Abstract
PPARγ is a nuclear receptor that regulates gene transcription associated with intermediary metabolism, adipocyte differentiation, as well as tumor suppression and proliferation. To understand the role of PPARγ in tumorigenesis, transgenic mice were generated with mammary gland-directed expression of the dominant-negative transgene, Pax8PPARγ. Transgenic mice were phenotypically indistinguishable from wild-type mice, but mammary epithelial cells expressed a greater a high percentage of CD29hi/CD24neg, CK5+ and double positive CK14/CK18 cells. These changes correlated with reduced PTEN and increased Ras, ERK and AKT activation. Although spontaneous tumorigenesis did not occur, transgenic animals were highly susceptible to progestin/DMBA-induced mammary carcinogenesis, which in contrast to wild-type mice, resulted in a high tumor multiplicity and most importantly, in the appearance of predominantly estrogen receptorα-positive (ER+) ductal adenocarcinomas. Tumors expressed a similar PTENlo/pERKhi/pAKThi phenotype as mammary epithelium, and exhibited high activation of ERE-dependent reporter gene activity. Tumorigenesis in MMTV-Pax8PPARγ mice was insensitive to the chemopreventive effect of a PPARγ agonist, but was profoundly inhibited by the ER antagonist fulvestrant. These results reveal important new insights into the previously unrecognized role of PPARγ in the specification of mammary lineage and the development of ER+ tumors.
Keywords: PPARγ, ERK, ER, mammary carcinogenesis, stem cells
Introduction
The PPAR5 nuclear receptor subfamily consists of the PPARα, PPARγ and PPARδ/β isotypes, which regulate multiple metabolic pathways controlling fatty acid β-oxidation, glucose utilization, cholesterol transport, energy balance and adipocyte differentiation (1–3). These and other receptor-mediated functions pertain to their use as targets for chemopreventive agents (2, 4–6). PPARγ agonists inhibit DMBA-induced preneoplastic lesions in the mammary gland (7) and delay the development of mammary carcinogenesis (8–10). Chemopreventive activity correlates, in part, with the ability of PPARγ to increase transcription of tumor suppressor genes, such as PTEN (11) and BRCA1 (12), through their respective PPRE promoter regions. In contrast, PPARγ haploinsufficiency increases mammary carcinogenesis (13), and the dominant-negative Pax8PPARγ fusion protein (14, 15) increases proliferation and transformation, and reduces expression of the Ras tumor suppressor gene, NORE1A, in thyroid tissue (16, 17).
These findings led us to examine the function of PPARγ in the mammary gland and in mammary carcinogenesis by generating transgenic mice that expressed Pax8PPARγ (14, 18). This animal model has lent itself to determining not only whether PPARγ agonists act directly or indirectly on mammary epithelium, but also whether negative regulation of PPARγ affects cell lineage within the normal gland and developing tumors (19). These studies have revealed a previously unknown linkage between PPARγ signaling, stem and progenitor cell expansion and tumor lineage specification pertaining to the induction of ER expression that may have significant implications for the treatment of breast cancer.
Materials and Methods
Transgenic mice
Transgenic MMTV-Pax8PPARγ mice were generated by pronuclear injection into FVB mouse embryos using standard techniques by the Transgenic Shared Resource, LCCC. The Pax8PPARγ cDNA (14) was provided by Dr. Todd Kroll, University of Chicago, and subcloned into the EcoRI site in plasmid MMTV-SV40-Bssk provided by Dr. William Muller, McMaster University. The MMTV-Pax8PPARγ construct was digested with SalI and SpeI, purified and used for microinjection. Founder lines were confirmed by Southern blot and western analysis (see Supplemental Fig. 1), two independent founders of similar phenotype were generated, and one was used for subsequent studies.
Whole mount preparation
Female mice were euthanized using carbon dioxide, and mammary glands were harvested, placed on a dry silanized glass slide and fixed overnight in 1 part glacial acetic acid:3 parts 100% ethanol. Tissues were rehydrated through successive incubation with 70% ethanol followed by distilled water, and stained with Carmine Red alum overnight. Tissues were then dehydrated by successive incubation in graded ethanol followed by mixed xylenes, and mounted in Permount (Fisher Scientific) (20).
Involution studies
On the day of weaning, nursing females were sacrificed and mammary glands were harvested for whole mount preparation on days 0, 3, 7 and 10 following induced involution by the teat sealing procedure (20).
Mammary carcinogenesis
Mammary carcinogenesis was initiated in MMTV-Pax8PPARγ and wild-type mice with medroxyprogesterone (Depo-Provera™) and DMBA as described previously (8, 21). GW7845 (provided by GlaxoSmithKline) was administered in Purina rodent chow 5001 at a concentration of 0.005% (w/w) immediately after the last dose of DMBA (8, 21).
Primary cell culture
Mammary glands or mammary tumors were excised and minced into 1 mm pieces under sterile conditions. Tissue was digested in DMEM/F12 medium supplemented with 5% FBS, 100 U/ml collagenase I, 100 U/ml hyaluronidase, 100 U/ml penicillin, 100 U/ml streptomycin, 10 µg/ml gentamicin and 2.5 µg/ml amphotericin B. After incubation at 37°C for 16 hr, tissue was harvested and centrifuged at 1,000 rpm for 5 min. The supernatant was discarded and the cell pellet was washed twice with PBS and resuspended in DMEM/F12 medium supplemented with 5% FBS, 10 ng/ml EGF (Upstate Biotechnology), 5 µg/ml insulin (Biofluids), 100 U/ml penicillin, 100 U/ml streptomycin, 10 µg/ml gentamicin and 2.5 µg/ml amphotericin B, and incubated at 37°C under 5% CO2. The cell culture was trypsinized for 1–2 min in 0.05% trypsin-EDTA (Biosource) every three days to remove fibroblast cells. Confluent cells were harvested between days 7 and 10 for further analysis. Mammary tumor cell lines MC and 437T were derived from medroxyprogesterone/DMBA-induced mammary adenocarcinomas formed in wild-type FVB and MMTV-Pax8PPARγ mice, respectively, and have maintained a stable phenotype for more than 30 passages.
FACS analysis
Cells were prepared as single-cell suspensions, washed twice with PBS and blocked for 15 min with PBS supplemented with 3% FBS. Cells were incubated for 1 hr at room temperature with the following antibodies: FITC-Sca-1 (1:200 dilution) (clone E13–161.7, BD Biosciences), PE-CD24 (1:200, BD Biosciences), biotinylated-CD29 (1:200, AbD Serotec), FITC-CD133 (1:200, BD Biosciences) and FITC-CD49f (1:200, BD Biosciences). PE-Cy5-conjugated streptavidin (eBioscience) was used as secondary antibody for biotinylated-CD29 at 1:200 dilution. IgG of the same isotype was used as a control. Cells were analyzed using a FACSort (BD Biosciences) and analyzed by FACExpress Denovo software by the Flow Cytometry/Cell Sorting Shared Resource, LCCC.
Western blot analysis
The cell pellet or pulverized tissue frozen in liquid nitrogen was mixed with lysis buffer containing: 0.1% SDS, 0.5% NP-40, PMSF, 1 mM sodium vanadate, 50 mM NaF, 10 mM βglycerophosphate, 5 mM sodium pyrophosphate and a protease inhibitor cocktail (Roche) (22). Following incubation on ice for 30 min, lysates were cleared by centrifugation at 13,000 × g at 4°C for 15 min. Protein concentration was determined by the BCA Protein Assay (Pierce), and 50 µg of lysate were separated in a 4–12% NuPAGE Bis-Tris gel (Invitrogen). After wet transfer, membranes were blocked for 1 hr at room temperature in 5% nonfat dry milk in Tris-buffered saline (pH 7.4) containing 0.1% Tween-20. Primary antibody was incubated for either 1.5 hr at room temperature or overnight at 4°C. Secondary antibody was incubated for 1 hr at room temperature, and proteins were visualized with either SuperSignal West Pico or SuperSignal West Dura (Pierce). The following antibodies and their dilution were used: PPARγ and PTEN (1:300, Santa Cruz Biotechnology); pβ-catenin, β-catenin, GSK3β, pGSK3β, pERK1/2, pT308Akt and Akt (1:1000, Cell Signaling), and β-actin (1:5,000, Sigma-Aldrich).
RT-PCR analysis
RNA was extracted from either mammary glands or cell cultures using Trizol reagent (Invitrogen). For reverse transcription, 2 µg total RNA was transcribed with SuperScript II reverse transcriptase (Invitrogen). Equal amounts of cDNA were used for the PCR reaction under the following conditions: 94°C 30 sec, 56°C 30 sec, 72°C 1 min for 30 cycles. For ER mRNA, 38 cycles of amplification were necessary to measure levels in wild-type mammary gland and MC tumor cells. The following primers were used for mRNA analysis: ERα: forward, 5’-TGA TCA ACT GGG CAA AGA-3’, reverse, 5’-CAG GAG CAG GTC ATA GAG-3’; Pax8PPARγ: forward, 5’-AAC CTC TCG ACT CAC CAG-3’, reverse, 5’-GAT GGC ATT ATG AGA CAT CCC-3’; β-actin: forward, 5’-AGA GGG AAA TCG TGC GTG AC-3’, reverse, 5’-CAA TAG TGA TGA CCT GGC CGT-3’. Samples were separated in a 1% agarose gel.
Gene microarray
RNA was prepared from MC and 437T tumor cells using Trizol (Invitrogen), and processed for array analysis as previously described (8, 21). Hybridization was carried out by the Macromolecular Analysis Shared Resource, LCCC, with an Affymetrix Mouse Genome 430A 2.0 GeneChip® representing 14,000 well-annotated genes. Gene array analysis was evaluated by comparing differences between paired samples and ranking changes by their log2 ratio. Only differences in signal ratio > log2 3.0 and < log2 −3.0 were ranked and are listed in Supplemental Table 1. Each cRNA was prepared from equal amounts of RNA pooled from three samples.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Genomic DNA was digested by incubation with RNase-free DNase for 15 min at room temperature. One µg of RNA was reverse transcribed in a total volume 20 µl using the Omniscript RT kit (Qiagen). PCR was performed in triplicate in an ABI-Prism 7700 instrument (Applied Biosystems) using SYBRGreen I detection (Qiagen) according to the manufacturer’s protocol (8, 21). Amplification using the appropriate primers (see Supplemental Table 2) was confirmed by ethidium bromide staining of the PCR products on an agarose gel. The expression of each target gene was normalized to the expression of 18S RNA and is presented as the ratio of the target gene to 18S RNA calculated by 2−ΔCt, where ΔCt = CtTarget-Ct18s (see Supplemental Fig. 2).
Immunohistochemical staining
Paraffin sections of mammary tissue and tumors were prepared by the Histopathology and Tissue Shared Resource, LCCC. Slides were baked at 60°C for 1 hr, deparaffinized in xylene for 15 min, and rehydrated in 100%, 95% and 70% ethanol for 5 min each. Antigen retrieval was achieved by steaming slides for 20 min in 10 mM citrate buffer, pH 6.0. Slides were washed three times in PBS and blocked for 1 hr in a buffer containing 10% goat serum in PBS, and then incubated at room temperature for 1 hr with the following antibodies: ERα (1:1000, Santa Cruz Biotechnology), CK5 (1:100, Chemicon), CK14 (1:25, Neomarkers), CK18 (1:25, Epitomics) and CK19 (1:100, NeoMarkers). Slides were washed three times in PBS and incubated with biotinylated secondary antibody for 30 min, washed three times in PBS and antigen visualized with ABC Vectastain and DAB as substrate (Vector Labs). Slides were counterstained with Harris-modified hematoxylin (Fisher Scientific) and mounted in Permount.
Reporter gene assays
Reporter assays were carried out with either 3XPPRE-TK-luciferase (provided by Dr. Mitchell Lazar, University of Pennsylvania) to measure PPARγ-dependent activity, pERE-luciferase (23) (provided by Dr. Anna Riegel, Georgetown University) to measure ERα-dependent activity or TopFlash (Upstate Biotechnology) to measure β-catenin/TCF-dependent activity. Cells were cultured in 24-well plates at 20,000 cells/well, and transfected using Lipofectamine Plus (Invitrogen). A plasmid expressing Renilla luciferase was co-transfected as an internal control. Cells were harvested after 48 h and luciferase activity determined using the Dual Luciferase Assay (Promega) according to the manufacturer’s protocol. All samples were run in triplicate and repeated three times. For PPARγ-dependent activity, cells were incubated overnight in medium containing delipidated stripped serum (Sigma-Aldrich) and cells treated for 24 hr with 1 µM GW7845. For ER-dependent activity, cells were grown for 24 hr in phenol red-free medium containing stripped serum, and transfected with pERE-luciferase. Cells were then treated with 10 nM 17-β-estradiol, and luciferase activity measured 24 hr later. TopFlash activity was determined in cells grown in 5% FBS in DMEM medium.
Ras activation assay
Ras activity was determined in MC and 437T cells grown in 5% FBS in DMEM medium for 24 hr in 100 mm dishes, and then incubated for 24 hr in serum-free DMEM medium. Cells were then incubated for 5 min with 10 ng/ml EGF (Invitrogen), and cell lysates assayed with the Ras Activation Assay Kit (Cytoskeleton, Inc.) according to the manufacturer’s protocol. Each reaction contained 250 µg protein, and the Raf-1 pull-down product was separated in a 4–12% NuPage gel (Invitrogen) and analyzed for activated Ras by western blotting.
DNA methylation assay
Genomic DNA was extracted from mammary gland and tumor cells in 600 µl DNA lysis buffer (10 mM Tris, 0.4 M NaCl, 0.6% SDS, 0.1 M EDTA, 20 µg/ml Protease K) at 56 °C for 5 hr. The lysate was mixed with 150 µl 6 M NaCl and centrifuged at 12,000 rpm for 10 min. The supernatant was removed and mixed with an equal volume of 100% ethanol, centrifuged at 12,000 rpm for 5 min, and the pellet washed once with 75% ethanol. DNA was dissolved in 300 µl nuclease-free water, extracted with phenol:chloroform:isoamyl alcohol (25:24:1) (Fluka), centrifuged at 12,000 rpm for 5 min and washed twice with 75% ethanol. DNA (2 µg) was then digested overnight at 37° with EcoRI and purified with the QIAquick Nucleotide Removal Kit (Qiagen). DNA (45 µl) was mixed with 5 µl freshly prepared 3M NaOH and incubated at room temperature for 10 min. Cytosine methylation was determined using the EZ DNA Methylation-Gold kit (Zymo Research Corp.) according to the manufacturer’s protocol. The ERα promoter region of untreated DNA and bisulfite-oxidized DNA was amplified by PCR and sequenced. The primers used were: bisulfite-treated DNA: sense, 5’-TTG GAG TTT TTT TTA GGA ATG TTG ATT TTA G-3’, antisense, 5’-AAC CAA TCC TAC CCT ACT AAT TCA AAA AC-3’; Untreated DNA: sense, 5’-CTG GAG TTT CTT CTA GGA ATG CTG ATT CTA G-3’, antisense, 5’-AGC CGA TCC TAC CCT GCT GGT TCA AGA GC-3’. The 345 bp PCR product was cloned into pCR2.1 (Invitrogen) and sequenced (see Supplemental Fig. 3).
Statistical analyses
Analysis of two variables was conducted by Student’s two-tailed t test, and of multiple independent variables by Fischer’s Exact test. Survival curves were analyzed by Wilcoxon’s rank test. P < 0.05 was considered to be statistically significant.
Results
MMTV-Pax8PPARγ transgenic mice
Mice expressing MMTV-Pax8PPARγ were developed to first assess the influence of endogenous PPARγ activation on mammary carcinogenesis, and secondly, to determine if the chemopreventive effects of PPARγ agonists were a result of their direct action on mammary epithelium. Pax8PPARγ was chosen as the transgene since it functions as an effective dominant-negative receptor that completely blocks ligand-dependent PPARγ activation (14, 18), in contrast to “dominant-negative” variants that act as low affinity receptors (24). Transgenic mice did not exhibit gross morphologic changes in mammary gland structure in either virgin or pregnant mice after lactation and involution (see Supplemental Fig. 1). To ascertain that Pax8PPARγ functioned as a true dominant-negative receptor, primary mammary epithelial cell cultures were transfected with PPRE-luciferase in the presence and absence of the PPARγ agonist, GW7845 (see Supplemental Fig. 1B). Transgenic mice did not express endogenous PPARγ-dependent reporter gene activity, in contrast to wild-type mice, which did express activity.
Gene profiling of mammary epithelium from MMTV-Pax8PPAR revealed changes in genes associated with transport, the cell cycle, adhesion and receptor signaling, such as PDK1, PI3Kp110α, Ras-activating protein, Fos, Jak1, RGS5, cyclin D2, ABCA1 and PGC-1α, a coactivator of ERα (25), ERRα (26) and PPARγ (27) (see Supplemental Table 1 and Supplemental Fig. 2). Upregulation of proliferative gene expression was complemented by a reduction of the cyclin kinase inhibitor, p27Kip1, the Notch inhibitor Deltex, and an increase in the antiapoptotic gene Birc4. Expression of the ER-responsive genes, XBP1, cyclin D1 and IL-6 signal transducer (28) were all significantly increased in MMTV-Pax8PPARγ mice.
Increased stem/progenitor cell expansion in MMTV-Pax8PPARγ
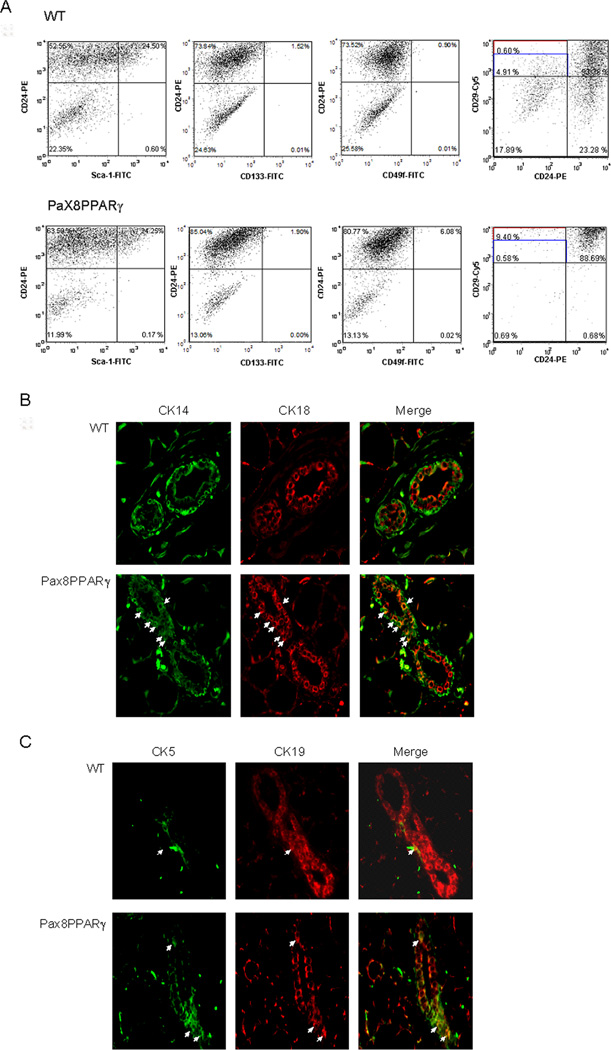
To determine if PPARγ affected cell lineage, primary cell cultures were evaluated for CD antigen expression (Fig. 1A). CD29hi/CD24neg cells were more abundant in transgenic mice in comparison to wild-type mice (9.4% vs 0.6%). In addition, immunostaining for cytokeratin expression revealed a greater percentage of CK5+ cells in the transgenic mammary gland (Fig. 1B), and an increased number of double-positive CK14/CK18 cells (fig. 1C). These results suggest increased stem and progenitor cell expansion downstream of Pax8PPARγ.
Figure 1.
MMTV-Pax8PPARγ mice exhibit an increase in mammary stem and progenitor cell markers. A, FACS analysis of primary mammary cell cultures. Cells from MMTV-Pax8PPARγ mice (Pax8PPARγ) express an increased percentage of CD29hi/CD24 cells vs. wild type (WT) mice. B, MMTV-Pax8PPARγ mice (Pax8PPARγ) express a greater percentage of CK5+ cells (arrows) than wild-type mice (WT); magnification 400X. C, MMTV-Pax8PPARγ mice express a greater percentage of double positive CK14/CK18 cells (arrows) vs. wild-type mice (WT); magnification 400X.
β-Catenin and ERK signaling are increased in transgenic mice
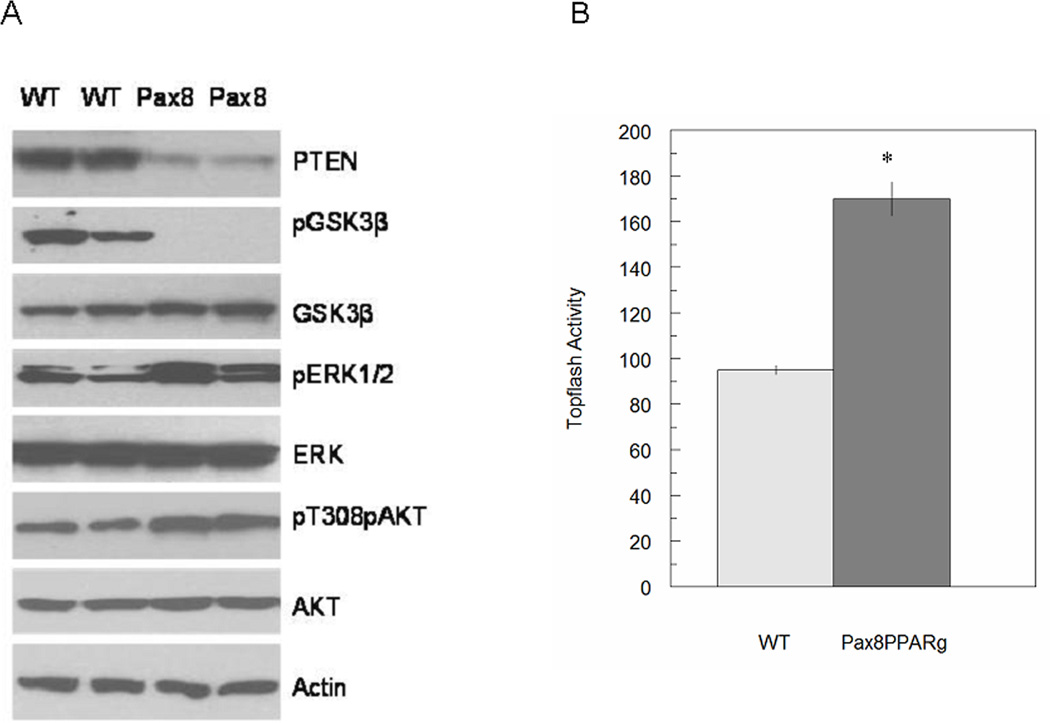
Tissue lysates from MMTV-Pax8PPARγ mice revealed a marked reduction in PTEN and pGSK3β, and an increase in pERK1 and pAKT (Fig. 2A). Measurement of β-catenin/TCF-dependent reporter gene activity in primary cell cultures indicated a moderate increase in activity in cells from transgenic mice in comparison to those from wild-type animals (Fig. 2B). These findings suggest that Pax8PPARγ increases Wnt signaling through reducing PTEN expression, increasing pERK and pAKT and reducing pGSK3β to promote β-catenin/TCF transactivation.
Figure 2.
MMTV-Pax8PPARγ mice exhibit an increase in Wnt pathway signaling. A, Mammary gland lysates analyzed by western blotting indicate that MMTV-PPARγ mice (Pax8) express less PTEN, no pGSK3β and greater pERK and pAKT vs. wild-type mice (WT). B, Primary mammary cell cultures transfected with the Topflash reporter plasmid indicate that cells from MMTV-Pax8PPARγ mice express greater β-catenin/TCF-dependent activity vs. wild-type mice. Each value is the mean ± SE; N=3 per assay; *, P < 0.01 by Student’s t test.
Pax8PPARγ reduces PTEN and upregulates pERK and pAKT in mammary tumors
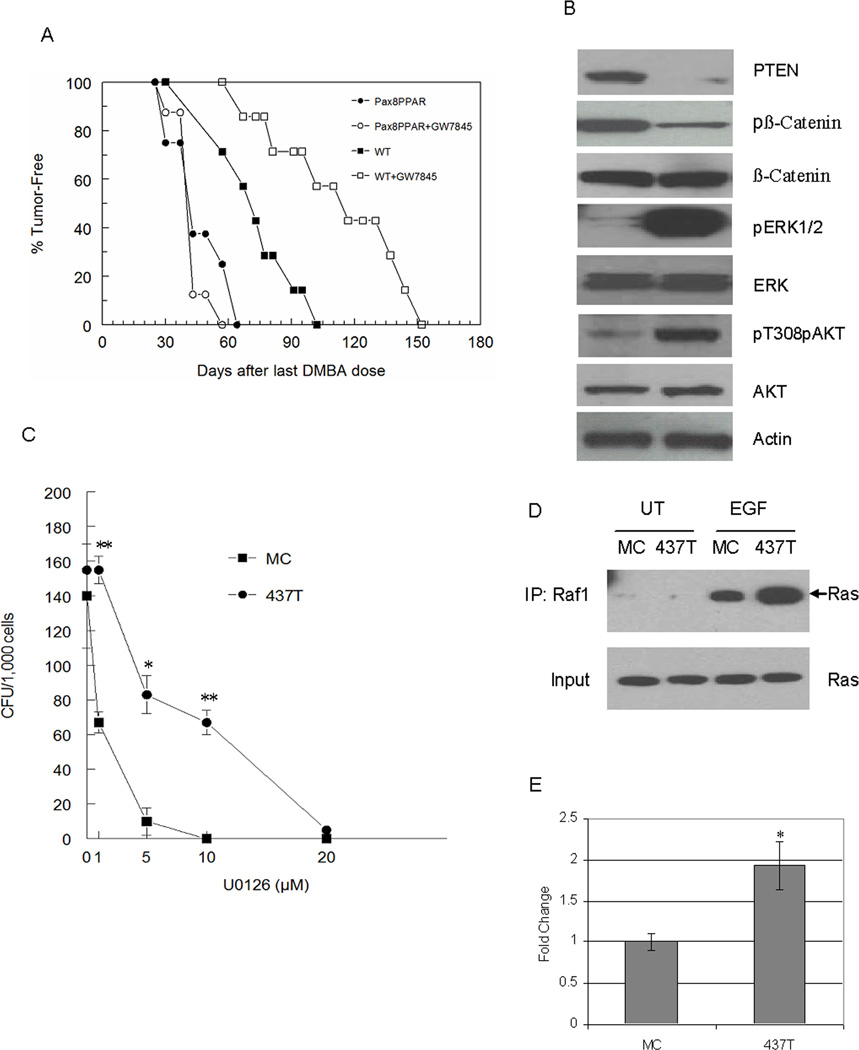
Since spontaneous tumorigenesis did not occur in MMTV-Pax8PPARγ mice over their life span, their sensitivity to mammary carcinogenesis (8) was determined (Fig. 3A). MMTV-Pax8PPARγ mice were more sensitive to carcinogenesis, and exhibited a median tumor latency of 40 days vs. 70 days in wild-type mice. Tumor multiplicity was greater in MMTV-Pax8PPARγ mice, and ductal adenocarcinoma formation was significantly greater, and squamous cell carcinoma formation significantly less than in wild-type mice (Table 1).
Figure 3.
MMTV-Pax8PPARγ mice are more susceptible to carcinogenesis and resistant to the chemopreventive effects of PPARγ agonist GW7845. A, Tumor latency was significantly reduced (P = 0.018) from 80 days in wild-type mice (WT) to 40 days in MMTV-Pax8PPARγ mice (Pax8PPAR). Administration of a diet containing 0.005% GW7845 significantly delayed (P = 0.024) tumor formation in wild-type mice, but not in transgenic mice (Pax8PPAR) (P = 0.551); N = 8 per group. Wilcoxon’s rank test was used for statistical analysis. B, Tumor cells from MMTV-Pax8PPARγ mice exhibit a reduction in PTEN and pβ-catenin and an increase in pERK and pAKT. Western analysis was carried out with lysates from progestin/DMBA-induced adenocarcinoma cell lines derived from MMTV-Pax8PPARγ mice (437T cells, 437T) and wild-type mice (MC cells, MC). C, Colony formation in 437T cells is less sensitive than MC cells to MEK inhibitor U0126. 437T cells (437T) and MC cells (MC) were treated with 1, 5 and 10 µM U0126, and colony formation determined. Each value is the mean ± standard error; N=3 per assay; *, P < 0.05, **, P < 0.01 by Student’s t test. D, Ras is activated to a greater extent in 437T cells vs. MC cells. Cells were serum-starved overnight and either left untreated (UT) or treated for 5 min with10 ng/ml EGF (EGF). Lysates were ‘pulled down’ with the Raf activated Ras-binding domain and western blotting carried out with a Ras antibody. E, 437T cells exhibit greater β-catenin/TCF reporter gene activity. 437T cells (437T) transfected with Topflash as a measure of β-catenin/TCF activation exhibit significantly greater activity than MC cells (MC). Each value is the mean ± SE; N=3 per assay; *, P < 0.05 by Student’s t test.
Table 1.
Tumor multiplicity and histopathology in MMTV-Pax8PPARγ mice. Wild-type FVB and MMTV-Pax8PPARγ mice fed standard rodent chow are the controls in fig. 3A. Each group consists of 8 mice. Tumor multiplicity, the average number of tumors per animal, was significantly greater in MMTV-PaxPPARγ mice than in wild-type mice, and transgenic mice exhibited a greater frequency of ≥3 tumors/animal. The total number of tumors in wild-type and MMTV-Pax8PPARγ mice were 12 and 23, respectively. The total number of adenocarcinomas in MMTV-Pax8PPARγ mice was significantly greater, and the total number of squamous carcinomas was significantly lower than in wild-type mice.
| Tumor Multiplicity |
Mice with ≥3 Tumors* |
Adenocarcinoma | Squamous | Myoepithelial | |
|---|---|---|---|---|---|
| WT | 1.50±0.16 | 0/8 | 3/12 (25.0%) | 5/12 (41.7%) | 4/12 (33.3%) |
| Pax8PPARγ | 2.88±0.48** | 4/8 | 18/23 (78.3%)*** | 2/23 (8.7%)*** | 3/23 (13.0%) |
no. of mice with ≥3 tumors/total no. of mice
P < 0.02 by Student’s two-tailed t test
P= 0.004; ***, P = 0.014 by Fischer’s Exact test
To assess the effect of PPARγ activation on mammary tumorigenesis, animals were maintained on a diet supplemented with PPARγ agonist GW7845 (Fig. 3A). While GW7845 delayed median tumor latency by approximately one month in wild-type mice as reported previously (16), transgenic mice were insensitive to the PPARγ agonist. These results show for the first time that a PPARγ agonist acts directly on mammary epithelium, and not indirectly through its effects on stromal tissue.
To determine if mammary adenocarcinomas recapitulated the signaling phenotype present in the mammary gland of transgenic mice, mammary adenocarcinoma cell lines were developed from wild-type (MC cells) and transgenic (437T cells) animals (Fig. 1B). PTEN was markedly reduced in 437T cells, and pERK and pAKT were increased coincident with reduced β-catenin phosphorylation in comparison to MC cells. 437T cells were also less sensitive to the MEK inhibitor, U0126, which was consistent with their greater pERK expression (Fig. 3C). To determine if ERK activation was associated with Ras activation, ‘pull-down’ assays were carried out with the Raf domain recognizing activated Ras-GTP (Fig. 3D). In response to EGF stimulation, Ras activation was greater in 437T vs. MC cells. In addition, β-catenin/TCF-dependent reporter gene activity was increased to a greater extent in 437T cells than in MC cells (Fig. 3E), which were consistent with the findings in primary cultures from transgenic mice (Fig. 2B).
Pax8PPARγ increases ER+ mammary carcinogenesis
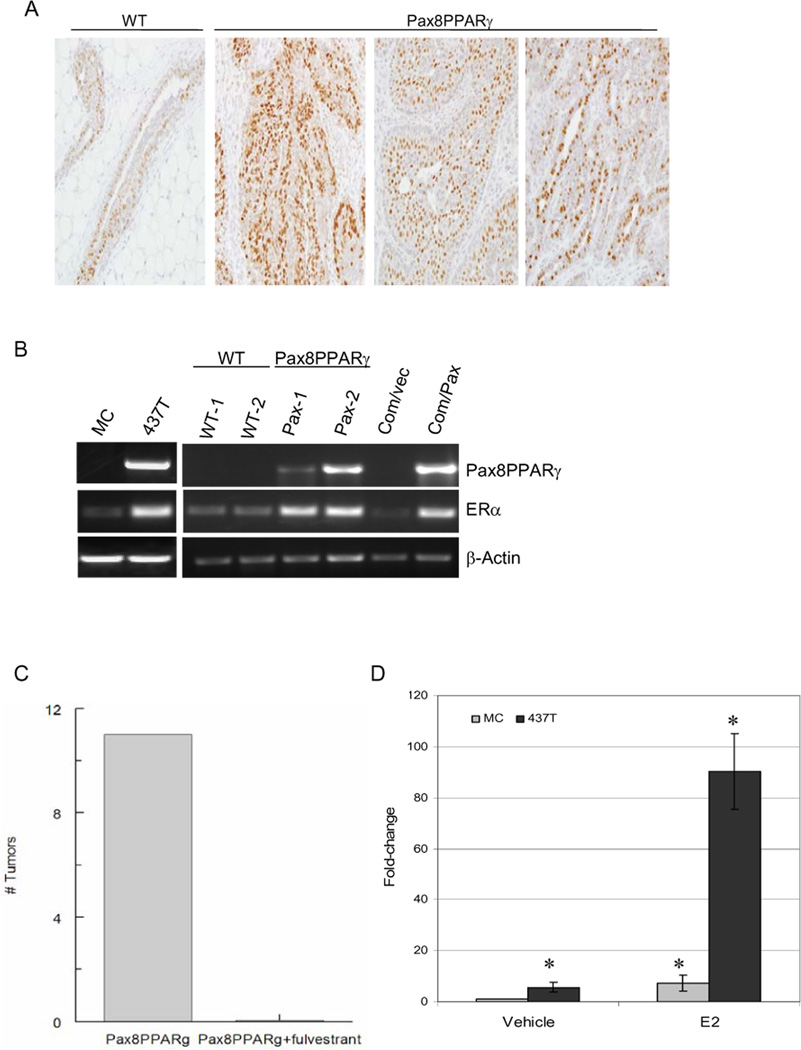
Since ductal adenocarcinomas were the predominant histopathologic phenotype in MMTV-Pax8PPARγ mice following progestin/DMBA-induced carcinogenesis (Table 1), tumors were analyzed for ER expression (Fig. 4A). Significantly, tumors from transgenic animals were predominantly ER+, whereas little or no ER expression was present in adenocarcinomas from wild-type animals (Fig. 4A). ER mRNA was increased in tumors and primary mammary gland cultures from transgenic animals (Fig. 4B). In this regard, transduction of Comma-1D mouse mammary epithelial cells with Pax8PPARγ induced ER expression (Fig. 4B). Importantly, tumor formation in transgenic mice was inhibited by the ER antagonist, fulvestrant, when administered on a weekly basis beginning after the last dose of DMBA (Fig. 4C), indicating that tumor growth is functionally dependent on ER signaling. ER-dependent reporter gene activity was further assessed in MC and 437T cells (Fig. 4D). 437T cells exhibited ~10-fold increase in reporter gene activity in comparison to MC cells. To determine if ER activation was associated with hypomethylation of the ER promoter, genomic analysis of the CpG motifs in the ER promoter region of MC and 437T cells was determined (see Supplemental Fig. 3). The ER promoter was not methylated in either tumor cell line, indicating that this mechanism is not responsible for ER induction in Pax8PPARγ-expressing cells.
Figure 4.
MMTV-Pax8PPARγ mice express ER+ adenocarcinomas after progestin/DMBA carcinogenesis. A, Tumors from MMTV-Pax8PPARγ mice (Pax8PPARγ) exhibit strong ER expression in ductal adenocarcinomas vs. the same tumor type from wild-type mice (WT); magnification 400X. B, ER mRNA is upregulated in the mammary gland and tumor cells from MMTV-Pax8PPARγ mice. RT-PCR reveals that ER mRNA is increased in 437T cells (437T) vs. MC cells (MC), in mammary epithelial cells from MMTV-Pax8PPARγ mice (Pax-1 and Pax-2) vs. wild-type mice (WT-1 and WT-2), and in Comma-1D cells transduced with Pax8PPARγ (Com/Pax) vs control cells (Com/Vec). Pax8PPARγ and β-actin mRNA amplification were carried out for 30 cycles, and ER mRNA amplification for 38 cycles to show the presence of ER mRNA in wild-type mammary gland and MC cells. C, Fulvestrant inhibits carcinogenesis in MMTV-Pax8PPARγ mice. Following progestin/DMBA carcinogenesis as in A, animals were treated s.c. with either vehicle (Pax8PPARγ) or fulvestrant (Pax8PPARγ+fulvestrant) at a dose of 200 mg/kg once per week for three months, and the number of mammary tumors determined; N=6 per group. D, ER-dependent reporter gene activity is increased in 437T cells. 437T and MC cells were grown in phenol red-free medium containing stripped serum for 24 hr, and transfected with ERE-luciferase. After 24 hr, cells were treated with 10 nM 17-β-estradiol (E2), and reporter gene activity determined 24 hr later in 437T cells (black bar) and MC cells (gray bar). Reporter activity is significantly increased by E2 in 437T and MC cells, and reporter activity in the absence or presence of E2 is significantly greater in 437T cells vs. MC cells. Each value is the mean ± SE; N=3 per assay; *, P < 0.001 by Student’s t test.
Discussion
Tumorigenesis involves the activation of oncogenic pathways in concurrence with repression of ‘tumor suppressor’ pathways that ultimately influence not only tumor initiation and progression, but also the lineage characteristics of malignant cells derived from transformed stem and/or early progenitor cells. Here we present evidence that inhibition of PPARγ through a dominant-negative receptor promotes a stem and progenitor cell phenotype that results in the development of ER+ ductal carcinomas following carcinogenesis (Fig. 5). These results suggest a previously unknown link between PPARγ suppression and cell fate determination in modulating mammary gland differentiation along the luminal lineage.
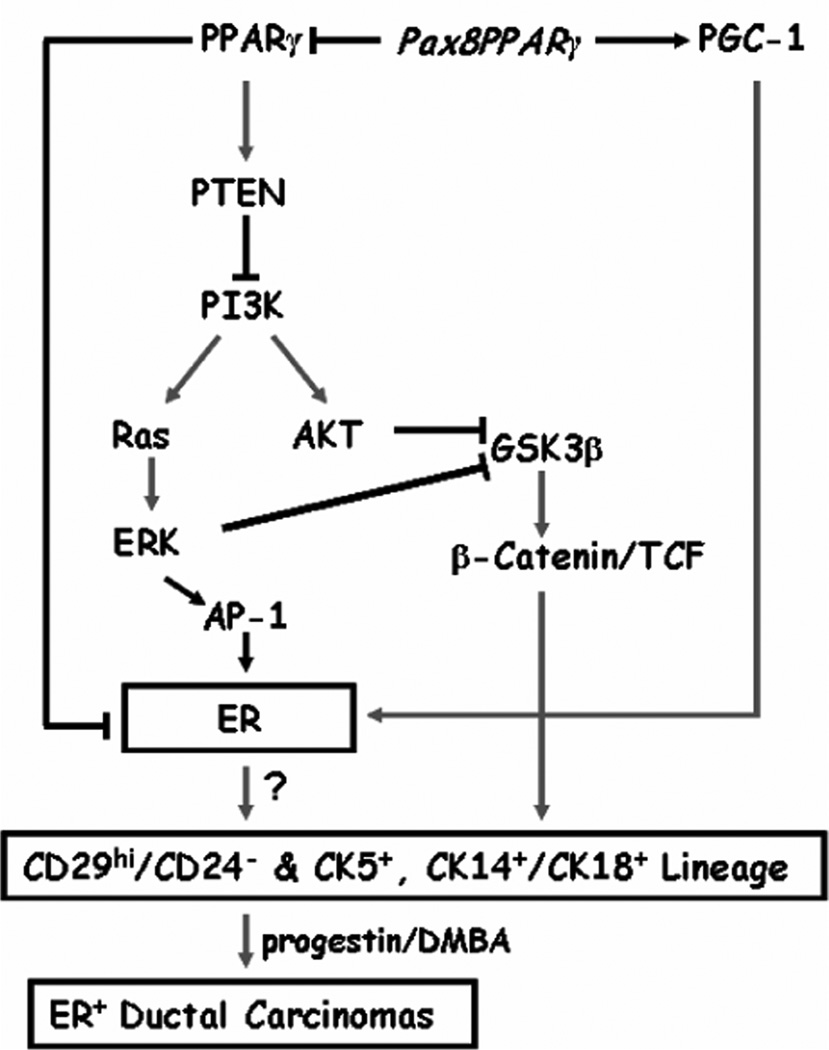
Figure 5.
Schematic of signal transduction in MMTV-Pax8PPARγ mice. Pax8PPARγ is envisioned to act in a dominant-negative fashion to block PPARγ-dependent inactivation of ER through the proteasomal pathway, promote PGC-1 coactivation of ER, and block PPARγ-mediated transactivation of PTEN. Reduced PTEN activates PI3K and Ras/ERK and AKT, which Inhibit GSK3β and activate β-catenin/TCF signaling. ERK may stimulate ER transcription through AP-1 downstream of ERK. Although, ERK is known to phosphorylate and stabilize ER, pSer104/106ER levels were not altered in the mammary gland of MMTV-Pax8PPARγ vs. wild-type mice. ER may also be phosphorylated by AKT and GSK3β (thin lines), but this has not been examined. Activation of ER and PI3K signaling are believed to increase stem and progenitor cell expansion and ER+ tumors following progestin/DMBA carcinogenesis. It is controversial whether mammary stem cells are ER+.
The increase in proliferation and transformation of thyroid cells and fibroblasts by Pax8PPARγ (17) is consistent with its putative role as an oncogene in follicular thyroid cancer (14). Although Pax8PPARγ was not oncogenic in the mammary gland, it increased the percentage of CD29hi/CD24neg, CK5+ and CK5/CK19 and CK14/CK18 double-positive cells. CD29hi/CD24lo/neg, which have been reported to lack ER expression (29, 30). CD29hi/CD24neg cells are enriched in multipotent stem cells, are regulated by the Wnt pathway (31), and have a high regenerative capacity in the mammary fat pad (30, 32). Thus, despite the absence of an overt developmental phenotype in MMTV-Pax8PPARγ mice, which resembles PPARγ null mice (33), this model indicates that PPARγ regulates mammary lineage specification, and likely accounts for the histopathologic differences in tumors arising in these animals, such as increased ductal carcinomas and a reduction in heterologous metaplasia, ie. squamous cell carcinomas (Table 1). These findings are consistent with the ability of PPARγ to act as a developmental switch to control differentiation, as shown previously in adipocytes and osteoblasts (34, 35).
The Wnt pathway increases expansion of Sca-1+ mammary tumor cells (36), progenitor cells (37) and stem cells (38). This phenotype is reminiscent of the association between PPARγ haploinsufficiency, increased β-catenin expression and colon carcinogenesis (39). PPARγ associates with and targets wild-type β-catenin (40), but not oncogenic variants of β-catenin (41), for proteasomal degradation. β-Catenin activation (42) and transformation (22) are closely regulated by PI3K signaling, where loss of PTEN suppression increases stem and progenitor cell expansion (43) (Fig. 4). The PI3K pathway is crucial for embryonic stem cell proliferation and tumorigenicity (44), and for the maintenance of stem cell pluripotency by AKT activation (45). It has also been shown that PI3K regulates ERK activation downstream of several oncogenes (46), that Ras and ERK mediate the transforming activity of the PI3K p110β and p110 γ catalytic subunits (47) and that PI3K inhibition blocks ERK activation mediated by integrin signaling (48). These results are concordant with our findings that loss of PTEN, inhibition of GSK3β, activation of ERK and AKT and increased β-catenin/TCF transactivation activity result from Pax8PPARγ expression in the mammary gland, and are consistent with the ability of the PI3K-Ras pathway to promote Wnt pathway activation (49). In this context, Pax8PPARγ is known to suppress expression of NORE1A (16), an inhibitor of the ERK pathway (50), and is in agreement with the ability of Pax8PPARγ to stimulate, and of wild-type PPARγ to inhibit ERK activation (51). Since ERK inhibits PPARγ and targets it for proteasomal degradation (52–55), this effect may also account for the reduction of PPARγ in the mammary gland of pregnant transgenic mice (Supplemental Fig. 1A).
MMTV-Pax8PPARγ mice expressed predominantly ER+ ductal adenocarcinomas. The high sensitivity of tumor formation to fulvestrant indicates that tumor growth is functionally dependent on ER signaling, although the precise mechanism by which inhibition of PPARγ induces ER expression is unknown. One possibility consistent with increased ER expression and ER-dependent transcriptional activity is the increase in expression of PGC-1 (Supplemental Table 1), a PPARγ co-activator (27), that preferentially co-activates ER (25), especially under conditions where PPARγ is repressed by Pax8PPARγ. Pax8PPARγ may also interfere with the ability of PPARγ to induce proteasomal degradation of ER and cyclin D (56, 57), an action that would be further sustained by Ras and ERK activation. An alternate, but not necessarily exclusive possibility, is that Pax8PPARγ interferes with the reported competition between PPARγ and ER for promoter occupation in ERE-responsive genes (58–60). However, we have not found this to be the case for ERE-dependent reporter gene activity when both receptors are co-expressed in 293T cells (unpublished results). Lastly, Pax8PPARγ could act as a dominant-negative regulator of Pax8 transcriptional activity. Pax8 transactivates the Wilm’s tumor suppressor gene, WT-1, which is known to inhibit ER function (61, 62); however, Pax8 and WT-1 expression were not detectable by RT-PCR in either the mammary gland or mammary tumors (unpublished results). Thus, by relieving the negative regulatory effects of PPARγ on ER stability and PTEN tumor suppressor activity, Pax8PPARγ would promote ER stability and coactivation by PGC-1 through ERK and AKT-dependent phosphorylation (Fig. 5).
In summary, Pax8PPARγ induced mammary stem and progenitor cell expansion in the context of reduced PTEN and increased Ras, ERK and AKT activation. Transgene expression resulted in a high tumor multiplicity and most significantly, predominantly ER+ ductal adenocarcinomas following carcinogenesis, which was insensitive to the chemopreventive effect of a PPARγ agonist, but profoundly inhibited by the ER antagonist, fulvestrant. These results reveal important new insights into the previously unrecognized role of PPARγ in the specification of mammary lineage and the development of ER+ tumors, including potential intervention modalities.
Supplementary Material
Acknowledgments
Grant support: Grant R01 CA11482 and contracts N01 CN43309 and N01 CN43330 from the National Cancer Institute, National Institutes of Health. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Grant C06 RR14567 from the National Center for Research Facilities, NIH. We thank Aaron Foxworth, Georgetown University Animal Research Resource, for assistance with the carcinogenesis studies.
Footnotes
The abbreviations used are: DMBA, dimethylbenz(a)anthracene; ER, estrogen receptorα; LCCC, Lombardi Comprehensive Cancer Center; PDK1, 3-phosphoinositide-dependent protein kinase-1; PI3K, phosphatidylinositol 3-kinase; PPAR, peroxisome proliferator-activated receptor; PPRE, PPAR responsive element.
References
- 1.Berger JP, Akiyama TE, Meinke PT. PPARs: therapeutic targets for metabolic disease. Trends Pharmacol Sci. 2005;26:244–251. doi: 10.1016/j.tips.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 3.Lehrke M, Lazar MA. The many faces of PPARgamma. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 4.Kopelovich L, Fay JR, Glazer RI, Crowell JA. Peroxisome proliferator-activated receptor modulators as chemopreventive agents. Mol Cancer Ther. 2002;1:357–363. [PubMed] [Google Scholar]
- 5.Stoll BA. Linkage between retinoid and fatty acid receptors: implications for breast cancer prevention. Eur J Cancer Prev. 2002;11:319–325. doi: 10.1097/00008469-200208000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Rosen ED, Spiegelman BM. PPARgamma: a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 7.Mehta RG, Williamson E, Patel MK, Koeffler HP. A ligand of peroxisome proliferator-activated receptor gamma, retinoids, and prevention of preneoplastic mammary lesions. J Natl Cancer Inst. 2000;92:418–423. doi: 10.1093/jnci/92.5.418. [DOI] [PubMed] [Google Scholar]
- 8.Yin Y, Bai R, Russell RG, et al. Characterization of medroxyprogesterone and DMBA-induced multilineage mammary tumors by gene expression profiling. Mol Carcinog. 2005;44:42–50. doi: 10.1002/mc.20119. [DOI] [PubMed] [Google Scholar]
- 9.Pighetti GM, Novosad W, Nicholson C, et al. Therapeutic treatment of DMBA-induced mammary tumors with PPAR ligands. Anticancer Res. 2001;21:825–829. [PubMed] [Google Scholar]
- 10.Suh N, Wang Y, Williams CR, et al. A new ligand for the peroxisome proliferator-activated receptor-gamma (PPAR-gamma), GW7845, inhibits rat mammary carcinogenesis. Cancer Res. 1999;59:5671–5673. [PubMed] [Google Scholar]
- 11.Patel L, Pass I, Coxon P, Downes CP, Smith SA, Macphee CH. Tumor suppressor and anti-inflammatory actions of PPARgamma agonists are mediated via upregulation of PTEN. Curr Biol. 2001;11:764–768. doi: 10.1016/s0960-9822(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 12.Pignatelli M, Cocca C, Santos A, Perez-Castillo A. Enhancement of BRCA1 gene expression by the peroxisome proliferator-activated receptor gamma in the MCF-7 breast cancer cell line. Oncogene. 2003;22:5446–54450. doi: 10.1038/sj.onc.1206824. [DOI] [PubMed] [Google Scholar]
- 13.Nicol CJ, Yoon M, Ward JM, et al. PPARgamma influences susceptibility to DMBA-induced mammary, ovarian and skin carcinogenesis. Carcinogenesis. 2004;25:1747–1755. doi: 10.1093/carcin/bgh160. [DOI] [PubMed] [Google Scholar]
- 14.Kroll TG, Sarraf P, Pecciarini L, et al. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma. Science. 2000;289:1357–1360. doi: 10.1126/science.289.5483.1357. [DOI] [PubMed] [Google Scholar]
- 15.Yin Y, Kristipati ST, Yuan H, Kopelovich L, Glazer RI. Proceedings of the American Association for Cancer Research. Washington, DC: 2006. Apr 1–4, Modulation of stem cell antigen (Sca-1) expression by the PDK1 and Stat pathways. [Google Scholar]
- 16.Foukakis T, Au AY, Wallin G, et al. The Ras effector NORE1A is suppressed in follicular thyroid carcinomas with a PAX8-PPARgamma fusion. J Clin Endocrinol Metab. 2006;91:1143–1149. doi: 10.1210/jc.2005-1372. [DOI] [PubMed] [Google Scholar]
- 17.Gregory Powell J, Wang X, Allard BL, et al. The PAX8/PPARgamma fusion oncoprotein transforms immortalized human thyrocytes through a mechanism probably involving wild-type PPARgamma inhibition. Oncogene. 2004;23:3634–3641. doi: 10.1038/sj.onc.1207399. [DOI] [PubMed] [Google Scholar]
- 18.Yin Y, Yuan H, Wang C, et al. 3-Phosphoinositide-Dependent Protein Kinase-1 Activates the Peroxisome Proliferator-Activated Receptor-{gamma} and Promotes Adipocyte Differentiation. Mol Endocrinol. 2006;20:268–278. doi: 10.1210/me.2005-0197. [DOI] [PubMed] [Google Scholar]
- 19.Glazer RI, Yuan H, Xie Z, Yin Y. PPARgamma and PPARdelta as Modulators of Neoplasia and Cell Fate. PPAR Res. 2008;2008:247379. doi: 10.1155/2008/247379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ackler S, Ahmad S, Tobias C, Johnson MD, Glazer RI. Delayed mammary gland involution in MMTV-AKT1 transgenic mice. Oncogene. 2002;21:198–206. doi: 10.1038/sj.onc.1205052. [DOI] [PubMed] [Google Scholar]
- 21.Yin Y, Russell RG, Dettin LE, et al. Peroxisome proliferator-activated receptor delta and gamma agonists differentially alter tumor differentiation and progression during mammary carcinogenesis. Cancer Res. 2005;65:3950–3957. doi: 10.1158/0008-5472.CAN-04-3990. [DOI] [PubMed] [Google Scholar]
- 22.Zeng X, Xu H, Park B-K, Glazer RI. Transformation of mammary epithelial cells by 3-phosphoinositide-dependent protein kinase-1 (PDK1) is associated with the induction of protein kinase Cα. Cancer Res. 2002;62:3538–3543. [PubMed] [Google Scholar]
- 23.Catherino WH, Jordan VC. Increasing the number of tandem estrogen response elements increases the estrogenic activity of a tamoxifen analogue. Cancer Lett. 1995;92:39–47. doi: 10.1016/0304-3835(95)03755-l. [DOI] [PubMed] [Google Scholar]
- 24.Agostini M, Schoenmakers E, Mitchell C, et al. Non-DNA binding, dominant-negative, human PPARgamma mutations cause lipodystrophic insulin resistance. Cell Metab. 2006;4:303–311. doi: 10.1016/j.cmet.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tcherepanova I, Puigserver P, Norris JD, Spiegelman BM, McDonnell DP. Modulation of estrogen receptor-alpha transcriptional activity by the coactivator PGC-1. J Biol Chem. 2000;275:16302–16308. doi: 10.1074/jbc.M001364200. [DOI] [PubMed] [Google Scholar]
- 26.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 27.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 28.Tozlu S, Girault I, Vacher S, et al. Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription-PCR approach. Endocr Relat Cancer. 2006;13:1109–1120. doi: 10.1677/erc.1.01120. [DOI] [PubMed] [Google Scholar]
- 29.Buono KD, Robinson GW, Martin C, et al. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–580. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 30.Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176:19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 32.Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui Y, Miyoshi K, Claudio E, et al. Loss of the peroxisome proliferation-activated receptor gamma (PPARgamma) does not affect mammary development and propensity for tumor formation but leads to reduced fertility. J Biol Chem. 2002;277:17830–17835. doi: 10.1074/jbc.M200186200. [DOI] [PubMed] [Google Scholar]
- 34.Moerman EJ, Teng K, Lipschitz DA, Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell. 2004;3:379–389. doi: 10.1111/j.1474-9728.2004.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takada I, Suzawa M, Matsumoto K, Kato S. Suppression of PPAR transactivation switches cell fate of bone marrow stem cells from adipocytes into osteoblasts. Ann N Y Acad Sci. 2007;1116:182–195. doi: 10.1196/annals.1402.034. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Welm B, Podsypanina K, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci U S A. 2004;101:4158–4163. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotgia F, Williams TM, Cohen AW, Minetti C, Pestell RG, Lisanti MP. Caveolin-1-deficient mice have an increased mammary stem cell population with upregulation of Wnt/beta-catenin signaling. Cell Cycle. 2005;4:1808–1816. doi: 10.4161/cc.4.12.2198. [DOI] [PubMed] [Google Scholar]
- 39.Girnun GD, Smith WM, Drori S, et al. APC-dependent suppression of colon carcinogenesis by PPARgamma. Proc Natl Acad Sci U S A. 2002;99:13771–137716. doi: 10.1073/pnas.162480299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma C, Pradeep A, Wong L, Rana A, Rana B. Peroxisome proliferator-activated receptor gamma activation can regulate beta-catenin levels via a proteasome-mediated and adenomatous polyposis coli-independent pathway. J Biol Chem. 2004;279:35583–35594. doi: 10.1074/jbc.M403143200. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Wang H, Zuo Y, Farmer SR. Functional interaction between peroxisome proliferator-activated receptor gamma and beta-catenin. Mol Cell Biol. 2006;26:5827–5837. doi: 10.1128/MCB.00441-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Z, Zeng X, Waldman T, Glazer RI. Transformation of mammary epithelial cells by 3-phosphoinositide- dependent protein kinase-1 activates beta-catenin and c-Myc, and down-regulates caveolin-1. Cancer Res. 2003;63:5370–5375. [PubMed] [Google Scholar]
- 43.Wang S, Garcia AJ, Wu M, Lawson DA, Witte ON, Wu H. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc Natl Acad Sci U S A. 2006;103:1480–1485. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi K, Murakami M, Yamanaka S. Role of the phosphoinositide 3-kinase pathway in mouse embryonic stem (ES) cells. Biochem Soc Trans. 2005;33:1522–1525. doi: 10.1042/BST0331522. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe S, Umehara H, Murayama K, Okabe M, Kimura T, Nakano T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene. 2006:1–11. doi: 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 46.Amundadottir LT, Leder P. Signal transduction pathways activated and required for mammary carcinogenesis in response to specific oncogenes. Oncogene. 1998;16:737–746. doi: 10.1038/sj.onc.1201829. [DOI] [PubMed] [Google Scholar]
- 47.Denley A, Kang S, Karst U, Vogt PK. Oncogenic signaling of class I PI3K isoforms. Oncogene. 2008;27:2561–2574. doi: 10.1038/sj.onc.1210918. [DOI] [PubMed] [Google Scholar]
- 48.King WG, Mattaliano MD, Chan TO, Tsichlis PN, Brugge JS. Phosphatidylinositol 3-kinase is required for integrin-stimulated AKT and Raf-1/mitogen-activated protein kinase pathway activation. Mol Cell Biol. 1997;17:4406–4418. doi: 10.1128/mcb.17.8.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Rath O, Kolch W, Cho KH. A hidden oncogenic positive feedback loop caused by crosstalk between Wnt and ERK Pathways. Oncogene. 2007 doi: 10.1038/sj.onc.1210230. [DOI] [PubMed] [Google Scholar]
- 50.Moshnikova A, Frye J, Shay JW, Minna JD, Khokhlatchev AV. The growth and tumor suppressor NORE1A is a cytoskeletal protein that suppresses growth by inhibition of the ERK pathway. J Biol Chem. 2006;281:8143–8152. doi: 10.1074/jbc.M511837200. [DOI] [PubMed] [Google Scholar]
- 51.Appel S, Mirakaj V, Bringmann A, Weck MM, Grunebach F, Brossart P. PPAR-gamma agonists inhibit toll-like receptor-mediated activation of dendritic cells via the MAP kinase and NF-kappaB pathways. Blood. 2005;106:3888–3894. doi: 10.1182/blood-2004-12-4709. [DOI] [PubMed] [Google Scholar]
- 52.Adams M, Reginato MJ, Shao D, Lazar MA, Chatterjee VK. Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen- activated protein kinase site. J Biol Chem. 1997;272:5128–5132. doi: 10.1074/jbc.272.8.5128. [DOI] [PubMed] [Google Scholar]
- 53.Hu E, Kim JB, Sarraf P, Spiegelman BM. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science. 1996;274:2100–2103. doi: 10.1126/science.274.5295.2100. [DOI] [PubMed] [Google Scholar]
- 54.Shao D, Rangwala SM, Bailey ST, Krakow SL, Reginato MJ, Lazar MA. Interdomain communication regulating ligand binding by PPAR-gamma. Nature. 1998;396:377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 55.Floyd ZE, Stephens JM. Interferon-gamma-mediated activation and ubiquitin-proteasome-dependent degradation of PPARgamma in adipocytes. J Biol Chem. 2002;277:4062–4068. doi: 10.1074/jbc.M108473200. [DOI] [PubMed] [Google Scholar]
- 56.Qin C, Burghardt R, Smith R, Wormke M, Stewart J, Safe S. Peroxisome proliferator-activated receptor gamma agonists induce proteasome-dependent degradation of cyclin D1 and estrogen receptor alpha in MCF-7 breast cancer cells. Cancer Res. 2003;63:958–964. [PubMed] [Google Scholar]
- 57.Gunin AG, Bitter AD, Demakov AB, Vasilieva EN, Suslonova NV. Effects of peroxisome proliferator activated receptors-alpha and -gamma agonists on estradiol-induced proliferation and hyperplasia formation in the mouse uterus. J Endocrinol. 2004;182:229–239. doi: 10.1677/joe.0.1820229. [DOI] [PubMed] [Google Scholar]
- 58.Keller H, Givel F, Perroud M, Wahli W. Signaling cross-talk between peroxisome proliferator-activated receptor/retinoid X receptor and estrogen receptor through estrogen response elements. Mol Endocrinol. 1995;9:794–804. doi: 10.1210/mend.9.7.7476963. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Gao H, Marstrand TT, et al. The genome landscape of ERalpha- and ERbeta-binding DNA regions. Proc Natl Acad Sci U S A. 2008;105:2604–2609. doi: 10.1073/pnas.0712085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonofiglio D, Gabriele S, Aquila S, et al. Estrogen receptor alpha binds to peroxisome proliferator-activated receptor response element and negatively interferes with peroxisome proliferator-activated receptor gamma signaling in breast cancer cells. Clin Cancer Res. 2005;11:6139–6147. doi: 10.1158/1078-0432.CCR-04-2453. [DOI] [PubMed] [Google Scholar]
- 61.Ferretti E, Arturi F, Mattei T, et al. Expression, regulation, and function of paired-box gene 8 in the human placenta and placental cancer cell lines. Endocrinology. 2005;146:4009–4015. doi: 10.1210/en.2005-0084. [DOI] [PubMed] [Google Scholar]
- 62.Sarfstein R, Maor S, Reizner N, Abramovitch S, Werner H. Transcriptional regulation of the insulin-like growth factor-I receptor gene in breast cancer. Mol Cell Endocrinol. 2006;252:241–246. doi: 10.1016/j.mce.2006.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.