Abstract
Background:
To investigate the association of FADS gene polymorphisms with age-related changes in polyunsaturated fatty acids (PUFAs) in serum phospholipids and oxidative stress markers.
Methods:
We genotyped 122 nonobese men aged 35–59 years without any known diseases at baseline for rs174537 near FADS1 (FEN1 rs174537G > T), FADS2 (rs174575, rs2727270), and FADS3 (rs1000778), and followed them for 3 years.
Results:
Among the four single-nucleotide polymorphisms, the minor variants of rs174537 and rs2727270 were significantly associated with lower concentrations of long-chain PUFAs. However, rs174537G > T showed stronger association. At baseline, men with the rs174537T allele had lower arachidonic acid (AA) and AA/linoleic acid (LA), and higher interleukin (IL)-6 levels than rs174537GG counterparts. After 3 years, rs174537GG men had significantly increased AA (P = 0.022), AA/dihomo-γ-linolenic acid (DGLA) (P = 0.007), docosapentaenoic acid (DPA), low-density lipoprotein (LDL) cholesterol, and oxidized LDL (ox-LDL), but decreased eicosatrienoic acid. The rs174537T group showed significantly increased γ-linolenic acid and ox-LDL, and decreased eicosadienoic acid, eicosapentaenoic acid (EPA)/α-linolenic acid (ALA), and IL-6. After 3 years, the rs174537T group had lower AA (P < 0.001), AA/DGLA (P = 0.019), EPA, DPA, EPA/ALA, and urinary 8-epi-prostaglandin F2α (8-epi-PGF2α) (P = 0.011) than rs174537GG. Changes in AA (P = 0.001), AA/DGLA (P = 0.017), EPA, DPA, EPA/ALA, and urinary 8-epi-PGF2α (P < 0.001) were significantly different between the groups after adjusting for baseline values. Overall, changes in AA positively correlated with changes in urinary 8-epi-PGF2α (r = 0.249, P = 0.007), plasma ox-LDL (r = 0.199, P = 0.045), and serum IL-6 (r = 0.289, P = 0.004).
Conclusion:
Our data show that FADS polymorphisms can affect age-associated changes in serum phospholipid long-chain PUFAs, Δ5-desaturase activity, and oxidative stress in middle-aged nonobese men. In particular, the rs174537T allele did not show the age-associated increases in AA and Δ5-desaturase activity seen with the rs174537GG genotype.
Keywords: FADS gene, age-related changes, serum phospholipid polyunsaturated fatty acids, oxidative stress markers, nonobese men
Introduction
The concentrations and ratios of fat types that people eat have shifted, with marked increases in saturated and ω6 polyunsaturated fatty acids (PUFAs).1 In South Korea, similar profound quantitative and qualitative changes in fat intake have occurred in the last four decades, particularly the rising intake of saturated fatty acids and ω6 PUFAs.2 Dietary PUFAs influence the fatty acid (FA) composition of tissue lipids.3 However, interindividual variability in serum phospholipid FA, as markers of the FA status of an individual, can be attributed to a combination of dietary factors and genetic variation. The key enzymes in PUFA metabolism are Δ5-desaturase and Δ6-desaturase, which are encoded by the FADS1 and FADS2 genes, respectively.4–6 These two genes are located in the desaturase gene cluster on chromosome 11q12–13.1. This cluster also includes FADS3, a gene that shares 52% and 62% sequence identity with the FADS1 and FADS2 genes, respectively, and encodes a desaturase of unknown activity.4 In addition, dietary PUFAs have been shown to suppress the activity of stearoyl-coenzyme A (CoA) desaturase (Δ9-desaturase),7 which is encoded by the stearoyl-CoA desaturase gene family and the rate-limiting enzyme in the cellular synthesis of monounsaturated FA (oleic acid; 18:1 n-9) from saturated FA (stearic acid; 18:0 n-9).8
A genome-wide association study for plasma PUFAs showed strong evidence for association with the region of chromosome 11 that encodes FADS1, FADS2, and FADS3.9 The most significant association was between the single-nucleotide polymorphism (SNP) rs174537 (flap structure-specific endonuclease [FEN1]) near FADS1 and arachidonic acid (AA, 20:4ω6). Recently, Mathias et al1 suggested that variants in the Δ5-desaturase enzymatic step likely regulate the efficiency of conversion of medium-chain PUFAs, such as dietary linoleic acid (LA, 18:ω6), to potentially inflammatory PUFAs, such as AA. In our previous study, we investigated the association of FADS polymorphism, including rs174537, with PUFAs in serum phospholipids and coronary artery disease-related biomarkers in South Koreans through a case-control study. We also determined the effect of these SNPs on lipid peroxides. We found that rs174537T was associated with a lower proportion of AA in serum phospholipids and reduced coronary artery disease risk, in association with reduced total and low-density lipoprotein cholesterol (LDL-C) and lipid peroxides.10 However, there are no reports on the effect of FADS polymorphisms on age-associated changes in serum phospholipid PUFA composition, proinflammatory cytokines, or oxidative stress markers. Therefore, we followed 122 nonobese men aged between 35 and 59 years without a history of known diseases at baseline for 3 years to investigate the association of FADS polymorphisms, including rs174537, with age-associated changes in serum phospholipid PUFA composition. We also examined the effects of these SNPs on lipid peroxides and oxidative stress markers, including oxidized-LDL (ox-LDL) and urinary 8-epi-prostaglandin F2α (8-epi-PGF2α).
Materials and methods
Study population
A total of 160 healthy nonobese (18.5 ≤ body mass index [BMI] < 30 kg/m2) men aged 35–59 years were recruited at a health-promotion center at the National Health Insurance Corporation Ilsan Hospital in South Korea between August and December 2006. Subjects were sedentary, had no history of known disease, and completed a personal health and medical history questionnaire, which served as a screening tool. Exclusion criteria were type 2 diabetes, cardiovascular disease, psychiatric problems, and use of any medication. Written informed consent was obtained from all participants, and the study protocol was approved by the Institutional Review Board of Yonsei University.
Anthropometric parameters and blood collection
Body weight and height were measured in the morning while participants were unclothed and not wearing shoes. BMI was calculated as body weight in kilograms divided by the square of the height in meters (kg/m2). Systolic and diastolic blood pressures (SBP and DBP, respectively) were obtained from the left arm of seated patients with an automatic blood pressure monitor (TM-2654; A&D, Tokyo, Japan) after 20 minutes of rest. After overnight fasting, venous blood samples were collected in ethylenediaminetetraacetic acid-treated or plain tubes, separated into plasma and serum, and then stored at −70°C until analysis.
Genotyping of FADS gene polymorphisms
Genomic DNA was extracted from 5 mL whole blood using a commercially available DNA isolation kit (Wizard genomic DNA purification kit; Promega, Fitchburg, WI, USA) according to the manufacturer’s protocol. Based on previous reports of genetic studies and public databases on the FADS gene cluster9,11 and the HapMap project (http://www.hapmap.org), eight relevant FADS SNPs were prescreened and four SNPs (FEN1 rs174537G > T, FADS2 rs174575C > G, FADS2 rs2727270C > T, FADS3 1000778C > T) were selected for further analysis (Table S1).
Serum lipid profile and fasting glucose
Fasting serum total cholesterol and triglyceride (TG) were measured using a 7150 Autoanalyzer (Hitachi, Tokyo, Japan). After precipitation of serum chylomicrons using dextran sulfate magnesium, HDL-cholesterol concentrations in the supernatants were enzymatically measured. LDL-C was estimated indirectly using the Friedewald formula for subjects with serum TG concentrations <400 mg/dL (4.52 mol/L). In subjects with serum TG concentrations ≥ 4.52 mol/L (400 mg/mL), LDL-C was directly measured by an enzymatic method on the 7150 Autoanalyzer. Fasting glucose was measured by the glucose oxidase method using a glucose analyzer (Beckman Coulter, Brea, CA, USA).
Plasma ox-LDL and serum high-sensitivity C-reactive protein
Plasma ox-LDL was measured using an enzyme immunoassay (Mercodia, Uppsala, Sweden). The resulting color reaction was read at 450 nm on a Wallac Victor2 multilabel counter (PerkinElmer, Waltham, MA, USA). High-sensitivity C-reactive protein (hs-CRP) levels were measured on an Express Plus™ autoanalyzer (Chiron Diagnostics Co., Walpole, MA, USA) using commercially available hs-CRP-Latex (II) X2 kits (Seiken Laboratories Ltd., Tokyo, Japan) that allowed detection of CRP in the range of 0.001–31 mg/dL.
Urinary 8-epi-PGF2α and serum cytokine levels
Urine was collected in polyethylene tubes containing 1% butylated hydroxytoluene after a 12-hour fast. The tubes were immediately covered with aluminum foil and stored at −70°C until analysis. 8-epi-PGF2α was measured using an enzyme immunoassay (Bioxytech Urinary 8-epi-PGF2α Assay Kit, Oxis International, Portland, OR, USA), and the resulting color reaction was read at 650 nm using a Wallac Victor2 multilabel counter. Urinary creatinine was determined by the alkaline picrate (Jaffe) reaction. Urinary 8-epi-PGF2α concentrations were expressed as pg/mg creatinine. Levels of interleukin (IL)-6 and tumor necrosis factor (TNF)-α in serum were measured using the Bio-Plex Reagent Kit on a Bio-Plex instrument (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions.
FA composition in serum phospholipids
Serum phospholipid FA composition was analyzed by gas chromatography (7890A; Hewlett Packard, Palo Alto, CA, USA) using a modification of a previously described method.12,13 Individual FAs were calculated as a relative percentage of the total of 26 FAs (set as 100%) using GC ChemStation software revision B.04.02 (Agilent Technologies, Santa Clara, CA, USA). The activities of Δ5 desaturase and Δ6 desaturase were estimated as the ratios of AA to DGLA and γ-linolenic acid (GLA; 18:3ω6) to LA, respectively.
Assessment of dietary intake and physical activity level
The duration of the study was 3 years. At baseline, usual dietary intake for each subject was assessed using a semi-quantitative food-frequency questionnaire and 24-hour recall method.14 Subjects were encouraged to maintain their body weight within ±2 kg and given general oral and written information about healthy food choices and exercise at baseline and at a subsequent visit (after 3 years). Subjects were instructed by trained dietitians and were also asked to keep food records for 3 days (2 weekdays and 1 weekend day) at each visit. Nutrient intake was determined and calculated as mean values from the 3-day food record using Can-Pro (Korean Nutrition Society, Seoul, South Korea), based on food-composition tables from the National Rural Living Science Institute in South Korea. Total energy expenditure (kcal/d) was calculated from activity patterns, including basal metabolic rate calculated with the Harris–Benedict equation, physical activity for 24 hours, and specific dynamic action of food.
Statistical analysis
Statistical analyses were performed using SPSS version 12.0 for Windows (IBM, Armonk, NY, USA). Hardy–Weinberg equilibrium and linkage disequilibrium tests were examined using Haploview 4.1 (Broad Institute, Cambridge, MA, USA). Frequencies were compared by the Chi-square test. We examined whether each variable was normally distributed before statistical testing, and logarithmic transformation was performed for skewed variables. Paired t-tests were used to test between baseline and follow-up values. Differences in clinical variables between the rs174537GG and T-allele carrier groups were tested by independent t-test, and a general linear model test was applied to adjust for baseline values. Pearson’s correlation coefficients were used to examine the relationships between variables. For descriptive purposes, mean values are presented using untransformed and unadjusted values. Results are expressed as means ± standard error or percentage. A two-tailed value of P < 0.05 was considered statistically significant.
Results
Clinical characteristics, serum phospholipid FA composition, and macronutrient intake at baseline and 3-year follow-up
Among the enrolled men (n = 160), 38 dropped out for personal reasons or poor compliance, leaving 122 men at 3 years. Clinical characteristics, serum phospholipid FA composition, and macronutrient intake at baseline and 3-year follow-up are shown in Table 1. After 3 years, subjects showed an increase in LDL-C (P = 0.018), ox-LDL (P < 0.001), serum phospholipid GLA (P = 0.024), and AA (P = 0.043). HDL-C (P = 0.001), IL-6 (P = 0.004), and serum phospholipid eicosadienoic acid (20:2ω6) (P = 0.016) decreased. There was no significant difference in total energy intake or macronutrient intake between baseline and 3-year follow-up (Table 1).
Table 1.
Clinical characteristics, PUFA composition in serum phospholipids, and macronutrition intake at baseline and at 3 years
| n = 122 | Baseline | 3-year follow-up | P |
|---|---|---|---|
| Age (years) | 46.7 ± 0.58 | 49.7 ± 0.60 | <0.001 |
| Body mass index (kg/m2) | 24.0 ± 0.20 | 24.1 ± 0.20 | 0.501 |
| Systolic blood pressure (mmHg) | 120.3 ± 1.10 | 119.6 ± 1.29 | 0.483 |
| Diastolic blood pressure (mmHg) | 74.5 ± 0.88 | 74.8 ± 1.05 | 0.679 |
| Fasting glucose (mg/dL)§ | 92.1 ± 0.86 | 93.5 ± 0.85 | 0.121 |
| Triglyceride (mg/dL)§ | 131.3 ± 6.96 | 133.4 ± 7.61 | 0.901 |
| Total cholesterol (mg/dL)§ | 191.3 ± 3.10 | 193.3 ± 3.07 | 0.367 |
| LDL cholesterol (mg/dL)§ | 117.0 ± 3.17 | 121.7 ± 2.85 | 0.018 |
| HDL cholesterol (mg/dL)§ | 49.6 ± 1.15 | 46.4 ± 1.08 | 0.001 |
| hs-CRP (mg/dL)§ | 1.64 ± 0.42 | 1.55 ± 0.29 | 0.601 |
| Serum TNF-α (pg/mL)§ | 11.4 ± 1.01 | 10.4 ± 0.88 | 0.856 |
| Serum IL-6 (pg/mL)§ | 4.58 ± 0.33 | 3.63 ± 0.26 | 0.004 |
| Oxidized LDL (U/L)§ | 34.8 ± 1.26 | 44.7 ± 1.15 | <0.001 |
| 8-epi-PGF2α (pg/mg creatinine)§ | 1318.6 ± 46.6 | 1363.7 ± 45.2 | 0.323 |
| PUFA composition (%) in serum PL | |||
| Total polyunsaturated | 19.1 ± 0.38 | 19.1 ± 0.46 | 0.983 |
| ω6 FA | |||
| 18:2 (ω6) | 12.0 ± 0.28 | 11.8 ± 0.32 | 0.621 |
| 18:3 (ω6) | 0.17 ± 0.01 | 0.20 ± 0.01 | 0.024 |
| 20:2 (ω6) | 0.67 ± 0.12 | 0.43 ± 0.08 | 0.016 |
| 20:3 (ω6) | 1.44 ± 0.04 | 1.43 ± 0.05 | 0.977 |
| 20:4 (ω6) | 4.36 ± 0.14 | 4.78 ± 0.19 | 0.043 |
| Total polyunsaturated | 4.66 ± 0.17 | 5.03 ± 0.20 | 0.123 |
| ω3 FA | |||
| 18:3 (ω3) | 0.17 ± 0.02 | 0.19 ± 0.01 | 0.425 |
| 20:3 (ω3) | 0.09 ± 0.01 | 0.07 ± 0.00 | 0.054 |
| 20:5 (ω3) | 1.12 ± 0.05 | 1.26 ± 0.07 | 0.115 |
| 22:5 (ω3) | 0.57 ± 0.03 | 0.58 ± 0.03 | 0.727 |
| 22:6 (ω3) | 2.71 ± 0.12 | 2.93 ± 0.13 | 0.171 |
| Estimates of daily nutrient intakes | |||
| Total energy intake (kcal) | 2440.2 ± 19.1 | 2425.7 ± 15.8 | 0.399 |
| Carbohydrate (% of energy) | 61.5 ± 0.13 | 61.5 ± 0.08 | 0.672 |
| Protein (% of energy) | 16.8 ± 0.12 | 16.5 ± 0.07 | 0.114 |
| Fat (% of energy) | 21.6 ± 0.14 | 21.7 ± 0.11 | 0.794 |
| Total energy expenditure (kcal) | 2361.5 ± 16.7 | 2333.0 ± 13.7 | 0.085 |
Notes:
Tested by logarithmic transformation, P-values derived from paired t-test. Values are means ± standard error.
Abbreviations: PUFA, polyunsaturated fatty acid; LDL, low-density lipoprotein; HDL, high-density lipoprotein; hs-CRP, high sensitivity C-reactive protein; TNF, tumor necrosis factor; IL, interleukin; 8-epi-PGF2α, urinary 8-epi-prostaglandin F2α.
Genotype distribution of four selected SNPs
Genotype distributions in Hardy–Weinberg equilibrium with 41. 0% GG, 47. 5% GT, and 11. 5% TT at position rs174537; 88.5% CC, 9.8% CG, and 1.6% GG at position rs174575; 48.4% CC, 44.3% CT, and 7.4% TT at position rs2727270; and 49.2% CC, 38.5% CT, and 12.3% TT at position rs1000778. The major alleles were G at position rs174537 (frequency 0.648, P = 0.647), C at rs174575 (frequency 0.934, P = 0.087), C at rs2727270 (frequency 0.705, P = 0.480), and C at position rs1000778 (frequency 0.684, P = 0.233). Because the FADS3 rs1000778C > T genotype-related PUFA was not significantly different among serum phospholipids, and the FADS2 rs174575 genotype only showed a trend toward an association with serum phospholipid PUFA (data not shown), we did not perform further analysis on FADS3 rs1000778C > T and FADS2 rs174575C > G. The genotype and haplotype distributions of FEN1 rs174537 and FADS2 rs2727270 were both associated with PUFAs in serum phospholipids. However, because haplotype analysis did not provide information beyond that revealed by each SNP (data not shown), we present only the results of FEN1 rs174537 and FADS2 rs2727270.
Serum phospholipid FA composition according to genotypes
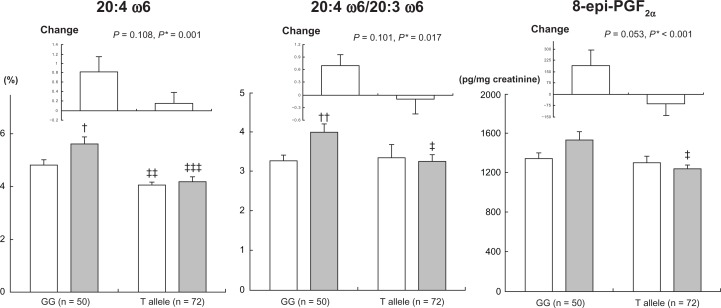
At baseline, men with the FEN1 rs174537T allele showed a lower proportion of AA (P = 0.007) and lower ratio of AA to LA (P = 0.007) in serum phospholipids than those with rs174537GG (Table 2). After 3 years, men with rs174537GG showed a significant increase in AA (P = 0.022), ratio of AA to DGLA (P = 0.007, Figure 1), and docosapentaenoic acid (DPA; 22:5ω3) (P = 0.044), but a significant decrease in eicosatrienoic acid (20:3ω3) (P = 0.037, Table 2). Carriers of the rs174537T allele showed a significant increase in GLA (P = 0.031) and a significant decrease in EDA (P = 0.030), and the ratio of eicosapentaenoic acid (EPA; 20:5ω3) to α-linolenic acid (ALA; 18:3ω3) (P = 0.024). At 3-year follow-up, men with the rs174537T allele showed lower AA (P > 0.001), AA/DGLA (P = 0.019, Figure 1), EPA (P = 0.010), DPA (P = 0.016), and EPA/ALA (P = 0.048) than rs174537GG carriers. Changes in AA (P = 0.001), AA/DGLA (P = 0.017, Figure 1), EPA (P = 0.004), DPA (P = 0.011), and EPA/ALA (P = 0.048) were significantly different between rs174537GG men and rs174537T allele carriers after adjusting for baseline values (Table 2). Similar but weaker associations of FADS2 rs2727270 with PUFAs were also found (Table S2).
Table 2.
Associations of FEN1 rs174537 genotypes with PUFA composition in serum phospholipids in men at baseline and 3-year follow-up
|
FEN1 rs174537
|
||||||
|---|---|---|---|---|---|---|
| GG (n = 50) | Pa | T allele (n = 72) | Pa | Pb | Pc | |
| PUFA composition (%) in serum PL | ||||||
| 18:2 (ω6) | ||||||
| Baseline | 11.9 ± 0.42 | 0.957 | 12.1 ± 0.38 | 0.518 | 0.685 | |
| Follow-up | 11.9 ± 0.52 | 11.8 ± 0.41 | 0.835 | |||
| Change | 0.03 ± 0.52 | −0.34 ± 0.53 | 0.627 | 0.753 | ||
| 18:3 (ω6) | ||||||
| Baseline | 0.18 ± 0.02 | 0.303 | 0.16 ± 0.01 | 0.031 | 0.413 | |
| Follow-up | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.584 | |||
| Change | 0.02 ± 0.02 | 0.03 ± 0.01 | 0.659 | 0.652 | ||
| 20:2 (ω6) | ||||||
| Baseline | 0.61 ± 0.19 | 0.158 | 0.72 ± 0.16 | 0.030 | 0.665 | |
| Follow-up | 0.33 ± 0.03 | 0.51 ± 0.14 | 0.287 | |||
| Change | −0.28 ± 0.19 | −0.21 ± 0.09 | 0.733 | 0.313 | ||
| 20:3 (ω6) | ||||||
| Baseline | 1.53 ± 0.07 | 0.926 | 1.37 ± 0.06 | 0.903 | 0.081 | |
| Follow-up | 1.53 ± 0.09 | 1.36 ± 0.06 | 0.101 | |||
| Change | 0.01 ± 0.10 | −0.01 ± 0.08 | 0.880 | 0.184 | ||
| 20:4 (ω6) | ||||||
| Baseline | 4.79 ± 0.23 | 0.022 | 4.04 ± 0.16 | 0.587 | 0.007 | |
| Follow-up | 5.61 ± 0.30 | 4.17 ± 0.21 | <0.001 | |||
| Change | 0.81 ± 0.34 | 0.14 ± 0.25 | 0.108 | 0.001 | ||
| 18:3 (ω3) | ||||||
| Baseline | 0.17 ± 0.03 | 0.742 | 0.17 ± 0.03 | 0.463 | 0.998 | |
| Follow-up | 0.19 ± 0.01 | 0.20 ± 0.02 | 0.589 | |||
| Change | 0.01 ± 0.03 | 0.02 ± 0.03 | 0.760 | 0.591 | ||
| 20:3 (ω3) | ||||||
| Baseline | 0.10 ± 0.02 | 0.037 | 0.08 ± 0.01 | 0.613 | 0.153 | |
| Follow-up | 0.07 ± 0.00 | 0.07 ± 0.00 | 0.392 | |||
| Change | −0.04 ± 0.02 | −0.01 ± 0.01 | 0.108 | 0.408 | ||
| 20:5 (ω3) | ||||||
| Baseline | 1.19 ± 0.08 | 0.090 | 1.07 ± 0.06 | 0.787 | 0.231 | |
| Follow-up | 1.49 ± 0.14 | 1.09 ± 0.07 | 0.010 | |||
| Change | 0.30 ± 0.18 | 0.02 ± 0.08 | 0.153 | 0.004 | ||
| 22:5 (ω3) | ||||||
| Baseline | 0.54 ± 0.04 | 0.044 | 0.59 ± 0.05 | 0.213 | 0.441 | |
| Follow-up | 0.66 ± 0.04 | 0.53 ± 0.03 | 0.016 | |||
| Change | 0.12 ± 0.06 | −0.06 ± 0.05 | 0.019 | 0.011 | ||
| 22:6 (ω3) | ||||||
| Baseline | 2.81 ± 0.20 | 0.134 | 2.64 ± 0.15 | 0.649 | 0.495 | |
| Follow-up | 3.20 ± 0.20 | 2.73 ± 0.16 | 0.071 | |||
| Change | 0.39 ± 0.26 | 0.09 ± 0.20 | 0.354 | 0.088 | ||
| 20:4 (ω6)/20:3 (ω6) | ||||||
| Baseline | 3.26 ± 0.15 | 0.007 | 3.33 ± 0.35 | 0.808 | 0.882 | |
| Follow-up | 3.96 ± 0.25 | 3.24 ± 0.19 | 0.019 | |||
| Change | 0.70 ± 0.25 | −0.09 ± 0.36 | 0.101 | 0.017 | ||
| 20:4 (ω6)/18:2 (ω6) | ||||||
| Baseline | 0.43 ± 0.03 | 0.202 | 0.34 ± 0.01 | 0.237 | 0.007 | |
| Follow-up | 0.53 ± 0.06 | 0.41 ± 0.05 | 0.146 | |||
| Change | 0.10 ± 0.07 | 0.06 ± 0.05 | 0.719 | 0.185 | ||
| 20:5 (ω3)/18:3 (ω3) | ||||||
| Baseline | 9.55 ± 0.88 | 0.328 | 9.06 ± 0.87 | 0.024 | 0.694 | |
| Follow-up | 8.42 ± 0.71 | 6.69 ± 0.53 | 0.048 | |||
| Change | −1.13 ± 1.14 | −2.37 ± 1.02 | 0.426 | 0.048 | ||
Notes:
Values derived from paired t-test;
values derived from independent t-test;
values derived after adjusting for baseline values. Values are means ± standard error.
Abbreviations: PUFA, polyunsaturated fatty acid; PL, phospholipid.
Figure 1.
Serum phospholipid AA (20:4 ω6), the ratio of 20:4 ω6/20:3 ω6, and urinary levels of 8-epi-PGF2α according to FEN1 rs174537 G > T in men at baseline (□) and 3-year follow-up (▪).
Notes:†P < 0.05; ††P < 0.01 for baseline vs 3 years, tested by paired t-test; ‡P < 0.05; ‡‡P < 0.01; and ‡‡‡P < 0.001 for GG vs T-allele group, tested by independent t-test; *after adjusting for baseline values. Changes are differences between baseline and 3 years; values are means ± standard error.
Abbreviation: 8-epi-PGF2α, urinary 8-epi-prostaglandin F2α.
LDL-cholesterol, hs-CRP, cytokines, and oxidative stress markers according to genotypes
At baseline, men with the FEN1 rs174537T allele showed higher serum IL-6 than those with rs174537GG (P = 0.038, Table 3). After 3 years, men with rs174537GG showed a significant increase in serum LDL-C (P = 0.043) and plasma ox-LDL (P < 0.001). Carriers of the rs174537T allele showed a significant decrease in IL-6 (P = 0.001) and a significant increase in ox-LDL (P < 0.001). At 3-year follow-up, men with the rs174537T allele showed lower urinary PGF2α levels (P = 0.011) than rs174537GG carriers. Changes in urinary PGF2α levels (P < 0.001) were significantly different between rs174537GG and rs174537T allele carriers after adjusting for baseline values (Figure 1). Additionally, changes in serum TNF-α (P = 0.089) tended to be different (Table 3). Similar associations were found between FADS2 rs2727270 and LDL-C, hs-CRP, cytokines, and oxidative stress markers (Table S3).
Table 3.
Associations of FEN1 rs174537 genotypes with LDL-cholesterol, hs-CRP, cytokines, and oxidative stress markers in men at baseline and 3-year follow-up
|
FEN1rs174537
|
||||||
|---|---|---|---|---|---|---|
| GG (n = 50) | Pa | T allele (n = 72) | Pa | Pb | Pc | |
| LDL cholesterol (mg/dL) | ||||||
| Baseline§ | 117.8 ± 4.61 | 0.043 | 116.5 ± 4.31 | 0.145 | 0.830 | |
| Follow-up§ | 123.3 ± 3.92 | 120.7 ± 3.99 | 0.498 | |||
| Change | 5.50 ± 3.11 | 4.21 ± 3.25 | 0.785 | 0.677 | ||
| hs-CRP (mg/dL) | ||||||
| Baseline§ | 1.38 ± 0.35 | 0.828 | 1.82 ± 0.67 | 0.424 | 0.751 | |
| Follow-up§ | 1.46 ± 0.35 | 1.62 ± 0.43 | 0.603 | |||
| Change | 0.08 ± 0.18 | −0.20 ± 0.79 | 0.769 | 0.623 | ||
| Serum IL-6 (pg/mL) | ||||||
| Baseline§ | 3.83 ± 0.42 | 0.557 | 5.07 ± 0.47 | 0.001 | 0.038 | |
| Follow-up§ | 3.90 ± 0.46 | 3.44 ± 0.30 | 0.702 | |||
| Change | 0.07 ± 0.49 | −1.63 ± 0.54 | 0.030 | 0.307 | ||
| Serum TNF-α (pg/mL) | ||||||
| Baseline§ | 11.7 ± 1.42 | 0.943 | 11.2 ± 1.40 | 0.863 | 0.467 | |
| Follow-up§ | 12.0 ± 2.02 | 9.26 ± 0.55 | 0.376 | |||
| Change | 0.33 ± 2.27 | −1.95 ± 1.40 | 0.369 | 0.089 | ||
| 8-epi-PGF2α (pg/mg creatinine) | ||||||
| Baseline§ | 1342.7 ± 60.4 | 0.156 | 1301.0 ± 67.9 | 0.995 | 0.349 | |
| Follow-up§ | 1531.8 ± 86.7 | 1240.1 ± 40.1 | 0.011 | |||
| Change | 189.2 ± 104.5 | −60.9 ± 78.2 | 0.053 | <0.001 | ||
| Oxidized LDL (U/L) | ||||||
| Baseline§ | 33.1 ± 1.77 | <0.001 | 36.1 ± 1.76 | <0.001 | 0.292 | |
| Follow-up§ | 45.2 ± 1.53 | 44.3 ± 1.64 | 0.373 | |||
| Change | 12.2 ± 1.83 | 8.19 ± 1.70 | 0.119 | 0.241 | ||
Notes:
Tested by logarithmic transformation;
values derived from paired t-test;
values derived from independent t-test;
values derived after adjusting for baseline value. Values are means ± standard error.
Abbreviations: PUFA, polyunsaturated fatty acid; LDL, low-density lipoprotein; hs-CRP, high-sensitivity C-reactive protein; TNF, tumor necrosis factor; IL, interleukin; 8-epi-PGF2α, urinary 8-epi-prostaglandin F2α.
Relation of serum phospholipid AA with oxidative stress markers and cytokines
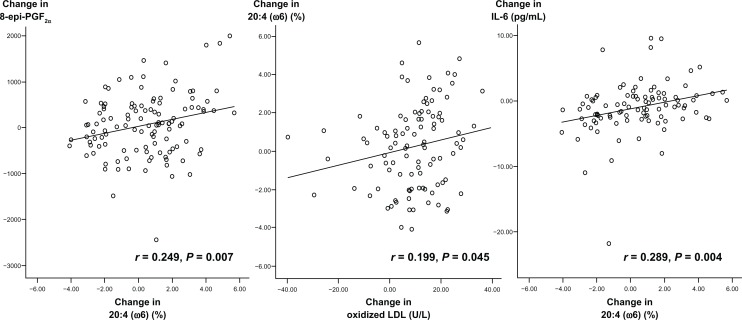
Pearson correlation analysis showed that the changes in the AA proportion in serum phospholipids were positively correlated with changes in urinary 8-epi-PGF2α levels (r = 0.249, P = 0.007), plasma ox-LDL (r = 0.199, P = 0.045), and serum IL-6 (r = 0.289, P = 0.004) in all subjects (Figure 2). There was a marginal correlation between changes in serum phospholipid AA and changes in serum TNF-α (r = 0.191, P = 0.058). Additionally, changes in urinary 8-epi-PGF2α levels were positively correlated with changes in serum IL-6 (r = 0.242, P = 0.015) and TNF-α (r = 0.246, P = 0.013). Changes in serum IL-6 were positively correlated with changes in serum TNF-α (r = 0.464, P < 0.001) and hs-CRP (r = 0.507, P < 0.001, Table 4).
Figure 2.
Relation of changes in serum phospholipid arachidonic acid with changes in oxidized LDL, urinary 8-epi-PGF2α and serum IL-6 in all male subjects.
Note: Changes are differences between baseline and 3 years.
Abbreviations: 8-epi-PGF2α, urinary 8-epi-prostaglandin F2α; LDL, low-density lipoprotein; IL, interleukin.
Table 4.
Correlations between 8-epi-PGF2α and cytokine
|
8-epi-PGF2α
|
IL-6
|
|||
|---|---|---|---|---|
| r | P | r | P | |
| IL-6 | 0.242 | 0.015 | – | – |
| TNF-α | 0.246 | 0.013 | 0.464 | <0.001 |
| hs-CRP | 0.085 | 0.364 | 0.507 | <0.001 |
Abbreviations: 8-epi-PGF2α, urinary 8-epi-prostaglandin F2α ; IL, interleukin; TNF, tumor necrosis factor; hsCRP, high-sensitivity C-reactive protein C-reactive protein.
Discussion
The major finding of this study is that FADS polymorphisms may affect age-associated changes in serum phospholipid long-chain PUFAs, Δ5-desaturase activity, and oxidative stress in middle-aged nonobese men. At 3-year follow-up, there were significant differences between men with FEN1 rs174537GG and those with the 174537T allele in changes in AA, and Δ5-desaturase activity (determined by the ratio of AA/DGLA), as well as urinary levels of PGF2α, one of the radical peroxides of AA, and an indicator of oxidative stress.15 In particular, the rs174537T allele did not show the age-associated increases in AA and Δ5-desaturase activity seen with the rs174537GG genotype. Therefore, our data show that FADS polymorphisms could affect age-associated changes in serum phospholipid long-chain PUFAs, Δ5-desaturase activity, and oxidative stress in middle-aged nonobese men.
Similar to previous studies,1,16 the minor variants of rs174537G > T and rs2727270 were significantly associated with lower concentrations of long-chain PUFAs. However, rs174537G > T showed stronger association with changes in AA, Δ5-desaturase activity, EPA, DPA, and EPA/ALA than rs2727270. Although numerous SNPs in the FADS gene cluster were reported to be significantly associated with FA alterations in tissues, such as serum and red blood cell membranes,1,9,11,17,18 a recent genome-wide association study9 found the most significant association to that of rs174537 with AA. However, rs174537 is located in an intron and in linkage disequilibrium with rs174546 (r2 = 0.99) and rs3834458 (r2 = 0.98), which are candidates for a direct influence on gene expression.11,19 Therefore, it is possible that this variant is a marker of other functional polymorphisms or is in linkage with currently unidentified causal variants affecting FA concentrations.
AA, a precursor of eicosanoids including prostaglandins and leukotrienes, is liberated from the hydrolysis of the sn-2 position of glycerophospholipids (phosphatidylcholine).20 Radical peroxidation of AA produces a family of prostaglandin F2-isomers called F2-isoprostanes.21 One such F2-isoprostane, 8-epi-PGF2α, is a sensitive and independent risk marker for coronary artery disease.15,22,23 It is probably released into biological fluids through a phospholipase-mediated pathway and consequently excreted in urine. In this study, the changes in the AA proportion in serum phospholipids were positively correlated with changes in urinary 8-epi-PGF2α levels as well as changes in serum IL-6. Interestingly, changes in urinary 8-epi-PGF2α also correlate with changes in IL-6 and TNF-α. This result is consistent with previous findings of a positive association between urinary 8-epi-PGF2α and circulating proinflammatory cytokines.24
High concentrations of AA may influence the levels of proinflammatory eicosanoids, which in turn appear to be associated with elevated markers of low-level systemic inflammation.25–27 Thus, low synthesis and availability of AA have been suggested to mitigate the inflammatory response by altering, for example, eicosanoid levels.28 At 3-year follow-up, men with the rs174537T allele showed lower AA, Δ5-desaturase activity, and urinary PGF2α levels compared to those with rs174537GG. In addition, men with the rs174537T allele also showed significant reduction in IL-6 at 3-year follow-up compared to baseline, and changes in serum TNF-α tended to be different between rs174537GG and rs174537T allele carriers.
Tekola Ayele et al conducted a genome-wide association study of IL-10, IL-1Ra, and IL-6 level in nondiabetic Africans. They reported that IL-6 levels showed genome-wide significant association with one SNP (RP11-314E23.1; chr6:133397598; P = 8.63 × 10−9), but did not confirm an association of IL-6 with FADS genotypes.29 In addition, Naitza et al investigated a genome-wide association scan on the levels of markers of inflammation including IL-6; however, they did not find an association of IL-6 with FADS genotypes either.30 In our result, at baseline, men with the FEN1 rs174537T allele showed higher serum IL-6 than those with rs174537GG (P = 0.038, Table 3). After adjusting for baseline, changes in IL-6 levels were not significantly different between rs174537GG and rs174537T allele carriers. Thus, we may need to consider further study with an increased number of study subjects in the future to confirm and clarify the result pattern.
Age is known to play an important role in increased LDL oxidation.31 After 3 years, LDL-C increased in men with rs174537GG, and ox-LDL, one of the products of oxidative stress, increased regardless of genotype. Although the change in ox-LDL in subjects with the rs174537T allele was 33% lower than that of men with GG, it failed to reach statistical significance. However, the present findings of direct correlation between serum phospholipid AA and both plasma ox-LDL and urinary 8-epi-PGF2α could suggest a possible role of AA in lipid peroxidation or oxidative stress during aging. More than 95% of the serum phospholipids are phosphatidylcholine, one of the major phospholipids in membranes. Thus, serum phospholipids mirror membrane composition in the body, as markers of the FA status of an individual.32
Several limitations of this study should be mentioned. First, the small sample size was not conducive to identification of weak associations due to low statistical power. Second, PUFA levels were expressed as a percentage of total FAs in serum phospholipids, not as an absolute concentration. Therefore, we were able to detect relative differences in PUFA levels and Δ5-desaturase activity, but unable to decipher the mechanisms, which depend on the absolute values. Third, dietary intake of PUFAs was not investigated in this study, but we confirmed dietary intake of total fat percentage, which did not change for the follow-up period. Finally, we specifically focused on a representative group of South Korean nonobese (18.5 ≤ BMI < 30 kg/m2) men aged 35–59 years. Our subjects were not taking any medications or vitamin/mineral supplementations. Therefore, our data cannot be generalized to other ethnic groups or other populations.
Conclusion
Our data show that FADS polymorphisms could affect age-associated changes in serum phospholipid long-chain PUFAs, Δ5-desaturase activity, and oxidative stress in middle-aged nonobese men. In particular, the rs174537T allele did not show age-associated increase in AA and Δ5-desaturase activity seen in the rs174537GG genotype. Because it has been suggested that appropriate dietary intake of FAs can obviously overcome genetic risk factors,33 these results provide good evidence for tailoring dietary intervention programs to individuals based on their genetic patterns.
Acknowledgments
This work was supported by the National Research Foundation, Ministry of Education, Science and Technology (Mid-career Researcher Program: 2012-0005604, M10642120002-06 N4212-00210, C00048, and 2012M3 A9C4048762), South Korea.
Footnotes
Disclosure
None of the authors had any personal or financial conflicts of interest.
Supplementary tables
Table S1.
Information for the prescreening of eight single-nucleotide polymorphisms (SNPs) and selection of four SNPs
| FEN1 10154G >T | FADS2 rs174575 | FADS3 rs1000778 | FADS2 rs2727270 | FADS2 rs174576 | FADS2 rs174570 | FADS2 rs174583 | FADS3 rs174456 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | |
| FEN1 −10154G > T | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| FADS2 rs174575 | 0.96 | 0.16 | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| FADS3 rs1000778 | 0.21 | 0.04 | 0.46 | 0.04 | – | – | – | – | – | – | – | – | – | – | – | – |
| FADS2 rs2727270 | 0.98 | 0.71 | 1.00 | 0.03 | 0.13 | 0.01 | – | – | – | – | – | – | – | – | – | – |
| FADS2 rs174576 | 1.00 | 0.98 | 1.00 | 0.17 | 0.22 | 0.04 | 1.00 | 0.72 | – | – | – | – | – | – | – | – |
| FADS2 rs174570 | 1.00 | 0.98 | 1.00 | 0.17 | 0.21 | 0.04 | 0.99 | 0.71 | 1.00 | 0.99 | – | – | – | – | – | – |
| FADS2 rs174583 | 0.99 | 0.95 | 0.96 | 0.16 | 0.22 | 0.04 | 1.00 | 0.71 | 0.99 | 0.98 | 0.99 | 0.97 | – | – | – | – |
| FADS3 rs174456 | 0.21 | 0.04 | 0.46 | 0.04 | 1.00 | 1.00 | 0.01 | 0.01 | 0.22 | 0.04 | 0.21 | 0.04 | 0.22 | 0.04 | – | – |
Table S2.
Associations of FADS2 rs 2727270 genotypes with polyunsaturated fatty acid (PUFA) composition in serum phospholipids in men at baseline and 3-year follow-up
|
FADS2 rs 2727270C>T
|
||||||
|---|---|---|---|---|---|---|
| CC (n = 59) | Pa | T allele (n = 63) | Pa | Pb | Pc | |
| PUFA composition (%) in serum PL | ||||||
| 18:2 (ω6) | ||||||
| Baseline | 12.1 ± 0.38 | 0.971 | 12.0 ± 0.42 | 0.511 | 0.880 | |
| Follow-up | 12.1 ± 0.47 | 11.6 ± 0.45 | 0.455 | |||
| Change | 0.02 ± 0.48 | −0.38 ± 0.58 | 0.594 | 0.466 | ||
| 18:3 (ω6) | ||||||
| Baseline | 0.18 ± 0.02 | 0.215 | 0.16 ± 0.01 | 0.051 | 0.452 | |
| Follow-up | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.693 | |||
| Change | 0.02 ± 0.02 | 0.03 ± 0.02 | 0.666 | 0.756 | ||
| 20:2 (ω6) | ||||||
| Baseline | 0.59 ± 0.17 | 0.172 | 0.75 ± 0.18 | 0.024 | 0.508 | |
| Follow-up | 0.36 ± 0.03 | 0.51 ± 0.16 | 0.357 | |||
| Change | −0.23 ± 0.17 | −0.24 ± 0.11 | 0.946 | 0.516 | ||
| 20:3 (ω6) | ||||||
| Baseline | 1.51 ± 0.06 | 0.974 | 1.36 ± 0.06 | 0.943 | 0.083 | |
| Follow-up | 1.52 ± 0.08 | 1.36 ± 0.07 | 0.123 | |||
| Change | 0.00 ± 0.09 | −0.01 ± 0.09 | 0.941 | 0.219 | ||
| 20:4 (ω6) | ||||||
| Baseline | 4.73 ± 0.20 | 0.027 | 3.99 ± 0.18 | 0.559 | 0.008 | |
| Follow-up | 5.42 ± 0.27 | 4.16 ± 0.23 | 0.001 | |||
| Change | 0.69 ± 0.30 | 0.16 ± 0.28 | 0.206 | 0.003 | ||
| 18:3 (ω3) | ||||||
| Baseline | 0.17 ± 0.02 | 0.319 | 0.18 ± 0.03 | 0.945 | 0.912 | |
| Follow-up | 0.21 ± 0.02 | 0.18 ± 0.01 | 0.267 | |||
| Change | 0.04 ± 0.03 | 0.00 ± 0.03 | 0.474 | 0.268 | ||
| 20:3 (ω3) | ||||||
| Baseline | 0.10 ± 0.02 | 0.029 | 0.08 ± 0.01 | 0.702 | 0.276 | |
| Follow-up | 0.06 ± 0.00 | 0.07 ± 0.00 | 0.123 | |||
| Change | −0.04 ± 0.02 | −0.01 ± 0.01 | 0.138 | 0.128 | ||
| 20:5 (ω3) | ||||||
| Baseline | 1.18 ± 0.07 | 0.134 | 1.06 ± 0.07 | 0.585 | 0.240 | |
| Follow-up | 1.41 ± 0.12 | 1.11 ± 0.07 | 0.033 | |||
| Change | 0.24 ± 0.16 | 0.05 ± 0.09 | 0.293 | 0.027 | ||
| 22:5 (ω3) | ||||||
| Baseline | 0.54 ± 0.03 | 0.088 | 0.60 ± 0.05 | 0.250 | 0.333 | |
| Follow-up | 0.63 ± 0.04 | 0.54 ± 0.04 | 0.085 | |||
| Change | 0.09 ± 0.05 | −0.06 ± 0.05 | 0.043 | 0.058 | ||
| 22:6 (ω3) | ||||||
| Baseline | 2.74 ± 0.18 | 0.130 | 2.69 ± 0.16 | 0.694 | 0.819 | |
| Follow-up | 3.10 ± 0.18 | 2.77 ± 0.18 | 0.205 | |||
| Change | 0.36 ± 0.23 | 0.09 ± 0.22 | 0.399 | 0.215 | ||
| 20:4(ω6)/20:3(ω6) | ||||||
| Baseline | 3.24 ± 0.13 | 0.008 | 3.36 ± 0.39 | 0.798 | 0.785 | |
| Follow-up | 3.84 ± 0.22 | 3.25 ± 0.21 | 0.052 | |||
| Change | 0.60 ± 0.22 | −0.11 ± 0.41 | 0.134 | 0.043 | ||
| 20:4(ω6)/18:2(ω6) | ||||||
| Baseline | 0.42 ± 0.03 | 0.206 | 0.34 ± 0.02 | 0.232 | 0.024 | |
| Follow-up | 0.50 ± 0.06 | 0.42 ± 0.06 | 0.314 | |||
| Change | 0.08 ± 0.06 | 0.07 ± 0.06 | 0.921 | 0.377 | ||
| 20:5(ω3)/18:3(ω3) | ||||||
| Baseline | 9.36 ± 0.79 | 0.148 | 9.18 ± 0.96 | 0.060 | 0.883 | |
| Follow-up | 7.88 ± 0.64 | 6.99 ± 0.58 | 0.306 | |||
| Change | −1.49 ± 1.01 | −2.19 ± 1.14 | 0.646 | 0.307 | ||
Notes:
Values derived from paired t-test;
values derived from independent t-test;
values derived after adjusting for baseline values. Values are means ± standard error.
Abbreviations: PUFA, polyunsaturated fatty acid; PL, phospholipids
Table S3.
Associations of FADS2 rs2727270 genotypes with LDL cholesterol, hs-CRP, cytokines, and oxidative stress markers in men at baseline and 3-year follow-up
|
FADS2rs2727270C>T
|
||||||
|---|---|---|---|---|---|---|
| CC (n = 59) | Pa | T allele (n ==63) | Pa | Pb | Pc | |
| LDL cholesterol (mg/dL) | ||||||
| Baseline§ | 119.3 ± 4.14 | 0.035 | 114.9 ± 4.76 | 0.184 | 0.447 | |
| Follow-up§ | 124.5 ± 3.46 | 119.2 ± 4.47 | 0.210 | |||
| Change | 5.22 ± 2.98 | 4.26 ± 3.51 | 0.837 | 0.533 | ||
| hs-CRP (mg/dL) | ||||||
| Baseline§ | 1.34 ± 0.30 | 0.972 | 1.92 ± 0.77 | 0.534 | 0.606 | |
| Follow-up§ | 1.45 ± 0.30 | 1.65 ± 0.49 | 0.947 | |||
| Change | 0.11 ± 0.15 | −0.27 ± 0.90 | 0.691 | 0.477 | ||
| Serum IL-6 (pg/mL) | ||||||
| Baseline§ | 4.27 ± 0.40 | 0.199 | 4.87 ± 0.52 | 0.006 | 0.407 | |
| Follow-up§ | 3.87 ± 0.39 | 3.39 ± 0.34 | 0.474 | |||
| Change | −0.41 ± 0.46 | −1.48 ± 0.61 | 0.168 | 0.268 | ||
| Serum TNF-α (pg/mL) | ||||||
| Baseline§ | 13.4 ± 1.73 | 0.765 | 9.53 ± 1.02 | 0.603 | 0.067 | |
| Follow-up§ | 11.9 ± 1.68 | 8.87 ± 0.57 | 0.122 | |||
| Change | −1.42 ± 2.24 | −0.66 ± 1.16 | 0.761 | 0.303 | ||
| 8-epi-PGF2α (pg/mg creatinine) | ||||||
| Baseline§ | 1313.0 ± 55.5 | 0.123 | 1324.3 ± 75.4 | 0.809 | 0.736 | |
| Follow-up§ | 1484.1 ± 76.2 | 1243.3 ± 43.9 | 0.022 | |||
| Change | 171.1 ± 93.2 | −80.9 ± 85.1 | 0.048 | 0.003 | ||
| Oxidized LDL (U/L) | ||||||
| Baseline§ | 34.0 ± 1.61 | <0.001 | 35.7 ± 1.96 | <0.001 | 0.663 | |
| Follow-up§ | 45.9 ± 1.49 | 43.4 ± 1.74 | 0.147 | |||
| Change | 11.9 ± 1.65 | 7.76 ± 1.87 | 0.097 | 0.092 | ||
Notes:
Tested by logarithmic transformation;
values derived from paired t-test;
values derived from independent t-test;
values derived after adjusting for baseline values. Values are means ± standard error.
Abbreviations: LDL, low-density lipoprotein; hs-CRP, high-sensitivity C-reactive protein; TNF, tumor necrosis factor; IL, interleukin; 8-epi-PGF2α, urinary 8-epi-prostaglandin F2α.
References
- 1.Mathias RA, Vergara C, Gao L, et al. FADS genetic variants and omega-6 polyunsaturated fatty acid metabolism in a homogeneous island population. J Lipid Res. 2010;51:2766–2774. doi: 10.1194/jlr.M008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ailhaud G, Guesnet P, Cunnane SC. An emerging risk factor for obesity: does disequilibrium of polyunsaturated fatty acid metabolism contribute to excessive adipose tissue development? Br J Nutr. 2008;100:461–470. doi: 10.1017/S0007114508911569. [DOI] [PubMed] [Google Scholar]
- 3.Karlsson M, Mårild S, Brandberg J, Lönn L, Friberg P, Strandvik B. Serum phospholipid fatty acids, adipose tissue, and metabolic markers in obese adolescents. Obesity. 2006;14:1931–1939. doi: 10.1038/oby.2006.225. [DOI] [PubMed] [Google Scholar]
- 4.Marquardt A, Stöhr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 5.Cho HP, Nakamura MT, Clarke SD. Cloning, expression, and nutritional regulation of the mammalian delta-6 desaturase. J Biol Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 6.Cho HP, Nakamura M, Clarke SD. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J Biol Chem. 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- 7.Ntambi JM. The regulation of stearoyl-CoA desaturase (SCD) Prog Lipid Res. 1995;34:139–150. doi: 10.1016/0163-7827(94)00010-j. [DOI] [PubMed] [Google Scholar]
- 8.Ntambi JM. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J Lipid Res. 1999;40:1549–1558. [PubMed] [Google Scholar]
- 9.Tanaka T, Shen J, Abecasis GR, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5:e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak JH, Paik JK, Kim OY, et al. FADS gene polymorphisms in Koreans: association with ω6 polyunsaturated fatty acids in serum phospholipids, lipid peroxides, and coronary artery disease. Atherosclerosis. 2011;214:94–100. doi: 10.1016/j.atherosclerosis.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Schaeffer L, Gohlke H, Müller M, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 12.Folch J, Lees M, Sloane, Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 13.Lepage G, Roy CC. Direct transesterification of all classes of lipids in a one-step reaction. J Lipid Res. 1986;27:114–120. [PubMed] [Google Scholar]
- 14.Shim JS, Oh KW, Suh I, et al. A study on validity of a 299 semiquantitative food frequency questionnaire of Korean adults. Korean J Community Nutr. 2002;7:484–494. [Google Scholar]
- 15.Schwedhelm E, Bartling A, Lenzen H, et al. Urinary 8-iso prostaglandin F2alpha as a risk marker in patients with coronary heart disease: a matched case-control study. Circulation. 2004;109:843–848. doi: 10.1161/01.CIR.0000116761.93647.30. [DOI] [PubMed] [Google Scholar]
- 16.Merino DM, Johnston H, Clarke S, et al. Polymorphisms in FADS1 and FADS2 alter desaturase activity in young Caucasian and Asian adults. Mol Genet Metab. 2011;103:171–178. doi: 10.1016/j.ymgme.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Malerba G, Schaeffer L, Xumerle L, et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43:289–299. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- 18.Bokor S, Dumont J, Spinneker A, et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010;51:2325–2333. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 20.Stafforini DM, Sheller JR, Blackwell TS, et al. Release of free F2-isoprostanes from esterified phospholipids is catalyzed by intracellular and plasma platelet-activating factor acetylhydrolases. J Biol Chem. 2006;281:4616–4623. doi: 10.1074/jbc.M507340200. [DOI] [PubMed] [Google Scholar]
- 21.Voss P, Siems W. Clinical oxidation parameters of aging. Free Radic Res. 2006;40:1339–1349. doi: 10.1080/10715760600953859. [DOI] [PubMed] [Google Scholar]
- 22.Wolfram R, Oguogho A, Palumbo B, Sinzinger H. Enhanced oxidative stress in coronary heart disease and chronic heart failure as indicated by an increased 8-epi-PGF(2alpha) Eur J Heart Fail. 2005;7:167–172. doi: 10.1016/j.ejheart.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Kim JY, Hyun YJ, Jang Y, et al. Lipoprotein-associated phospholipase A2 activity is associated with coronary artery disease and markers of oxidative stress: a case-control study. Am J Clin Nutr. 2008;88:630–637. doi: 10.1093/ajcn/88.3.630. [DOI] [PubMed] [Google Scholar]
- 24.Kim OY, Chae JS, Paik JK, et al. Effects of aging and menopause on serum interleukin-6 levels and peripheral blood mononuclear cell cytokine production in healthy nonobese women. Age (Dordr) 2012;34:415–425. doi: 10.1007/s11357-011-9244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cesari M, Penninx BW, Newman AB, et al. Inflammatory markers and onset of cardiovascular events: results from the Health ABC study. Circulation. 2003;108:2317–2322. doi: 10.1161/01.CIR.0000097109.90783.FC. [DOI] [PubMed] [Google Scholar]
- 26.Chilton FH, Rudel LL, Parks JS, Arm JP, Seeds MC. Mechanisms by which botanical lipids affect inflammatory disorders. Am J Clin Nutr. 2008;87:498S–503S. doi: 10.1093/ajcn/87.2.498S. [DOI] [PubMed] [Google Scholar]
- 27.Poudel-Tandukar K, Nanri A, Matsushita Y, et al. Dietary intakes of alpha-linolenic and linoleic acids are inversely associated with serum C-reactive protein levels among Japanese men. Nutr Res. 2009;29:363–370. doi: 10.1016/j.nutres.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Vessby B, Uusitupa M, Hermansen K, et al. Substituting dietary saturated for monounsaturated fat impairs insulin sensitivity in healthy men and women: the KANWU Study. Diabetologia. 2001;44:312–319. doi: 10.1007/s001250051620. [DOI] [PubMed] [Google Scholar]
- 29.Tekola Ayele F, Doumatey A, Huang H, et al. Genome-wide associated loci influencing interleukin (IL)-10, IL-1Ra, and IL-6 levels in African Americans. Immunogenetics. 2012;64:351–359. doi: 10.1007/s00251-011-0596-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naitza S, Porcu E, Steri M, et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012;8:e1002480. doi: 10.1371/journal.pgen.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holvoet P, Vanhaecke J, Janssens S, Van de Werf F, Collen D. Oxidized LDL and malondialdehyde-modified LDL in patients with acute coronary syndromes and stable coronary artery disease. Circulation. 1998;98:1487–1494. doi: 10.1161/01.cir.98.15.1487. [DOI] [PubMed] [Google Scholar]
- 32.Stubbs CD, Smith AD. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochim Biophys Acta. 1984;779:89–137. doi: 10.1016/0304-4157(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 33.Lattka E, Illig T, Heinrich J, Koletzko B. Do FADS genotypes enhance our knowledge about fatty acid related phenotypes? Clin Nutr. 2010;29:277–287. doi: 10.1016/j.clnu.2009.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Information for the prescreening of eight single-nucleotide polymorphisms (SNPs) and selection of four SNPs
| FEN1 10154G >T | FADS2 rs174575 | FADS3 rs1000778 | FADS2 rs2727270 | FADS2 rs174576 | FADS2 rs174570 | FADS2 rs174583 | FADS3 rs174456 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | |
| FEN1 −10154G > T | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| FADS2 rs174575 | 0.96 | 0.16 | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| FADS3 rs1000778 | 0.21 | 0.04 | 0.46 | 0.04 | – | – | – | – | – | – | – | – | – | – | – | – |
| FADS2 rs2727270 | 0.98 | 0.71 | 1.00 | 0.03 | 0.13 | 0.01 | – | – | – | – | – | – | – | – | – | – |
| FADS2 rs174576 | 1.00 | 0.98 | 1.00 | 0.17 | 0.22 | 0.04 | 1.00 | 0.72 | – | – | – | – | – | – | – | – |
| FADS2 rs174570 | 1.00 | 0.98 | 1.00 | 0.17 | 0.21 | 0.04 | 0.99 | 0.71 | 1.00 | 0.99 | – | – | – | – | – | – |
| FADS2 rs174583 | 0.99 | 0.95 | 0.96 | 0.16 | 0.22 | 0.04 | 1.00 | 0.71 | 0.99 | 0.98 | 0.99 | 0.97 | – | – | – | – |
| FADS3 rs174456 | 0.21 | 0.04 | 0.46 | 0.04 | 1.00 | 1.00 | 0.01 | 0.01 | 0.22 | 0.04 | 0.21 | 0.04 | 0.22 | 0.04 | – | – |
Table S2.
Associations of FADS2 rs 2727270 genotypes with polyunsaturated fatty acid (PUFA) composition in serum phospholipids in men at baseline and 3-year follow-up
|
FADS2 rs 2727270C>T
|
||||||
|---|---|---|---|---|---|---|
| CC (n = 59) | Pa | T allele (n = 63) | Pa | Pb | Pc | |
| PUFA composition (%) in serum PL | ||||||
| 18:2 (ω6) | ||||||
| Baseline | 12.1 ± 0.38 | 0.971 | 12.0 ± 0.42 | 0.511 | 0.880 | |
| Follow-up | 12.1 ± 0.47 | 11.6 ± 0.45 | 0.455 | |||
| Change | 0.02 ± 0.48 | −0.38 ± 0.58 | 0.594 | 0.466 | ||
| 18:3 (ω6) | ||||||
| Baseline | 0.18 ± 0.02 | 0.215 | 0.16 ± 0.01 | 0.051 | 0.452 | |
| Follow-up | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.693 | |||
| Change | 0.02 ± 0.02 | 0.03 ± 0.02 | 0.666 | 0.756 | ||
| 20:2 (ω6) | ||||||
| Baseline | 0.59 ± 0.17 | 0.172 | 0.75 ± 0.18 | 0.024 | 0.508 | |
| Follow-up | 0.36 ± 0.03 | 0.51 ± 0.16 | 0.357 | |||
| Change | −0.23 ± 0.17 | −0.24 ± 0.11 | 0.946 | 0.516 | ||
| 20:3 (ω6) | ||||||
| Baseline | 1.51 ± 0.06 | 0.974 | 1.36 ± 0.06 | 0.943 | 0.083 | |
| Follow-up | 1.52 ± 0.08 | 1.36 ± 0.07 | 0.123 | |||
| Change | 0.00 ± 0.09 | −0.01 ± 0.09 | 0.941 | 0.219 | ||
| 20:4 (ω6) | ||||||
| Baseline | 4.73 ± 0.20 | 0.027 | 3.99 ± 0.18 | 0.559 | 0.008 | |
| Follow-up | 5.42 ± 0.27 | 4.16 ± 0.23 | 0.001 | |||
| Change | 0.69 ± 0.30 | 0.16 ± 0.28 | 0.206 | 0.003 | ||
| 18:3 (ω3) | ||||||
| Baseline | 0.17 ± 0.02 | 0.319 | 0.18 ± 0.03 | 0.945 | 0.912 | |
| Follow-up | 0.21 ± 0.02 | 0.18 ± 0.01 | 0.267 | |||
| Change | 0.04 ± 0.03 | 0.00 ± 0.03 | 0.474 | 0.268 | ||
| 20:3 (ω3) | ||||||
| Baseline | 0.10 ± 0.02 | 0.029 | 0.08 ± 0.01 | 0.702 | 0.276 | |
| Follow-up | 0.06 ± 0.00 | 0.07 ± 0.00 | 0.123 | |||
| Change | −0.04 ± 0.02 | −0.01 ± 0.01 | 0.138 | 0.128 | ||
| 20:5 (ω3) | ||||||
| Baseline | 1.18 ± 0.07 | 0.134 | 1.06 ± 0.07 | 0.585 | 0.240 | |
| Follow-up | 1.41 ± 0.12 | 1.11 ± 0.07 | 0.033 | |||
| Change | 0.24 ± 0.16 | 0.05 ± 0.09 | 0.293 | 0.027 | ||
| 22:5 (ω3) | ||||||
| Baseline | 0.54 ± 0.03 | 0.088 | 0.60 ± 0.05 | 0.250 | 0.333 | |
| Follow-up | 0.63 ± 0.04 | 0.54 ± 0.04 | 0.085 | |||
| Change | 0.09 ± 0.05 | −0.06 ± 0.05 | 0.043 | 0.058 | ||
| 22:6 (ω3) | ||||||
| Baseline | 2.74 ± 0.18 | 0.130 | 2.69 ± 0.16 | 0.694 | 0.819 | |
| Follow-up | 3.10 ± 0.18 | 2.77 ± 0.18 | 0.205 | |||
| Change | 0.36 ± 0.23 | 0.09 ± 0.22 | 0.399 | 0.215 | ||
| 20:4(ω6)/20:3(ω6) | ||||||
| Baseline | 3.24 ± 0.13 | 0.008 | 3.36 ± 0.39 | 0.798 | 0.785 | |
| Follow-up | 3.84 ± 0.22 | 3.25 ± 0.21 | 0.052 | |||
| Change | 0.60 ± 0.22 | −0.11 ± 0.41 | 0.134 | 0.043 | ||
| 20:4(ω6)/18:2(ω6) | ||||||
| Baseline | 0.42 ± 0.03 | 0.206 | 0.34 ± 0.02 | 0.232 | 0.024 | |
| Follow-up | 0.50 ± 0.06 | 0.42 ± 0.06 | 0.314 | |||
| Change | 0.08 ± 0.06 | 0.07 ± 0.06 | 0.921 | 0.377 | ||
| 20:5(ω3)/18:3(ω3) | ||||||
| Baseline | 9.36 ± 0.79 | 0.148 | 9.18 ± 0.96 | 0.060 | 0.883 | |
| Follow-up | 7.88 ± 0.64 | 6.99 ± 0.58 | 0.306 | |||
| Change | −1.49 ± 1.01 | −2.19 ± 1.14 | 0.646 | 0.307 | ||
Notes:
Values derived from paired t-test;
values derived from independent t-test;
values derived after adjusting for baseline values. Values are means ± standard error.
Abbreviations: PUFA, polyunsaturated fatty acid; PL, phospholipids
Table S3.
Associations of FADS2 rs2727270 genotypes with LDL cholesterol, hs-CRP, cytokines, and oxidative stress markers in men at baseline and 3-year follow-up
|
FADS2rs2727270C>T
|
||||||
|---|---|---|---|---|---|---|
| CC (n = 59) | Pa | T allele (n ==63) | Pa | Pb | Pc | |
| LDL cholesterol (mg/dL) | ||||||
| Baseline§ | 119.3 ± 4.14 | 0.035 | 114.9 ± 4.76 | 0.184 | 0.447 | |
| Follow-up§ | 124.5 ± 3.46 | 119.2 ± 4.47 | 0.210 | |||
| Change | 5.22 ± 2.98 | 4.26 ± 3.51 | 0.837 | 0.533 | ||
| hs-CRP (mg/dL) | ||||||
| Baseline§ | 1.34 ± 0.30 | 0.972 | 1.92 ± 0.77 | 0.534 | 0.606 | |
| Follow-up§ | 1.45 ± 0.30 | 1.65 ± 0.49 | 0.947 | |||
| Change | 0.11 ± 0.15 | −0.27 ± 0.90 | 0.691 | 0.477 | ||
| Serum IL-6 (pg/mL) | ||||||
| Baseline§ | 4.27 ± 0.40 | 0.199 | 4.87 ± 0.52 | 0.006 | 0.407 | |
| Follow-up§ | 3.87 ± 0.39 | 3.39 ± 0.34 | 0.474 | |||
| Change | −0.41 ± 0.46 | −1.48 ± 0.61 | 0.168 | 0.268 | ||
| Serum TNF-α (pg/mL) | ||||||
| Baseline§ | 13.4 ± 1.73 | 0.765 | 9.53 ± 1.02 | 0.603 | 0.067 | |
| Follow-up§ | 11.9 ± 1.68 | 8.87 ± 0.57 | 0.122 | |||
| Change | −1.42 ± 2.24 | −0.66 ± 1.16 | 0.761 | 0.303 | ||
| 8-epi-PGF2α (pg/mg creatinine) | ||||||
| Baseline§ | 1313.0 ± 55.5 | 0.123 | 1324.3 ± 75.4 | 0.809 | 0.736 | |
| Follow-up§ | 1484.1 ± 76.2 | 1243.3 ± 43.9 | 0.022 | |||
| Change | 171.1 ± 93.2 | −80.9 ± 85.1 | 0.048 | 0.003 | ||
| Oxidized LDL (U/L) | ||||||
| Baseline§ | 34.0 ± 1.61 | <0.001 | 35.7 ± 1.96 | <0.001 | 0.663 | |
| Follow-up§ | 45.9 ± 1.49 | 43.4 ± 1.74 | 0.147 | |||
| Change | 11.9 ± 1.65 | 7.76 ± 1.87 | 0.097 | 0.092 | ||
Notes:
Tested by logarithmic transformation;
values derived from paired t-test;
values derived from independent t-test;
values derived after adjusting for baseline values. Values are means ± standard error.
Abbreviations: LDL, low-density lipoprotein; hs-CRP, high-sensitivity C-reactive protein; TNF, tumor necrosis factor; IL, interleukin; 8-epi-PGF2α, urinary 8-epi-prostaglandin F2α.