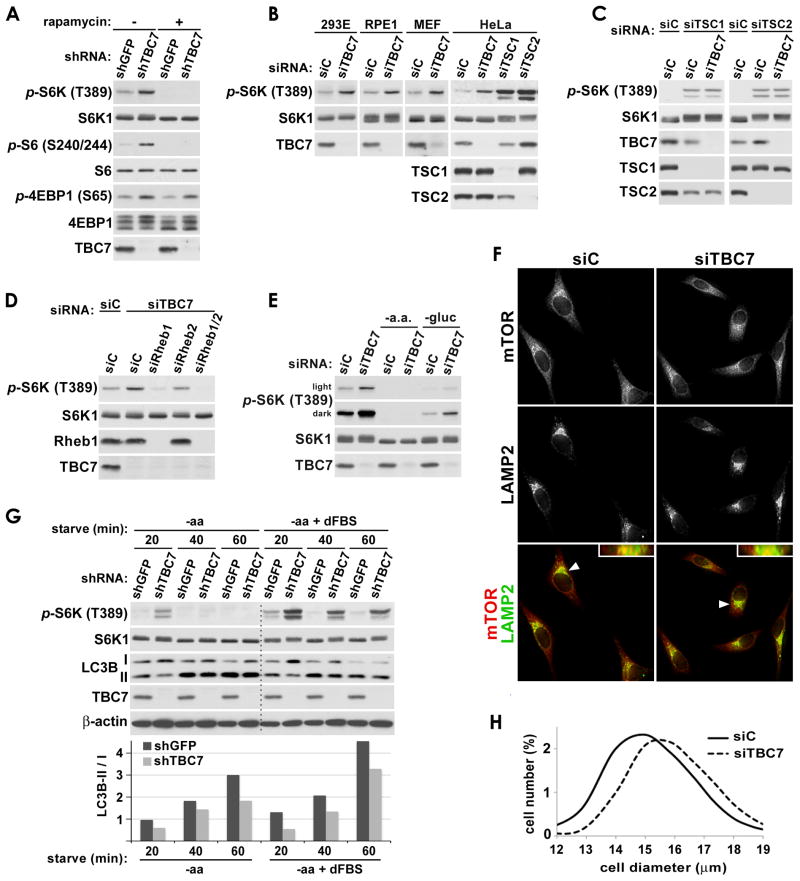
Loss of TBC1D7 results in growth factor-independent activation of mTORC1 signaling
The collective findings above that TBC1D7 is a core component of the functional Rheb-GAP complex that responds to upstream signals known to regulate Rheb, raised the possibility that, like the other members of the complex, TBC1D7 might be required for proper regulation of mTORC1 signaling by specific growth cues. In particular, TSC1 and TSC2 are required to suppress mTORC1 signaling upon growth factor withdrawal, with their loss resulting in sustained mTORC1 signaling in the absence of growth factors (Jaeschke et al., 2002; Kwiatkowski et al., 2002). Indeed, cells with stable knockdown of TBC1D7 exhibited growth factor-independent activation of mTORC1 signaling under serum starvation conditions, as detected by the increased phosphorylation of its direct downstream target S6K and the S6K substrate S6, both of which were sensitive to the mTORC1 inhibitor rapamycin (Figure 5A). TBC1D7 knockdown also increased the phosphorylation of 4E-BP1, another direct target of mTORC1, as scored by phospho-specific antibodies and electrophoretic mobility shifts, effects that are only partially sensitive to rapamycin, as reported by others (Choo et al., 2008). Transient knockdown of TBC1D7 in various mouse and human cell lines similarly led to increased growth factor-independent mTORC1 signaling (Figure 5B). When directly comparing the relative effects of knocking down the three subunits of the TSC-TBC complex, a graded effect on mTORC1 signaling is observed, with knockdown of TBC1D7 yielding a moderate increase, the scaffolding subunit TSC1 yielding an intermediate increase, and the Rheb-GAP TSC2 yielding the largest increase (Figure 5B, HeLa). Importantly, knockdown of TBC1D7 is not additive with knockdown of TSC1 or TSC2 for stimulation of S6K phosphorylation, consistent with the effects of TBC1D7 loss on mTORC1 signaling being through effects on the trimeric complex (Figure 5C). In support of the established mechanism by which the TSC1-TSC2 complex inhibits mTORC1 by acting as a GAP for Rheb, the stimulation of mTORC1 signaling by TBC1D7 knockdown was inhibited by siRNAs targeting Rheb1, with a minor contribution from Rheb2 (also known as RhebL1; Figure 5D).
Figure 5. TBC1D7 negatively regulates mTORC1 signaling under poor growth conditions.
(A) Growth factor-independent mTORC1 signaling in cells with stable knockdown of TBC1D7. HeLa cells with stable shRNA-mediated knockdown of TBC1D7 (shTBC7) or a GFP control (shGFP) were serum starved for 16 h and, where indicated, treated with rapamycin (20 nM) for the final 15 min.
(B) Growth factor-independent activation of mTORC1 signaling in a variety of cell lines with knockdown of TBC1D7. The indicated cell lines with siRNA-mediated knockdown of human (293E, RPE1, HeLa) or mouse (MEF) TBC1D7 compared to controls (siC) were serum starved for 16 h prior to lysis.
(C) Dual knockdown of TBC1D7 with TSC1 or TSC2 does not have an additive effect on mTORC1 signaling. HeLa cells were co-transfected with the indicated siRNAs and treated as in (B).
(D) mTORC1 signaling in TBC1D7-deficient cells remains Rheb-dependent. HeLa cells were co-transfected with the indicated siRNAs and treated as in (B).
(E) mTORC1 signaling in TBC1D7-deficient cells is resistant to growth factor and glucose-withdrawal but not amino acid withdrawal. MEFs were treated as in (B) but following serum starvation, the indicated samples were either starved of all amino acids or glucose for an additional 3 h. Light and dark exposures of the same phospho-S6K blot are shown.
(F) Co-localization of mTOR and LAMP2 is not affected by loss of TBC1D7. HeLa cells were serum starved for 16 h and immunofluorescently labeled with mTOR and LAMP2 antibodies. In merged images of mTOR (red) and LAMP2 (green), co-localization is represented by yellow and orange pixels. Insets in each merged image show planes perpendicular to the primary image that were constructed from z-stacks. Arrows indicate the cells shown in these insets.
(G) Autophagy induction is delayed in TBC1D7-deficient cells. HeLa cells with stable shRNA-mediated knockdown of TBC1D7 (shTBC7) or GFP as a control (shGFP) were grown in complete growth media and then starved of all amino acids (-aa) with or without dialyzed FBS (dFBS) for the indicated duration (min). LC3B-II to -I ratios were quantified with ImageJ software and are shown normalized to the 20 min shGFP samples.
(H) Knockdown of TBC1D7 increases cell size. HeLa cells were transfected with control (siC) or TBC1D7 (siTBC7) siRNAs and, 24-h post-transfection, were serum starved for 48 h. The distribution of cell diameters (in μm) within the population of cells was measured using a Z2 Coulter counter. p<0.0001 for comparison of siC and siTBC7 size distributions (average of 5 replicates, with 28,400 cells measured in each). See supporting data in Figure S5.
In addition to growth factors, the activity of mTORC1 is dependent on the presence of nutrients, such as glucose and amino acids, which are sensed through both TSC1-TSC2 complex-dependent and -independent pathways upstream of mTORC1. Consistent with previous findings that amino acid-sensing by mTORC1 occurs largely independent of the TSC1-TSC2 complex (Kim et al., 2008; Sancak et al., 2010; Sancak et al., 2008; Smith et al., 2005), amino acid withdrawal eliminates mTORC1 signaling in both control and TBC1D7 knockdown MEFs (Figure 5E). While withdrawal of glucose similarly reduces mTORC1 signaling, the level of S6K phosphorylation was elevated in TBC1D7 knockdowns relative to controls. These results are consistent with glucose withdrawal and energy stress acting through both TSC-TBC complex-dependent and independent mechanisms (Gwinn et al., 2008; Inoki et al., 2003b; Kalender et al., 2010; Shaw et al., 2004). These data suggest that TBC1D7 does not influence amino acid sensing by mTORC1, which involves the Rag GTPases recruiting mTORC1 to the LAMP2-containing compartment for activation by Rheb (Kim et al., 2008; Sancak et al., 2010; Sancak et al., 2008; Zoncu et al., 2011). That TBC1D7 acts upstream of Rheb rather than the Rag GTPases was further demonstrated by a lack of change in mTOR co-localization with LAMP2 upon knockdown of TBC1D7 (Figure 5F). Therefore, like TSC1 and TSC2, TBC1D7 participates in the proper sensing of both growth factors and glucose, but not amino acids, by mTORC1.
In order to determine whether the effects of TBC1D7 on mTORC1 signaling influence known cellular processes regulated by mTORC1, we examined autophagy and cell size, which are known to be affected by loss of function of the TSC1-TSC2 complex (Gao and Pan, 2001; Menon et al., 2012; Parkhitko et al., 2011; Potter et al., 2001; Tapon et al., 2001). While prolonged (>2 h) starvation of amino acids strongly inhibits mTORC1 signaling in TBC1D7 knockdown cells, a delay in mTORC1 inhibition upon amino acid withdrawal is evident in these cells, especially in the presence of dialyzed serum (Figure 5G). This is consistent with findings reported previously for Tsc2−/− MEFs (Smith et al., 2005). To detect the onset of autophagy induced by amino acid withdrawal, we examined the conversion of LC3B-I to its lipidated form (LC3B-II) in control versus TBC1D7 knockdown cells. As predicted from the delayed inhibition of mTORC1 signaling, TBC1D7 knockdown cells exhibited a delay in the initiation of autophagy over a time course of amino acid withdrawal (Figure 5G). Finally, we found that the growth factor-independent activation of mTORC1 signaling upon TBC1D7 knockdown (Figures 5A–E) coincides with an increase in cell growth, as both transient (Figure 5H) and stable (Figure S5A) knockdowns of TBC1D7 result in a significant increase in cell size. Collectively, these data support a model in which TBC1D7, as a core component of the TSC-TBC complex, regulates Rheb and mTORC1 signaling in response to alterations in specific cellular growth conditions to control downstream cellular processes (Figure 6).
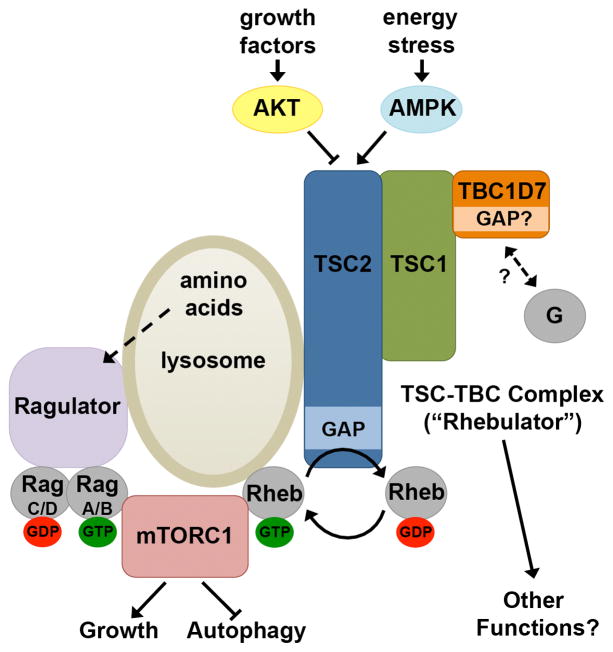
Figure 6. Model of the TSC-TBC Complex.
Schematic of the integrated regulation of mTORC1 by the Rag GTPases through the Ragulator, and Rheb through the TSC-TBC complex (or Rhebulator). See text for details.
DISCUSSION
Our biochemical studies demonstrate that TBC1D7 is a third functional subunit of the TSC1-TSC2 complex, distinguishing it from dozens of previously identified, but poorly characterized, binding partners of the complex (Rosner et al., 2008). Two previous independent studies have also reported an interaction between endogenous TBC1D7 and TSC1 (Nakashima et al., 2007; Sato et al., 2010), but these studies did not recognize this protein as a component of the TSC1-TSC2 complex. In the first study, TBC1D7 was found to co-immunoprecipitate with the complex through an interaction with TSC1 (Nakashima et al., 2007), which we confirm in our characterization. However, TBC1D7 overexpression experiments led to the conclusion that this interaction inhibits the TSC1-TSC2 complex and activates mTORC1 by promoting the ubiquitination of TSC1. Contradictory to these findings, we demonstrate that loss of TBC1D7 partially disrupts the association between TSC1 and TSC2, resulting in a decrease in Rheb-GAP activity. Given our biochemical and cell biological data that the endogenous TSC-TBC complex is the functional unit negatively regulating Rheb and mTORC1, it seems likely that overexpression of this single core subunit could disrupt the stoichiometric complex and its downstream functions, possibly explaining the conclusions of this other study. Our data also indicate that the binding of TBC1D7 to the complex is not affected by changes in cellular growth conditions known to control the activity of the complex, consistent with TBC1D7 being a component, rather than a transient regulator, of the complex. The second study found that TBC1D7 was stabilized by TSC1, which we confirm and expand upon to demonstrate the existence of an unstable free pool of TBC1D7 in cells. These investigators suggested that in lung cancer cells a TBC1D7-TSC1 complex promotes cell proliferation independently of mTORC1 (Sato et al., 2010). While we find that such a complex can exist when TSC2 is absent from the cell, our biochemical experiments on the endogenous TSC-TBC complex indicate that complexes between TBC1D7 and TSC1 without TSC2 are rare or nonexistent in cells and tissues. Finally, while the authors of both studies propose that this association may be cell and tissue-specific, we demonstrate a broad tissue-distribution of endogenous TSC-TBC complexes and have yet to identify a mammalian setting without this complex. This finding is consistent with the ubiquitous nature of its downstream targets, Rheb and mTORC1.
The molecular function of TBC1D7 is unknown but likely depends on its TBC domain, which spans nearly two-thirds of this small protein. TBC domains in other proteins possess GAP activity toward specific Rab family members, suggesting that the function of TBC1D7 will also be tied to a small G-protein. Importantly, TBC1D7 belongs to a subgroup of “unconventional” TBC-domain family members and lacks two out of three conserved motifs, which in classical TBC domains are essential for GAP activity (Frasa et al., 2012). Unconventional TBC domains generally lack known targets, and some have been described to be devoid of Rab-GAP activity while maintaining the ability to bind other small G proteins (Frittoli et al., 2008; Martinu et al., 2004). Surprisingly, in a high-throughput screen for targets of TBC proteins, recombinant TBC1D7 was found to have specific in vitro GAP activity towards Rab17, compared to over 40 other Rabs (Yoshimura et al., 2007). Overexpression of these two proteins was also found to affect formation of the primary cilium in RPE1 cells. However, we have found no significant effect of knocking down TBC1D7 on cilia formation or length using the same cell line (data not shown), and a specific Rab17 ortholog does not appear to be conserved in arthropods, which express clear orthologs of TBC1D7, TSC1, and TSC2. Further studies are required to determine whether TBC1D7 has physiologically relevant GAP activity towards, or binds to, members of the Rab or other small G-protein families. In addition, it will be important to determine whether such activities are altered by its complex formation with TSC1 and TSC2.
Our findings further confirm and expand upon current models of mTORC1 regulation by nutrients and growth factors (Figure 6; Laplante and Sabatini, 2012). Loss of any of the three components of the TSC-TBC complex results in the sustained activation of mTORC1 signaling, albeit to different degrees, upon growth factor withdrawal, and this is driven by elevated levels of GTP-bound Rheb. However, mTORC1 signaling remains partially sensitive to glucose starvation, consistent with the existence of additional regulatory mechanisms stemming from energy stress (Gwinn et al., 2008; Kalender et al., 2010). While we detect a delay in mTORC1 inhibition upon amino acid withdrawal in cells lacking TBC1D7, amino acids remain essential for mTORC1 signaling. This delayed inhibition is likely to reflect an aberrant stimulatory effect of elevated Rheb-GTP levels sustaining mTORC1 activity, as seen when Rheb is overexpressed in cells (reviewed in Huang and Manning, 2008). While the amino acid-sensing mechanism has not been fully defined, amino acids activate mTORC1 by stimulating its recruitment to the lysosomal surface in a manner dependent on the Rag GTPases and the scaffolding complex referred to as the Ragulator (Kim et al., 2008; Sancak et al., 2010; Sancak et al., 2008; Zoncu et al., 2011). The recruitment of mTORC1 to the lysosome is not sufficient to activate it but is believed to bring the kinase complex in contact with Rheb at this location, which when GTP-bound potently stimulates mTORC1 activation (Sancak et al., 2007). Importantly, we find that a substantial amount of cellular TSC2, which controls the levels of Rheb-GTP, is also localized at the lysosome. Therefore, these two sets of GTPases, the Rags and Rheb, and their regulatory complexes, the Ragulator and the TSC-TBC complex (or Rhebulator), are spatially poised to integrate diverse signals to properly turn mTORC1 activity on or off (Figure 6).
Further delineating the function of TBC1D7 within the TSC-TBC complex will provide molecular insights into the regulation of Rheb and mTORC1 and facilitate the characterization of putative mTORC1-independent functions of this key signal-integrating node. Although, to date, we have not found germline point mutations in the coding region of TBC1D7 in TSC patients, as a core subunit of the TSC-TBC complex, its molecular function will be aberrantly affected in both the TSC and LAM diseases, where the other components of the complex are mutated. Finally, given that the upstream regulatory network controlling the TSC-TBC complex is often perturbed through environmental or genetic events, the misregulation of this complex is also likely to underlie pathological features of a diverse set of human diseases, including other tumor syndromes, sporadic cancers, metabolic diseases, and neurological disorders (Ehninger and Silva, 2011; Laplante and Sabatini, 2012; Menon and Manning, 2009).
EXPERIMENTAL PROCEDURES
Cell culture, RNAi, and cell size measurements
Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS; Sigma), unless otherwise noted. Tsc1−/− and Tsc2−/− MEFs and their littermate-derived wild-type counterparts were described previously (Kwiatkowski et al., 2002; Zhang et al., 2003a). For siRNA knockdowns, cells were transfected with 20 nM ON-TARGETplus siRNA pools (ThermoScientific) and 2.5 μL (per ml media) RNAiMAX Lipofectamine (Life Technologies), except in Figure 5D where siRNAs were used at 40 nM and RNAiMAX Lipofectamine at 3.5 μL (per ml media), according to the manufacturer’s instructions. Cells were harvested 72 h post-transfection, unless otherwise noted. The TSC2 shRNA construct was described previously (Huang et al., 2008). For cell size distributions, freshly trypsinized cells were diluted in isotonic buffer and analyzed with a Z2 Coulter Counter (Beckman).
Cell and tissue lysis, immunoprecipitations, and sucrose density gradient fractionation
Cells and tissues were lysed/homogenized in NP-40 lysis buffer (40 mM HEPES, pH 7.4, 120 mM NaCl, 1 mM EDTA, 1% NP-40 [Igepal CA-630], 5% glycerol, 10 mM sodium pyrophosphate, 10 mM glycerol 2-phosphate, 50 mM NaF, 0.5 mM sodium orthovanadate, and protease inhibitors (Sigma)), unless otherwise noted. Flag-tagged TSC1-TSC2 complexes were immunopurified from HEK-293 cells as described previously (Huang et al., 2009) but with the 3xFlag peptide (Sigma) elution performed in lysis buffer lacking detergent. The 3xFlag-peptide eluate was processed for MS/MS analysis, as detailed in the supplemental materials. The TBC1D7 rabbit polyclonal antibody used for these studies was generated by Covance using the peptide CKVGFRGVEEKKSLEI. Antibodies used for the immunoprecipitations in Figure 1B: control IgG (Cell Signaling Technology (CST); TSC1 #1 (CST #4906), TSC1 #2 (Life Technologies (LT) #37-0400), TSC1 #3 (Santa Cruz Biotechnology (SCB) #SC-12082), TSC2 #1 (CST #3612), TSC2 #2 (SCB #SC-892), TSC2 #3 (SCB #SC-893). TSC1 (LT #37-0400) and TSC2 (LT #37-0500) antibodies were used for all other immunoprecipitations except for endogenous GAP assays (see below). Antibodies for immunoblotting: TSC1 (CST #6935), TSC2 (CST #4308), Rictor (Bethyl Laboratories), and all others were from CST. For density gradients, sucrose dissolved in NP-40 lysis buffer lacking glycerol was used to create gradient layers of 15% (0.5 ml), 20% (1 ml), 25% (1 ml), 27.5% (1 ml), 30% (0.5 ml), and 35% (0.5 ml) in 5 ml ultracentrifuge tubes. Cells from a near-confluent 10 cm plate were lysed in 700 μl NP-40 lysis buffer lacking glycerol, and 0.5 ml lysate was centrifuged over gradients at 175,000 x g for 16-17 h at 4 C. Ten 400 ml fractions were sequentially removed from the top of the gradient.
Immunofluorescence microscopy
HeLa cells plated on glass coverslips were serum starved for 16–18 h, fixed in 4% paraformaldehyde in PBS for 15 min, permeabilized in 0.2% Triton X-100 in PBS for 10 min, blocked with Odyssey blocking buffer diluted 1:1 in PBS (Licor Biosciences) for 1 h, incubated with primary antibodies (in blocking buffer) overnight at 4 C, and secondary antibodies for 1 h at room temperature. Primary antibodies were LAMP2 (Abcam #ab25631, 1:100), TSC2 (CST #4301, 1:1000), and mTOR (CST #2983, 1:100). Secondary antibodies were anti- mouse Alexa488 (Molecular Probes, 1:1000) and anti- rabbit Cy3 (Jackson ImmunoResearch, 1:1000). Pseudoconfocal images (~0.3 μm planes) were acquired with a Zeiss Axiotome fluorescence microscope with Axiovision software and the Axiotome feature engaged. Perpendicular images (representing single planes) were constructed from z-stacks of ten contiguous focal planes. Identical exposure times were used for comparative analyses.
GAP Assays
In vitro Rheb-GAP assays were performed as previously described (Tee et al., 2003), with the following modifications. Endogenous complexes were immunopurified with antibodies for TBC1D7 (Sigma #SAB1400543), TSC1 (CST #6935), and TSC2 (SCB #sc-892) from lysates combined from two (Figure 4C) or four (Figure 4D) 15 cm dishes of serum-starved (6 h) HeLa cells per reaction. Approximately 500 ng of recombinant GST-Rheb was used per reaction and assays were incubated at 30 C for 1 h.
Statistical Analyses
Significance of differences in cell size distributions was determined using an unpaired two-tailed Student’s T-test assuming equal variance.
Supplementary Material
Highlights.
TBC1D7 is a ubiquitous, stably-associated third core subunit of the TSC1-TSC2 complex.
TSC1 binds to and stabilizes both TSC2 and TBC1D7 within the trimeric complex.
The TSC1-TSC2-TBC1D7 (TSC-TBC) complex is the functional Rheb-GAP upstream of mTORC1.
As part of the TSC-TBC complex, TBC1D7 inhibits mTORC1 and downstream functions.
Acknowledgments
We thank Jordan Gallinetti, Bernie Boback, and Min Yuan for technical assistance and Dr. Danielle Manning for technical advice. This work was supported in part by NIH grants P01-CA120964 (B.D.M., D.J.K., and J.M.A.), R01-CA122617 (B.D.M.), and Dana Farber/Harvard Cancer Center Support Grant P30-CA006516 (J.M.A.).
Footnotes
Supplemental information includes Supplemental Experimental Procedures and References, and five supporting figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benvenuto G, Li S, Brown SJ, Braverman R, Vass WC, Cheadle JP, Halley DJ, Sampson JR, Wienecke R, DeClue JE. The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene. 2000;19:6306–6316. doi: 10.1038/sj.onc.1204009. [DOI] [PubMed] [Google Scholar]
- Camposano SE, Greenberg E, Kwiatkowski DJ, Thiele EA. Distinct clinical characteristics of tuberous sclerosis complex patients with no mutation identified. Ann Hum Genet. 2009;73:141–146. doi: 10.1111/j.1469-1809.2008.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- Ehninger D, Silva AJ. Rapamycin for treating Tuberous sclerosis and Autism spectrum disorders. Trends Mol Med. 2011;17:78–87. doi: 10.1016/j.molmed.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasa MA, Koessmeier KT, Ahmadian MR, Braga VM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- Frittoli E, Palamidessi A, Pizzigoni A, Lanzetti L, Garre M, Troglio F, Troilo A, Fukuda M, Di Fiore PP, Scita G, et al. The primate-specific protein TBC1D3 is required for optimal macropinocytosis in a novel ARF6-dependent pathway. Mol Biol Cell. 2008;19:1304–1316. doi: 10.1091/mbc.E07-06-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- Guo L, Ying W, Zhang J, Yuan Y, Qian X, Wang J, Yang X, He F. Tandem affinity purification and identification of the human TSC1 protein complex. Acta Biochim Biophys Sin (Shanghai) 2010;42:266–273. doi: 10.1093/abbs/gmq014. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Wu CL, Manning BD. Signaling events downstream of mammalian target of rapamycin complex 2 are attenuated in cells and tumors deficient for the tuberous sclerosis complex tumor suppressors. Cancer Res. 2009;69:6107–6114. doi: 10.1158/0008-5472.CAN-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003a;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003b;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jaeschke A, Hartkamp J, Saitoh M, Roworth W, Nobukuni T, Hodges A, Sampson J, Thomas G, Lamb R. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J Cell Biol. 2002;159:217–224. doi: 10.1083/jcb.jcb.200206108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalender A, Selvaraj A, Kim SY, Gulati P, Brule S, Viollet B, Kemp BE, Bardeesy N, Dennis P, Schlager JJ, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski D. TSC1, TSC2, TSC3? Or mosaicism? Eur J Hum Genet. 2005;13:695–696. doi: 10.1038/sj.ejhg.5201412. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, el-Hashemite N, Onda H. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- Martinu L, Masuda-Robens JM, Robertson SE, Santy LC, Casanova JE, Chou MM. The TBC (Tre-2/Bub2/Cdc16) domain protein TRE17 regulates plasma membrane-endosomal trafficking through activation of Arf6. Mol Cell Biol. 2004;24:9752–9762. doi: 10.1128/MCB.24.22.9752-9762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2009;27:S43–S51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Yecies JL, Zhang HH, Howell JJ, Nicholatos J, Harputlugil E, Bronson RT, Kwiatkowski DJ, Manning BD. Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Sci Signal. 2012;5:ra24. doi: 10.1126/scisignal.2002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Yoshino K, Miyamoto T, Eguchi S, Oshiro N, Kikkawa U, Yonezawa K. Identification of TBC7 having TBC domain as a novel binding protein to TSC1-TSC2 complex. Biochem Biophys Res Commun. 2007;361:218–223. doi: 10.1016/j.bbrc.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Nellist M, van Slegtenhorst MA, Goedbloed M, van den Ouweland AM, Halley DJ, van der Sluijs P. Characterization of the cytosolic tuberin-hamartin complex. Tuberin is a cytosolic chaperone for hamartin. J Biol Chem. 1999;274:35647–35652. doi: 10.1074/jbc.274.50.35647. [DOI] [PubMed] [Google Scholar]
- Parkhitko A, Myachina F, Morrison TA, Hindi KM, Auricchio N, Karbowniczek M, Wu JJ, Finkel T, Kwiatkowski DJ, Yu JJ, et al. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc Natl Acad Sci U S A. 2011;108:12455–12460. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank TL, Yeung RS, Henske EP. Hamartin, the product of the tuberous sclerosis 1 (TSC1) gene, interacts with tuberin and appears to be localized to cytoplasmic vesicles. Cancer Res. 1998;58:4766–4770. [PubMed] [Google Scholar]
- Potter CJ, Huang H, Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001;105:357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- Qin W, Kozlowski P, Taillon BE, Bouffard P, Holmes AJ, Janne P, Camposano S, Thiele E, Franz D, Kwiatkowski DJ. Ultra deep sequencing detects a low rate of mosaic mutations in tuberous sclerosis complex. Hum Genet. 2010;127:573–582. doi: 10.1007/s00439-010-0801-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner M, Hanneder M, Siegel N, Valli A, Hengstschlager M. The tuberous sclerosis gene products hamartin and tuberin are multifunctional proteins with a wide spectrum of interacting partners. Mutat Res. 2008;658:234–246. doi: 10.1016/j.mrrev.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sato N, Koinuma J, Ito T, Tsuchiya E, Kondo S, Nakamura Y, Daigo Y. Activation of an oncogenic TBC1D7 (TBC1 domain family, member 7) protein in pulmonary carcinogenesis. Genes Chromosomes Cancer. 2010;49:353–367. doi: 10.1002/gcc.20747. [DOI] [PubMed] [Google Scholar]
- Serfontein J, Nisbet RE, Howe CJ, de Vries PJ. Evolution of the TSC1/TSC2-TOR signaling pathway. Sci Signal. 2010;3:ra49. doi: 10.1126/scisignal.2000803. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M, Nellist M, Nagelkerken B, Cheadle J, Snell R, van den Ouweland A, Reuser A, Sampson J, Halley D, van der Sluijs P. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet. 1998;7:1053–1057. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest. 2003a;112:1223–1233. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003b;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.